Abstract
Intercellular adhesion molecule-4 (ICAM-4, syn. LW glycoprotein) interacts with the integrins αLβ2, αMβ2, A4β1, the αV family, and αIIbβ3. Systematic mutagenesis of surface-exposed residues conserved between human and murine ICAM-4 defined 12 single amino-acid changes that affect the interaction of ICAM-4 with αV integrins. Mutation of 10 of these residues, 8 of which are spatially close on the surface of the molecule, led to a reduction in adhesion. Moreover, peptides corresponding to regions of ICAM-4 involved in its interaction with αV integrins inhibited these interactions. The other 2 mutations increased the extent of interaction of ICAM-4 with αV integrins. These mutations appear to prevent glycosylation of N160, suggesting that changes in glycosylation may modulate ICAM-4–αV integrin interactions. The region of ICAM-4 identified as the binding site for αV integrins is adjacent to the binding sites for αLβ2 and αMβ2. Selective binding of ICAM-4 to different integrins may be important for a variety of normal red cell functions and also relevant to the pathology of thrombotic disorders and vasoocclusive events in sickle cell disease. Our findings suggest the feasibility of developing selective inhibitors of ICAM-4–integrin adhesion of therapeutic value in these diseases.
Introduction
Intercellular adhesion molecule-4 (ICAM-4), a glycoprotein carrying the LW blood group, is a member of the immunoglobulin superfamily (IgSF) of proteins.1 On the basis of the crystal structure of ICAM-2,2 we have constructed a molecular model of ICAM-4 that represents ICAM-4 as 2 Ig-like domains; N-terminal domain 1 and membrane-anchored domain 2.3 Each domain has 2 faces (or sides), the ABE and the CFG faces. Previously ICAM-4 has been shown to bind αV integrins (notably αVβ1, αVβ3, and αVβ5) on nonhemopoietic cells; α4β1 (VLA-4) on hemopoietic cells3,4 ; and αIIbβ3 (GpIIb/IIIa) on platelets.5 Like all other members of the ICAM family,6-9 ICAM-4 binds the β2 integrin αLβ2 (LFA-1).10,11 Additionally, ICAM-4 adheres to αMβ2 (Mac-1),10,11 as do ICAM-1 and ICAM-2.12,13 Half of all integrin α subunits contain an “inserted” or I domain of about 200 amino acids that, where present, is thought to play a significant role in ligand binding.14,15 The β2 integrin αL and αM subunits contain I domains that have an ICAM-4–binding region16 ; however, the αV, A4, and αIIb subunits do not contain I domains.17 Thus, ICAM-4 appears unique among the ICAM family in particular and cell adhesion molecules in general, in that it binds a diverse array of integrins. This diverse array of binding partners suggests multiple functions for ICAM-4.
The functional roles in inflammation and immune responses of various members of the ICAM family of proteins (ICAM-1, -2, and -3) have been documented.7 However, no clear biologic role(s) has been defined for ICAM-4, whose expression appears limited to erythroid cells (with the possible exception of placenta).18,19 We have previously suggested that ICAM-4–α4β1 interactions may be relevant to differentiation of healthy erythroblasts in bone marrow, whereas interactions with αV integrins may be relevant to erythroblast and red cell interactions with macrophages and/or endothelial cells.20 Selective binding of ICAM-4 to different integrins may also be pertinent to the pathology of vasoocclusive events in sickle cell disease.21
To identify critical sites on ICAM-4 required for its interaction with αV integrins, we performed comprehensive targeted mutagenesis of surface-exposed amino-acid residues that are conserved between murine and human ICAM-4. The ability of these mutant proteins to interact with αV integrins was tested in cell adhesion assays. On the basis of these findings, we identified the area, or footprint, on ICAM-4 important for its interaction with αV integrins. Interestingly, this footprint is adjacent to but distinct from the binding sites previously identified for 2 other ICAM-4 binding partners, αLβ2 and αMβ2.
Materials and methods
Cell adhesion assay
Cell adhesion assays were performed as described previously.3 Immulon-4 96-well plates (Dynes Technologies, Billingshurst, United Kingdom) were coated with 1 μg/well goat antihuman-Fc (Sigma, Poole, United Kingdom) for 24 hours at 4°C, washed 3 times with phosphate-buffered saline (PBS), and coated with Fc fusion ICAM-4 protein (ICAM-4Fc) for 18 hours at 4°C before blocking with 0.4% bovine serum albumin (BSA) PBS for 2 hours at 22°C. Cells were labeled with 10 μg/mL 2′,7′-bis-(2-carboxyethyl)-5-(and-6-)carboxyfluorescein acetoxymethyl ester (Sigma) in assay buffer (Iscoves modified Eagle medium, 2 mM EGTA [ethylene glycol tetraacetic acid], 10 μg/mL immune globulin intravenous [human; Cutter Biological, Newbury, Berks, United Kingdom]) for 15 minutes at 37°C. HT1080 cells were activated with 80 μM phorbol myristate acetate before being washed with assay buffer containing 2 mM Mn2+. Cells, at 5 × 104 cells per well, were added to the ICAM-4Fc–coated plates for 30 minutes at 37°C prior to being cyclically read on a fluorescence microplate reader (excitation 485 nm, emission 530 nm) and washed in assay buffer. The percentage of bound cells was calculated after each wash. Peptide inhibition was performed by incubating the cells with 500 μM peptide at 0°C for 15 minutes before their addition to the ICAM-4Fc–coated plates. A titration of each cell type with native ICAM-4Fc was performed prior to peptide inhibition assays to obtain the optimum concentration (the lowest concentration of ICAM-4 that allows maximum binding of cells) of ICAM-4Fc to use.
Preparation of ICAM-4 Fc fusion proteins
cDNA clones encoding the extracellular domains of ICAM-4 and neural cell adhesion molecule (NCAM) in pIg vector were gifts from Dr D. Simmons (Glaxo SmithKline, Harlow, United Kingdom). Point mutations were inserted into ICAM-4 in pIg vector by polymerase chain reaction (PCR) amplification over 2 stages.22 Oligonucleotides containing mismatched bases together with 5′-AGAACCCACTGCTTACTGGCT and 3′-TGAGCCTGCTTCCAGCAGCA primers were used to generate 2 overlapping products using standard PCR. Following gel purification the 2 overlapping PCR products were annealed together before final amplification by using the 5′ and 3′ primers. The final PCR product was restricted and ligated into pIg vector. All mutant clones were verified by sequence analysis. Native and mutant ICAM-4Fc proteins and NCAMFc protein were expressed in COS-7 cells as described previously23 and purified from culture supernatant on protein A–Sepharose. The concentration of the ICAM-4Fc proteins was obtained by enzyme-linked immunosorbent assay (ELISA) against a known standard curve of human immunoglobulin G (IgG).
SDS-PAGE analysis
The Mr of the N160A and T162V ICAM-4Fc fusion proteins was calculated from the migration of 2.5 μg of each protein on a reduced 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel in comparison with the mobility of standard proteins (prestained SDS-PAGE standards; Broad range; BioRad, Hemel Hempstead, United Kingdom). Purified Fc fusion proteins were dissolved in equal volumes of 20% (vol/vol) glycerol, 10% (wt/vol) SDS, 40 mM Tris (tris(hydroxymethyl)aminomethane)–HCL, pH 8, 10% (vol/vol) β-mercaptoethanol, and 0.2 mg/mL bromophenol blue, and were heated at 60°C for 10 minutes. Stacking and 7.5% separating SDS polyacrylamide gels were made as described.24 The gel was then run at 200V until the tracking dye reached the end of the separating gel. The gel was stained with 2 g l–1 Coomassie brilliant blue in a mixture of 20% (vol/vol) methanol, 7.5% (vol/vol) acetic acid and then de-stained until the background was clear.
Results
Effect of mutations on ICAM-4 function
Fifty-one residues in ICAM-4 were mutated (Table 1). The residues targeted for mutation were all surface exposed, and most are conserved in murine ICAM-4, which also interacts with human αV integrins.20 Each mutant was tested for adherence to HT1080 cells that express αV integrins. Like FLY cells, which are derived from the HT1080 cell line, antibody inhibition experiments showed that under the conditions of the cell adhesion assay used, HT1080 cells bound to ICAM-4 by way of αVβ1 and αVβ5 (data not shown and Spring et al3 ). We found that 10 of the 51 targeted residues of ICAM-4, at a concentration of 20 μg/mL, caused a reduction in cell adhesion, by more than 55% (Figure 1). Surprisingly, 2 mutations were found to increase the adhesion of ICAM-4 to αV integrins by more than 50% (Figure 1). None of the other 39 mutations caused either a significant increase or decrease in adhesion (Figure 1).
ICAM-4 mutations and their positions
Domain 1 . | . | Domain 2 . | . | Domain 2 . | . | |||
|---|---|---|---|---|---|---|---|---|
| Mutation . | Strand . | Mutation . | Strand . | Mutation . | Strand . | |||
| F18A | A | I108T | A | E166K | E | |||
| W19A | A | E110K | A | F167A | E | |||
| V20T | A | K118E | B | D173K | F | |||
| R21Q | A | F129A | B | R183E | F | |||
| E25K | A | G132V | C | N185A | F | |||
| F26A | A | Y133A | C | L186K | F | |||
| S34A | B | V135T | C | R192E | G | |||
| W66A | E | T137V | C | |||||
| Q70A | E | R139E | C | |||||
| L72K | E | R143E | C′ | |||||
| R75E | E | V144T | C′ | |||||
| W77A | F | I145T | C′ | |||||
| S79A | F | Y146A | C′ | |||||
| L80K | F | S147A | C′ | |||||
| K90E | G | E148K | C′ | |||||
| R92E | G | S149A | C′ | |||||
| W93A | G | R152E | C′ | |||||
| A94L | G | F153A | ′ | |||||
| T95V | G | N160A | E | |||||
| S96A | G | T162V | E | |||||
| R97E | G | T164V | E | |||||
| T99V | G | Y165A | E | |||||
Domain 1 . | . | Domain 2 . | . | Domain 2 . | . | |||
|---|---|---|---|---|---|---|---|---|
| Mutation . | Strand . | Mutation . | Strand . | Mutation . | Strand . | |||
| F18A | A | I108T | A | E166K | E | |||
| W19A | A | E110K | A | F167A | E | |||
| V20T | A | K118E | B | D173K | F | |||
| R21Q | A | F129A | B | R183E | F | |||
| E25K | A | G132V | C | N185A | F | |||
| F26A | A | Y133A | C | L186K | F | |||
| S34A | B | V135T | C | R192E | G | |||
| W66A | E | T137V | C | |||||
| Q70A | E | R139E | C | |||||
| L72K | E | R143E | C′ | |||||
| R75E | E | V144T | C′ | |||||
| W77A | F | I145T | C′ | |||||
| S79A | F | Y146A | C′ | |||||
| L80K | F | S147A | C′ | |||||
| K90E | G | E148K | C′ | |||||
| R92E | G | S149A | C′ | |||||
| W93A | G | R152E | C′ | |||||
| A94L | G | F153A | ′ | |||||
| T95V | G | N160A | E | |||||
| S96A | G | T162V | E | |||||
| R97E | G | T164V | E | |||||
| T99V | G | Y165A | E | |||||
Strand positions determined using molecular model of ICAM-4.
Mutations to ICAM-4 affect adhesion of HT1080 cells. Adhesion of HT1080 cells to native and mutated ICAM-4Fc in a microplate adhesion assay. (A) Representative binding assays depicting various mutant ICAM-4Fc (♦) and native ICAM-4Fc (□) are shown, 2 from the A strand of domain 1 (F18A and V20T), 2 from the G strand of domain 1 (R92E and T95V), 2 other mutations resulting in a reduction of adhesion (W66A and K118E), 2 mutations leading to an increase in ICAM-4 adhesion (N160A and T162V), and 2 mutations that had no affect on adhesion (W77A and T162V). The proteins were used at 20, 10, 5, 1, or 0.25 μg/mL, and the results are shown as the percentage of input cells bound □ SD (n = 3 [mutants] or n = 2 [native protein]). (B) Adhesion of HT1080 cells to all 51 mutants. Results are shown as the percentage of HT1080 cell binding relative to native ICAM-4Fc used at 20 μg/mL (100%).
Mutations to ICAM-4 affect adhesion of HT1080 cells. Adhesion of HT1080 cells to native and mutated ICAM-4Fc in a microplate adhesion assay. (A) Representative binding assays depicting various mutant ICAM-4Fc (♦) and native ICAM-4Fc (□) are shown, 2 from the A strand of domain 1 (F18A and V20T), 2 from the G strand of domain 1 (R92E and T95V), 2 other mutations resulting in a reduction of adhesion (W66A and K118E), 2 mutations leading to an increase in ICAM-4 adhesion (N160A and T162V), and 2 mutations that had no affect on adhesion (W77A and T162V). The proteins were used at 20, 10, 5, 1, or 0.25 μg/mL, and the results are shown as the percentage of input cells bound □ SD (n = 3 [mutants] or n = 2 [native protein]). (B) Adhesion of HT1080 cells to all 51 mutants. Results are shown as the percentage of HT1080 cell binding relative to native ICAM-4Fc used at 20 μg/mL (100%).
Eight of the 10 residues that when mutated caused a reduction in cell adhesion (F18, W19, V20, R92, A94, T95, S96, R97) were found in a patch, or “footprint” that spans the interface between the ABE and CFG faces of domain 1 (Figure 1; Table 1). The residues found within this footprint comprise runs of amino acids on 2 strands within the Ig-like domain. Residues F18, W19, and V20 are located on the A strand of domain 1, whereas R92, A94, T95, S96, and R97 are all on the G strand of domain 1. These 8 spatially close residues on the surface of ICAM-4 that are vital for binding to αV integrins are in close physical proximity to the patch on ICAM-4 that has previously been shown to be important for interaction with αLβ2 and αMβ2.11 The other 2 mutations that caused a reduction in adhesion (W66 and K118) were found on the ABE face of domain 1 and the ABE face of domain 2, respectively (Figure 1; Table 1).
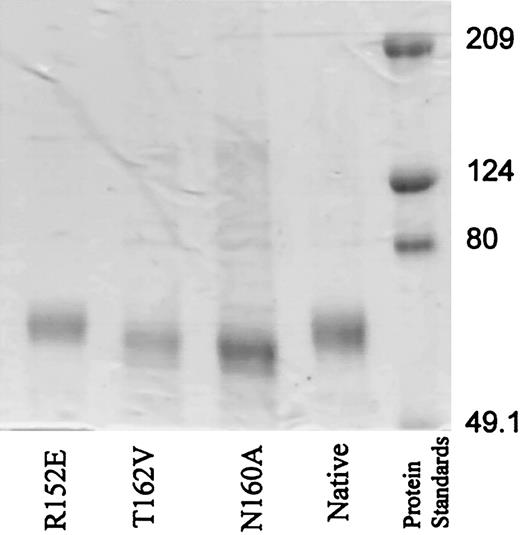
The 2 mutations, N160A and T162V, which caused a striking increase in adhesion of ICAM-4 to αV integrins, were considered “super-adhesive” (Figure 1). These 2 residues in ICAM-4 form part of a consensus asparagine-X-threonine/serine N-glycosylation site (X is any amino acid other than proline). Analysis of the mutant proteins by SDS-PAGE revealed that the N160A and T162V mutant ICAM-4Fc proteins migrated further than both native ICAM-4Fc and the R152E mutation (Figure 2). The native and R152E ICAM-4Fc were estimated to have a relative molecular mass (Mr) of 71 kDa, whereas the N160A and T162V mutations had a Mr of 65.5 and 66.6 kDa, respectively. This 5-kDa difference in Mr can be accounted for by the absence of N-glycan chains on mutant ICAM-4Fc. This suggestion is consistent with the approximately 5-kDa size of N-glycans on ICAM-225 and the predicted size of N-glycan chains on ICAM-4.26-28 On the basis of these observations we suggest that N160 in ICAM-4 is normally occupied by an N-glycan chain.
SDS-PAGE demonstrates increased migration of N160A and T162V mutants in comparison with native or R152E ICAM-4Fc. Mutant and native ICAM-4Fc proteins and prestained standard proteins (2.5 μg) were separated on a 7.5% reduced SDS-PAGE gel as described in “Material and methods.”
SDS-PAGE demonstrates increased migration of N160A and T162V mutants in comparison with native or R152E ICAM-4Fc. Mutant and native ICAM-4Fc proteins and prestained standard proteins (2.5 μg) were separated on a 7.5% reduced SDS-PAGE gel as described in “Material and methods.”
Inhibition of cell adhesion by peptides
Three synthetic peptides with amino-acid sequences corresponding to the parts of the A, G, and F strands of ICAM-4 covering most of the area involved in integrin binding were tested singly and in combination for inhibition of binding between ICAM-4 and αV integrins (Figure 3). The A, G, and F strand peptides were V(16)PFWVRMS (FWV), T(91)RWATSRI (ATSR), and A(76)WSSLAHC (AWSS), respectively (residues in bold face type are important for the binding of various integrins). A control peptide was also constructed, R(56)QGKTLRG (Cpep), which had the sequence of the D strand of domain 1, the strand on the opposite side to the proposed binding site in ICAM-4. Peptides FWV and ATSR corresponded to sequences containing residues F18, W19, and V20 and R92, A94, T95, S96, and R97, respectively. The AWSS peptide (together with the ATSR peptide) corresponded to the area of the ICAM-4 molecule, which has been reported to be important in binding the β2 integrins αLβ2 and αMβ2.11
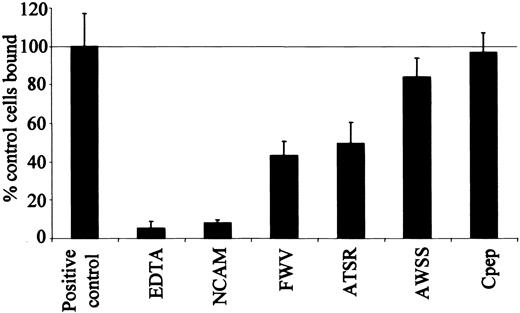
ICAM-4 peptides inhibit HT1080 cell binding to ICAM-4Fc. Adhesion of HT1080 cells to native ICAM-4 in the presence of assay buffer (positive control), defined peptides or EDTA (ethylenediaminetetraacetic acid), along with HT1080 adhesion to a NCAM-negative control. The results are expressed as a percentage of control cells bound □ SD (n = 3); 100% was equal to 41% of input cells bound, ICAM-4Fc was coated at a concentration of 7.5 μg/mL, and peptides were used at 500 μM.
ICAM-4 peptides inhibit HT1080 cell binding to ICAM-4Fc. Adhesion of HT1080 cells to native ICAM-4 in the presence of assay buffer (positive control), defined peptides or EDTA (ethylenediaminetetraacetic acid), along with HT1080 adhesion to a NCAM-negative control. The results are expressed as a percentage of control cells bound □ SD (n = 3); 100% was equal to 41% of input cells bound, ICAM-4Fc was coated at a concentration of 7.5 μg/mL, and peptides were used at 500 μM.
Both the FWV and ATSR peptides inhibited the interaction between ICAM-4 and αV integrins (Figure 3). The FWV peptide inhibited HT1080 cell binding to ICAM-4 by 60% and the ATSR peptide by 50%. When tested together, there was no additional inhibition of binding (data not shown). Similar results were obtained when binding of HT1080 cells to murine ICAM-4 was studied, the peptides inhibiting adhesion by 40% (data not shown).
The AWSS peptide was significantly less effective at inhibiting HT1080 cell binding (inhibition of only 20%) (Figure 3). These data suggest that the area of ICAM-4 represented by the AWSS peptide is less important for the ICAM-4 interaction with αV integrin than for its interaction with αLβ2 and αMβ2. The control peptide (Cpep) had no effect on the adhesion of HT1080 cells to ICAM-4. These results suggest that the FWV and ATSR peptides are specific inhibitors of ICAM-4 adhesion to αV integrins.
Discussion
Cell-cell adhesive interactions are commonly mediated by members of the IgSF, interacting with the integrin family of proteins.14 The binding site on ICAM-1, -2, and -3 for αLβ2 has been previously defined and involves a critical glutamic acid residue on the flat surface of the CFG face of the molecule.14,15,29-32 However, unlike ICAM-1, -2, -3, and -5, ICAM-4 does not contain the αLβ2 consensus binding motif of (L/I)ET(P/S)L and instead it has LR(52)TPL within the C strand of domain 1. It has been shown that this unusual LETS motif is not critical for the interaction between ICAM-4 and αV integrin.3 In addition, whereas an RGD peptide inhibits ICAM-4 adhesion to αV integrins,3 ICAM-4 contains no RGD motif. Therefore, ICAM-4 must have a novel integrin binding motif(s).
In the present study, a comprehensive analysis using an extensive set of mutant proteins has enabled us to document that one small region of ICAM-4 mediates its interaction with αV integrins. This region comprises a run of 3 residues (F18, W19, and V20) on the domain 1 A strand, and 5 of a run of 6 residues (R92, A94, T95, S96, and R97) on the domain 1 G strand. The close proximity of these residues in the structure of the protein suggests that this area is vital for the binding of ICAM-4 to its αV integrin ligands (Figure 4A). Of these 8 residues, only 4 residues (F18, V20, A94, and S96) are homologous between human ICAM-4 and other members of the human ICAM family. The lack of homology in this area could explain why none of the other ICAMs have been shown to interact with αV integrins. In marked contrast, murine ICAM-4, which is much more homologous to human ICAM-420 with conservation of 7 of the 8 residues in the area identified in this study, interacts with the αV integrins. Of these 8 residues, it is tempting to speculate that either of the 2 positively charged amino acids R92 and R97 are most likely to be crucial in the integrin interaction. These are the only charged residues within this set, and the arginine of an RGD peptide in complex with αVβ3 has previously been shown to be central in the interaction between the peptide and the integrin by forming salt bridges with D218 and D150 of the integrin.33
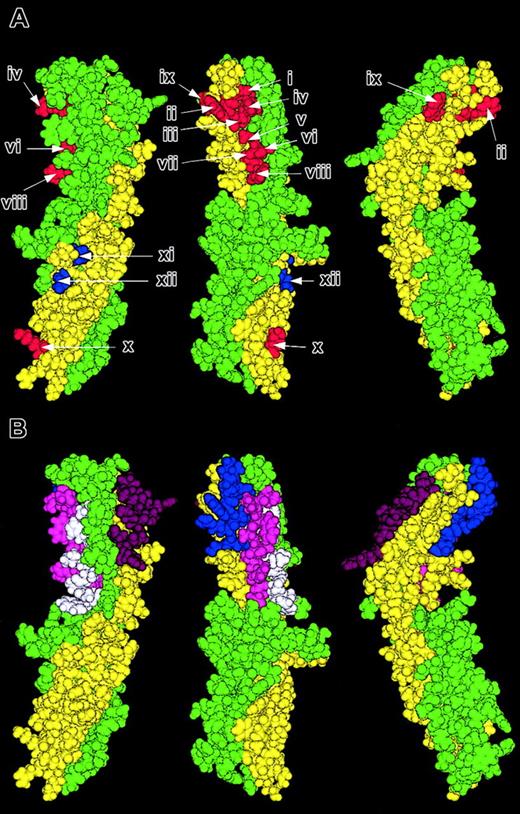
Location of important areas on ICAM-4 that mediate adhesion to αV integrins. Molecular model of ICAM-4 displayed in 3 orientations 120 degrees to one another. Residues within the ABE face are colored yellow and those in the CFG face green. (A) Residues that when mutated cause a decrease in adhesion to αV integrins are depicted in red and the super-adhesive residues in blue (i, F18; ii, W19; iii, V20; iv, R92; v, A94; vi, T95; vii, S96; viii, R97; ix, W66; x, K118; xi, N160; and xii, T162). (B) The sections of the A, G, F, and D strands of domain 1 that comprise the sequence of the peptides SVPFWVRMS (FWV) residues 15-23 (blue), TRWATSRIT (ATSR) residues 91-99 (magenta), AWSSLAHCL (AWSS) residues 76-84 (white), and RQGKTLRGP (Cpep) residues 56-64 (purple).
Location of important areas on ICAM-4 that mediate adhesion to αV integrins. Molecular model of ICAM-4 displayed in 3 orientations 120 degrees to one another. Residues within the ABE face are colored yellow and those in the CFG face green. (A) Residues that when mutated cause a decrease in adhesion to αV integrins are depicted in red and the super-adhesive residues in blue (i, F18; ii, W19; iii, V20; iv, R92; v, A94; vi, T95; vii, S96; viii, R97; ix, W66; x, K118; xi, N160; and xii, T162). (B) The sections of the A, G, F, and D strands of domain 1 that comprise the sequence of the peptides SVPFWVRMS (FWV) residues 15-23 (blue), TRWATSRIT (ATSR) residues 91-99 (magenta), AWSSLAHCL (AWSS) residues 76-84 (white), and RQGKTLRGP (Cpep) residues 56-64 (purple).
In addition to the 8 residues in this footprint, the mutation of 2 other residues, W66, on the E strand of domain 1, and K118, on the B strand of domain 2 also caused a reduction in the binding of HT1080 cells to ICAM-4. When their position is viewed on the model of ICAM-4, it can be seen that W66 and K118 are, in fact, spatially distant from the footprint (Figure 4A). We conclude that when interacting with the footprint on ICAM-4, integrins may contact a large surface area of ICAM-4. This is consistent with the relative size of the 2 protein components: integrins are far larger proteins than the 2 Ig domain ICAM-4. Integrins are approximately 280 kDa in size and αVβ3 has head dimensions of about 9.0 × 6.0 × 4.5 nm, with an extended tail of 16.0 × 2.0 nm.34 ICAM-4 is 42 kDA glycosylated (25 kDa deglycosylated).27,28 Our model of ICAM-4 has approximate dimensions of 2.5 × 2.0 × 8.0 nm, which is consistent with the typical average size of 2.0 × 2.5 × 4.0 nm dimension for a single Ig domain.14 We speculate that residues W66 and K118 might form peripheral contacts for the ICAM-4/integrin footprint area, stabilizing the overall interaction. In this context it is interesting to note that ICAM-4 binding to the platelet integrin αIIbβ3 has been reported to involve residues G65-V74 of ICAM-4.5 This peptide sequence corresponds to the opposite face of the protein from the footprint described here (strand E as opposed to the A, F, and G strand) but it contains W66.
Our data also strongly suggest that N160 is a site for N-linked glycosylation in ICAM-4 and that without this N-glycan chain ICAM-4 appears to be “super-adhesive.” Of the 4 potential N-linked glycosylation sites in human ICAM-4, N160 is one of only 2 that is also present in murine ICAM-4,20 adding more credibility to the suggestion that the N-glycan chain from N160 may have an affect on the avidity of the interaction between ICAM-4 and αV integrins. In the absence of a crystal structure of the complex it is not clear how this effect is moderated. It is possible that the presence of carbohydrate at this site may sterically influence the positioning of ICAM-4 domains relative to one another, or that the carbohydrate itself may participate directly in the binding of the integrin. This finding raises the intriguing possibility that changes in glycosylation could potentially be used to regulate the adhesive properties of ICAM-4.
Synthetic peptides were made that had the sequence of the various strands of ICAM-4 shown to be involved in adhesion to αV integrins (Figure 4B). Our data on inhibition of cell adhesion by these various peptides provided independent support for the role of the proposed footprint area in the binding to αV integrins. It is likely that the peptides can fit into the binding site of the integrin and prevent the docking of ICAM-4. Comparison of these findings with those of Hermand et al5,11 suggests that there are areas within ICAM-4 that are more or less important for binding specific integrins. Our data indicate that αV integrins on HT1080 cells tend to bind more avidly to the A strand (F18-V20) and the G strand (R92-R97) and with less avidity to the F strand (A76-L84).
Previously, a mutagenesis study has defined a binding site for αLβ2 and αMβ2 on ICAM-4, comprising 8 residues (T91, R52, E151, T154, W93, L80, R97, and W77).11 All of these residues are involved in binding αMβ2; however, only W93, L80, R97, and W77 comprise the αLβ2 binding site. These residues are mainly located on the F and G strands of domain 1, and the CC′ loop of domain 2. It has also been recently reported that residues 65-74 on the E strand of domain 1 of ICAM-4 comprise the binding site of platelet integrin αIIb β35 . The 8 spatially close residues on the surface of ICAM-4 that we identified as vital for binding to αV integrins are on the A and G strands of domain 1 and are in close physical proximity to the footprint on ICAM-4 important for its interaction with αLβ2 and αMβ2.11 R97 is the only residue that is shared between the 2 sites. Taken together, these findings imply that strands A, D, F, and G of ICAM-4 are all involved in integrin binding, although to different integrins.
In contrast to all other members of the ICAM family which interact with a limited number of specific integrin ligands, erythroid-specific ICAM-4 is an unusual ICAM in that it interacts with multiple integrins. Thus, we postulate that ICAM-4 may be involved in many dynamic cell-cell interactions during the life cycle of red cells. Binding to αV integrins on either macrophages or endothelium could be important from erythropoiesis to erythrocyte senescence. We suggest that during erythropoiesis erythroblasts could adhere to macrophages in “erythroblastic islands” in bone marrow through an αV–ICAM-4 interaction.20,21 An αV–ICAM-4 interaction could also participate in tethering of red cells to capillary endothelium to facilitate gas exchange.35 Finally, interaction of ICAM-4 on senescent red cells with αV integrins on splenic macrophages could play a role in red cell destruction. In addition, ICAM-4 binding to platelet αIIbβ3 and neutrophil αMβ2 may be part of the process whereby red cells participate in normal hemostatic processes5,36,37 and may also be relevant to thrombotic conditions such as deep vein thrombosis37 and vaso-occlusion in sickle cell disease.38 Indeed, it has recently been shown that during sickle cell crisis neutrophils that express β2 integrins, αLβ2 and αMβ2, bind not only inflamed endothelium but also adhere to erythrocytes.37,38 ICAM-4 is a likely candidate for mediating this erythrocyte adhesion with β2 integrins. ICAM-4 may also be of importance in erythrocyte adhesion observed in other diseases such as thalassemia, diabetes mellitus, and malaria.21,36 It has previously been reported that anti-αVβ3 antibodies inhibit sickle red cell adherence to rat endothelium activated by platelet activating factor.39 This finding, along with our peptide inhibition results, suggest it may be possible to selectively modulate ICAM-4–integrin interactions so that unwanted interactions associated with pathology can be ameliorated without compromising other functions. Further studies are required to define more precisely the integrin binding sites on ICAM-4 and to explore the effects of inhibitors in animal models. Ultimately, this information may allow the design of new drugs for the treatment for sickle cell crisis and other major thrombotic diseases that include an ICAM-4 adhesive interaction.
Prepublished online as Blood First Edition Paper, October 16, 2003; DOI 10.1182/blood-2003-08-2792.
Supported in part by National Institutes of Health grants (grants DK56267and HL31579); by the Director, Office of Health and Environment Research Division, US Department of Energy (contract DE-AC03-76SF00098); and by the United Kingdom NHS R&D Directorate.
Presented in part in abstract form to the 44th annual meeting of the American Society of Hematology, Philadelphia, PA, December 6-10, 2002.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Molecular graphics of ICAM-4 were performed by using the DS ViewerPro software from Accelrys, The Quorum, Barnwell Road, Cambridge, CB5 8RE, United Kingdom. We thank P. Martin for DNA sequence analysis and Dr W. Mawby for peptide synthesis.

![Figure 1. Mutations to ICAM-4 affect adhesion of HT1080 cells. Adhesion of HT1080 cells to native and mutated ICAM-4Fc in a microplate adhesion assay. (A) Representative binding assays depicting various mutant ICAM-4Fc (♦) and native ICAM-4Fc (□) are shown, 2 from the A strand of domain 1 (F18A and V20T), 2 from the G strand of domain 1 (R92E and T95V), 2 other mutations resulting in a reduction of adhesion (W66A and K118E), 2 mutations leading to an increase in ICAM-4 adhesion (N160A and T162V), and 2 mutations that had no affect on adhesion (W77A and T162V). The proteins were used at 20, 10, 5, 1, or 0.25 μg/mL, and the results are shown as the percentage of input cells bound □ SD (n = 3 [mutants] or n = 2 [native protein]). (B) Adhesion of HT1080 cells to all 51 mutants. Results are shown as the percentage of HT1080 cell binding relative to native ICAM-4Fc used at 20 μg/mL (100%).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-08-2792/6/m_zh80040456680001.jpeg?Expires=1766010500&Signature=Y1T1LYn3uj-QkgVXFFD7NyxvkOfEE1-cxoCrt2lerkcPX0s6EshTREtK4a~5bnSUMZ64wxi0MJUwqXrjKSeXXyp7v0pNMV4A-Ric12gCxwnne-UwI8dv1zqwHqOQaFRDw0XLZuYYPFTliPd23xZ9upAJNd-Z8manNnhurLXrLWo68PWwlIdcceFt9cpilX311ClPLgkVWN9IbJ1phQblx8a05sLkXxDu0LxOUFn~6G5AEm5FMxuJ9FSgvtzDPMnTd0iD59VPf7snmz-w~kqWQs-WXUr~yeOFX6sDkP92FiUh-P9mXyGpoPbPy8-JP3kIniINncsXtAmhi1RdFmAo7Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal