Abstract
Heterogeneity in the clinical behavior of patients with chronic lymphocytic leukemia (CLL) makes it difficult for physicians to accurately identify which patients may benefit from an early or more aggressive treatment strategy and to provide patients with relevant prognostic information. Given the potential efficacy of newer therapies and the desire to treat patients at “optimum” times, it is more important than ever to develop sensitive stratification parameters to identify patients with poor prognosis. The evolution of risk stratification models has advanced from clinical staging and use of basic laboratory parameters to include relevant biologic and genetic features. This article will review the dramatic progress in prognostication for CLL and will propose statistical modeling techniques to evaluate the utility of these new measures in predictive models to help determine the optimal combination of markers to improve prognostication for individual patients. This discussion will also elaborate which markers and tools should be used in current clinical practice and evaluated in ongoing clinical trials.
Introduction: the problem with being “indolent”
The B-cell malignancies share a common cell lineage but a wide range of clinical, laboratory, molecular, and genetic features. Once grouped together as “subtypes” under the broad term of non-Hodgkin lymphoma, the classification systems for these diseases have gone through extensive refinement to better distinguish their clinical and biologic characteristics.1,2 Traditional teaching labeled diseases that were fatal in weeks to months if untreated as “aggressive” and those that progressed over months to years as “indolent.”
Through a major, collaborative, international effort, the World Health Organization (WHO) published a consensus classification system that distinguished lymphoid disorders on the basis of a combination of clinical syndromes, morphology, immunophenotype, and genetic features.2 The WHO classification added the concept that each disorder represented a separate and distinct disease with chronic lymphocytic leukemia (CLL) and small lymphocytic lymphoma grouped into a single entity (CLL).2
Although this categorization no longer distinguishes these diseases as indolent or aggressive, the low-grade B-cell disorders are still evaluated along these lines.3 These disorders are often considered chronic diseases of the elderly, and patients are thought likely to die of unrelated causes. These assumptions are incorrect and, to a certain extent, have hampered progress in this field.
For CLL, the median age of diagnosis is 65 to 68 years old.4 Some patients experience a slowly progressive clinical course, but most will eventually progress and have fatal outcomes.4,5 Importantly, significant subsets of patients with early stage CLL become active,4,6-8 are refractory to treatment,7 experience infectious or autoimmune complications, and have more rapidly fatal outcomes than anticipated.
This clinical heterogeneity emphasizes the problem of considering CLL indolent and, in the era of more efficacious therapy, highlights the need to improve our ability to identify patients with poor prognosis. Recent trials of monoclonal antibodies in combination with chemotherapy have reported previously unattained response rates.9,10 These therapies frequently reduce disease burden to levels detectable only by flow cytometry or molecular methods and raise the possibility of cures.11 Additionally, progress in the field of autologous and allogeneic stem cell transplantation have led to studies of transplantation in younger patients with refractory or aggressive disease.12
For decades, “watchful waiting” has been the standard of care for patients with early stage CLL. This approach has focused on minimizing toxicity for the minority of patients whose disease may never evoke clinical symptoms. The recent progress in treatment raises critical new questions for patients with CLL. Which patients in early stage should be treated? Which patients should receive more aggressive therapy? Who should undergo stem cell transplantation?
These questions cannot be answered without accurate and precise prognostication for individual patients. For the purpose of this review, we term laboratory measures used to predict outcomes as “prognostic markers” (eg, β-2 microglobulin) and classification systems built on multiple measures or calculations as “prognostic tools” (eg, staging, lymphocyte doubling time). Risk stratification models will refer to the use of statistical modeling to validate integration of multiple markers and tools to improve prognostic precision.
Historic perspective and update
Staging
Clinical staging systems based on disease burden were the initial prognostic tool in CLL. In 1975, Rai et al13 developed a seminal set of clinical “staging” criteria for CLL based on the presence of lymphadenopathy, organomegaly (spleen and liver), and cytopenias. Importantly, Rai et al13 demonstrated a correlation between Rai stage and survival. The original Rai staging system was later revised from a 5-tier system (0-IV) to a 3-tier system that categorized patients into low-risk (original Rai stage 0), intermediate-risk (original Rai stages I-II), and high-risk (original Rai stages III-IV) groups.14 The Binet staging system was devised on the basis of a retrospective analysis that identified measures of disease burden similar to those of Rai as the most significant prognostic variables for survival.15,16 These 2 systems have delineated the clinical presentation and natural history of CLL and have allowed stratification of patients into similar groups for clinical research. Median survival from diagnosis in the original series using the Rai and Binet staging criteria is presented in Table 1. Other demographic and clinical factors, including age,5,13 sex,5,13,15 and performance status,17 have been shown to affect prognosis, although the magnitude and significance of age and sex independent of stage are inconsistent across series.18 Bone marrow histologic pattern (diffuse involvement versus all other patterns) is an accepted adjunct to staging in CLL and has the potential to identify patients with greater likelihood of progression and shorter survival independent of stage.19,20
. | Median survival, mo . |
|---|---|
| Rai stage | |
| 0 | 150 |
| I | 101 |
| II | 71 |
| III | 19 |
| IV | 19 |
| Binet stage | |
| A | Comparable to age-matched controls |
| B | 84 |
| C | 24 |
. | Median survival, mo . |
|---|---|
| Rai stage | |
| 0 | 150 |
| I | 101 |
| II | 71 |
| III | 19 |
| IV | 19 |
| Binet stage | |
| A | Comparable to age-matched controls |
| B | 84 |
| C | 24 |
Median survival as reported in initial publications of these staging systems for CLL.
Over the past 30 years, significant changes in the pattern of diagnosis of CLL have occurred with a shift from patients presenting with bulky nodes and cytopenias, to the detection of asymptomatic individuals identified by an elevated lymphocyte count on automated differential blood counts.4 With the use of immunophenotyping and flow cytometry, the diagnosis of CLL has also become more precise. We are now able to more easily distinguish CLL from mantle cell lymphoma and T-cell disorders. Furthermore, it is now recognized that a subset of patients with early stage CLL have a disease that will rapidly evolve to a more advanced, refractory, and fatal disease.4-8 Rai and Binet staging lack the ability to prospectively identify the rapidly evolving patient from patients destined to remain with early stage for decades. For patients with more advanced disease, these staging systems do not distinguish patients who will be stable from those with the more aggressive cohort.
Lymphocyte doubling time
One calculation that may be associated with meaningful differences in disease kinetics is the lymphocyte doubling time (LDT). The LDT is calculated by determining the number of months it takes the absolute lymphocyte count (ALC) to double in number. One study reported patients with an LDT of 12 months or less had a median survival of 61 months, whereas median survival was not reached for patients with a LDT more than 12 months (median follow-up, 118 months).21 For patients in early stage (Binet stage A or B) the estimated median survival was 66 months for those with LDT of 12 months or less, while no patients with an LDT of more than 12 months had died after a median follow-up of 48 months.
Although other reports have confirmed the usefulness of the LDT for patients in early stage,22 there are significant problems with this tool. The LDT is inherently retrospective, and its temporal change can be quite variable. It is also confounded by a host of factors that may transiently affect the ALC. Treatment decisions based on this parameter may delay treatment for some patients with characteristics of aggressive disease.23
β-2 Microglobulin
Serum β-2 microglobulin (B2M) is a serum marker that correlates with tumor burden and disease stage in patients with CLL.24,25 A retrospective series of 302 untreated patients from the MD Anderson Cancer Center found B2M was the strongest predictor of 5-year survival on multivariate analysis which controlled for age, stage, and performance status (5-year survival for Rai stages I-II with elevated B2M = 54 months versus 116 months for patients without elevated B2M).26 Although similar results were reported in other retrospective series that included large numbers of previously treated patients,27,28 a prospective trial of 106 untreated patients did not find B2M alone to be a significant predictor of survival in multivariate analysis that controlled for stage and LDT.29 Thus, although B2M has some prognostic relevance in CLL, additional studies are needed to demonstrate the independence of this marker from stage and LDT and to define its role in the management of patients with early stage CLL.
Recently identified biologic markers
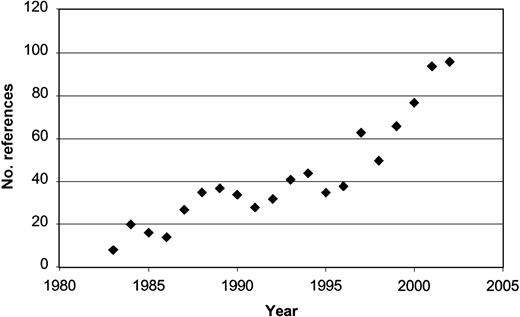
Technologic advancements have led to developments in diagnosis and targeted therapy for several hematologic malignancies.30,31 The application of fluorescent in situ hybridization (FISH) and the recent development of robust methods for genetic analysis are rapidly increasing our understanding of malignancy. Microarray analysis allows genomic assessment that can improve categorization of tumor type (class prediction) and can identify previously unrecognized tumor subtypes (class discovery).32 Global genetic analysis can also reveal clues to the early biologic defects underlying malignant transformation and identify novel targets for treatment. These techniques have generated a period of linear growth in the study of CLL biology, particularly in the search for risk stratification markers (Figure 1). The latter now include assays for cytokines, enzymes, soluble proteins, cellular receptors, markers of angiogenesis, and recurring genetic defects. These parameters define differences in underlying disease biology that can be used with clinical tools to identify patients with more aggressive disease.
Publications evaluating prognosis in CLL by year since 1983. Results of PubMed search by year using the key words “CLL” and “prognosis.”
Publications evaluating prognosis in CLL by year since 1983. Results of PubMed search by year using the key words “CLL” and “prognosis.”
Immunoglobulin variable heavy chain (IgVH) mutation status
Historically, CLL B cells were believed to represent a leukemic transformation of naive B lymphocytes that had not undergone germinal center antigen exposure and subsequent somatic mutation of their immunoglobulin (Ig) genes. Several studies have found that approximately 50% of CLL clones exhibit somatic mutation of their immunoglobulin chains, which suggests that some CLL clones arise from postgerminal center, “memory” B cells.33 In 1999, 2 publications simultaneously reported on the prognostic significance of IgVH mutational status in CLL.7,8 These studies defined patients with more than 2% difference in nucleotide sequences from germ line cells as having a mutated clone and those with 2% or less as nonmutated. When patients with Binet stage A disease were stratified according to IgVH gene mutational status, significant survival differences were observed. The median survival was 8 to 9 years for patients with nonmutated IgVH genes (germ line cells) but beyond 24 years for patients with somatically mutated IgVH genes. These reports also noted a relationship between nonmutated IgVH genes and the need for chemotherapy,7 response to chemotherapy,7 and unfavorable cytogenetic abnormalities detected by FISH.8
Although the biologic significance of IgVH gene mutation status is unclear,34 the prognostic significance of IgVH mutation status has been confirmed,23,35-41 Additionally, patients with CLL with nonmutated IgVH genes have a higher risk of relapse after stem cell transplantation.12 Others have identified subgroups of patients with somatic mutation using specific IgVH sequences (Vh3-21) with a prognosis similar to patients with nonmutated genes.35,37,42 This finding implies response to specific antigens that select the Vh3-21 sequence may confer different biologic behavior among patients with mutated IgVH genes. What percentage of nucleotide mutation in the CDR3 region of the IgVH gene optimally defines a “mutated” type clone is unknown.7,8,35,38-40 Currently, IgVH gene mutation testing is expensive, technically difficult, and not widely available for clinical use.
Mutational status and CD38
One early report found CLL B cells with higher CD38 expression were more likely to have nonmutated IgVH genes, and CD38 status possessed a prognostic value similar to IgVH mutation status with regard to median survival.7 The median survival for patients with intermediate Rai stage with 30% or more of cells expressing CD38 was 10 years, whereas no patient with less than 30% of CD38 cells died during the follow-up period. In this initial report, the CD38 status of CLL B cells was stable and unaffected by chemotherapy. Thus, CD38 status was proposed as a possible surrogate for IgVH mutation status.
Subsequent studies did not confirm the predictive power of CD38 as a surrogate for IgVH status35,36,38,43 but substantiated the independent prognostic significance of both markers.35,36,38,40,41,43 In one study of 168 patients with CLL, most of whom had low (n = 45) or intermediate (n = 116) Rai stage, progression-free survival 5 years after diagnosis was 75% for CD38– patients but only 37% for patients who were CD38+.44 Eight-year survival was 92% for CD38– patients and 50% for CD38+ patients, and on multivariate analysis, CD38 status was a significant prognostic variable for overall survival.44 Other investigators have confirmed the ability of CD38 status to predict disease progression, response to therapy, and overall survival.45-50
In 145 patients with predominantly Binet stage A disease (122 of 145), the predictive value of IgVH status, CD38 status, and disease stage were evaluated, and all 3 variables were significant on multivariate analysis.38 The combination of CD38 and IgVH mutational status had even greater prognostic power than either marker alone. Patients whose IgVH genes were nonmutated and had 30% or more CD38 expression had a median survival of 8 years, whereas those with mutated IgVH genes and less than 30% CD38 expression had a median survival of 26 years. Three patterns of CD38 expression are found in patients with CLL: homogenous negative, homogenous positive, and bimodal.36,50 In a recent report the presence of any CD38+ clone, rather than a numerically defined cutoff, identified patients who developed progressive disease and had worse overall survival independent of stage.50
The constancy of CD38 status in CLL remains a controversial issue. Although some investigators report no change or infrequent alteration in CD38 status through the course of disease,7,48,50,51 other investigators report changes from CD38+ to CD38– or CD38– to CD38+ in as many as 10% to 25% of cases.35,38,45,46 It is not known if the change in CD38 expression signals evolution to a more aggressive disease. Additionally, the 30% cutoff used to classify patients as CD38+ or CD38– is an arbitrary threshold. Some investigators have proposed lower cutoffs, and the optimal threshold by which to classify CD38 status is uncertain.35,40,45,46
On balance, CD38 appears to be a useful and readily measurable prognostic marker. However, it has limitations because of differences in CD38 expression for individual patients over time,35,38,45,46 and it is not a surrogate for IgVH mutation status. Confirmation of the prognostic significance of bimodal CD38 expression may further refine the ability of this marker to identify patients with aggressive disease.50 Tables 2 and 3 review the median survival of patients with CLL by IgVH and CD38 status in aggregate and by disease stage.
Median survival of patients with CLL by IgVH gene mutation and CD38 status
. | . | IgVH status . | . | CD38 status . | . | ||
|---|---|---|---|---|---|---|---|
. | N . | Nonmutated, n . | Mutated, n . | More than 30% positive, n . | Less than 30% positive, n . | ||
| Damle et al7 | 47 | 9 | Not reached | 10 | Not reached | ||
| Hamblin et al8 | 84 | 9.8 | 24.4 | — | — | ||
| Hamblin et al43 | 61 | — | — | 8.8 | 24.4 | ||
| Hamblin et al38 | 145 | 9.1 | 24.4 | 9.1 | 24.4 | ||
| Oscier et al39 | 205 | 9.9 | 25.8 | ‡ | ‡ | ||
| Matrai et al41 | 40 | 9 | Not reached | 5 | 13 | ||
| Jelinek et a36 * | 66 | 5.7 | Not reached | 4.5 | Not reached | ||
| Del Poeta et al44 | 168 | — | — | 7.5 | Not reached | ||
| Durig et al48 † | 129 | — | — | 10 | Not reached | ||
| D'Arena et al49 | 61 | — | — | 7.5 | Not reached | ||
| Chevallier et al45 | 111 | — | — | 6.4 | Not reached | ||
| Krober et al35 | 325 | 6.6 | 12.7 | 6.3‡ | 9.5‡ | ||
| Lin et al40 § | 71 | 7.5 | Not reached | 7.5 | Not reached | ||
| Ibrahim et al46 | 218 | — | — | 2.9 | Not reached | ||
| Mainou-Fowler et al47 | 81 | — | — | 9.7 | Not reached | ||
| Tobin et al37 | 119 | 5.9 | 10.2 | — | — | ||
| Magnac et al23 | 38 | 7 | Not reached | — | — | ||
| Ghia et al50 ¶ | 108 | 15.3 | Not reached | 15.3 | Not reached | ||
. | . | IgVH status . | . | CD38 status . | . | ||
|---|---|---|---|---|---|---|---|
. | N . | Nonmutated, n . | Mutated, n . | More than 30% positive, n . | Less than 30% positive, n . | ||
| Damle et al7 | 47 | 9 | Not reached | 10 | Not reached | ||
| Hamblin et al8 | 84 | 9.8 | 24.4 | — | — | ||
| Hamblin et al43 | 61 | — | — | 8.8 | 24.4 | ||
| Hamblin et al38 | 145 | 9.1 | 24.4 | 9.1 | 24.4 | ||
| Oscier et al39 | 205 | 9.9 | 25.8 | ‡ | ‡ | ||
| Matrai et al41 | 40 | 9 | Not reached | 5 | 13 | ||
| Jelinek et a36 * | 66 | 5.7 | Not reached | 4.5 | Not reached | ||
| Del Poeta et al44 | 168 | — | — | 7.5 | Not reached | ||
| Durig et al48 † | 129 | — | — | 10 | Not reached | ||
| D'Arena et al49 | 61 | — | — | 7.5 | Not reached | ||
| Chevallier et al45 | 111 | — | — | 6.4 | Not reached | ||
| Krober et al35 | 325 | 6.6 | 12.7 | 6.3‡ | 9.5‡ | ||
| Lin et al40 § | 71 | 7.5 | Not reached | 7.5 | Not reached | ||
| Ibrahim et al46 | 218 | — | — | 2.9 | Not reached | ||
| Mainou-Fowler et al47 | 81 | — | — | 9.7 | Not reached | ||
| Tobin et al37 | 119 | 5.9 | 10.2 | — | — | ||
| Magnac et al23 | 38 | 7 | Not reached | — | — | ||
| Ghia et al50 ¶ | 108 | 15.3 | Not reached | 15.3 | Not reached | ||
— indicates not evaluated.
From time of enrollment on trial.
Used cutoff of 20% rather than 30% to determine CD38 positivity.
Nonsignificant difference between groups.
Used 3 cutoffs to classify IgVH gene mutation status (<2%, 2%-5%, >5%) data shown for less than 2% and more than 5%.
for IgVH analysis, N = 108; for CD38 analysis N = 148. Study reported 3 categories of CD38 status: CD38-, CD38+, and bimodal CD38 expression. Data presented for CD38- and CD38+ groups are based on 30% cutoff. Discussed in “Mutational status and CD38.”
Median survival of patients in early stage CLL based on IgVH gene mutation and CD38 status
. | . | IgVH status . | . | CD38 status . | . | ||
|---|---|---|---|---|---|---|---|
. | Stage . | Nonmutated, n . | Mutated, n . | Less than 30% positive, n . | More than 30% positive, n . | ||
| Damle et al7 | Rai I or II | 9 | 17 | 10 | Not reached | ||
| Hamblin et al8 | Binet A | 7.9 | 24.4 | — | — | ||
| Hamblin et al38 | Binet A | 12.1 | 27 | Not reached | 25.8 | ||
| Oscier et al39 | Binet A | 10 | 25 | * | * | ||
| Chevallier et al45 | Binet A | — | — | 9.1 | Not reached | ||
| Krober et al35 | Binet A | 6.6 | 12.7 | — | — | ||
| Ibrahim et al46 | Rai 0-II | — | — | 3.7 | Not reached | ||
| Ghia et al50 † | Rai 0 | — | — | 15.3 | Not reached | ||
. | . | IgVH status . | . | CD38 status . | . | ||
|---|---|---|---|---|---|---|---|
. | Stage . | Nonmutated, n . | Mutated, n . | Less than 30% positive, n . | More than 30% positive, n . | ||
| Damle et al7 | Rai I or II | 9 | 17 | 10 | Not reached | ||
| Hamblin et al8 | Binet A | 7.9 | 24.4 | — | — | ||
| Hamblin et al38 | Binet A | 12.1 | 27 | Not reached | 25.8 | ||
| Oscier et al39 | Binet A | 10 | 25 | * | * | ||
| Chevallier et al45 | Binet A | — | — | 9.1 | Not reached | ||
| Krober et al35 | Binet A | 6.6 | 12.7 | — | — | ||
| Ibrahim et al46 | Rai 0-II | — | — | 3.7 | Not reached | ||
| Ghia et al50 † | Rai 0 | — | — | 15.3 | Not reached | ||
— indicates not evaluated.
Nonsignificant difference.
Study reported 3 categories of CD38 status: CD36-, CD38+, and bimodal CD38 expression. Data presented for CD38- and CD38+ groups are based on 30% cutoff. Discussed in “Mutational status and CD38.”
Chromosome analysis
Most CLL cells are G0 phase cells which accumulate primarily through prolonged survival because of defects in apoptosis.52 The small number of dividing leukemic cells in CLL makes conventional cytogenetic testing problematic, and mitogen stimulation of CLL B cells is required to achieve an adequate number of metaphases for analysis.53 Chromosome abnormalities can be detected in 30% to 50% of patients with CLL by using this technique, and early studies identified trisomy 12, 11q–, and 17p–as poor prognostic markers.54-56
In 1990, a pooled analysis reported on the prognostic significance of chromosome analysis with the use of conventional cytogenetics in 433 patients with CLL.56 Patients with a normal karyotype or 13q–abnormalities had a better survival than patients with trisomy 12 or a complex karyotype. After controlling for age and stage, the prognostic significance of chromosome abnormalities by conventional cytogenetic testing remained significant.
With the development of interphase FISH techniques, it became possible to detect selected chromosome abnormalities in nondividing cells. Although more sensitive and specific than conventional cytogenetics, FISH does not completely evaluate all chromosomes, so one must “know what one's looking for.” FISH probes in CLL found chromosomal abnormalities were more common than detected with the use of conventional cytogenetics and had a different distribution (13q–, 11q–more common and +12 less common).56-58
The prognostic significance of chromosome analysis by FISH was studied by using a comprehensive set of probes in 325 patients with CLL.58 Chromosome abnormalities were identified in 82% of patients: 55% of patients had a 13q–, 18% 11q–, 16% trisomy 12, 7% 17p–, 6% 6q–, and 29% of patients had more than one chromosome abnormality. On multivariate analysis 17p– and 11q– were identified as variables associated with shorter overall survival. A hierarchical survival model constructed after regression analysis was used to assign patients to risk categories (Table 4). Comprehensive reviews of the significance of chromosome abnormalities in CLL have recently been published.59
Chromosome analysis: use of FISH data for prognosis58
Category . | % of Patients . | Median treatment-free interval, mo* . | Median survival, mo* . |
|---|---|---|---|
| 17p- | 7 | 9 | 32 |
| 11q- without 17p- | 17 | 13 | 79 |
| Trisomy 12 without 11q- or 17p- | 14 | 33 | 114 |
| Normal karyotype | 18 | 49 | 111 |
| 13q- as sole abnormality | 36 | 92 | 133 |
| Other abnormalities | 8 | — | — |
Category . | % of Patients . | Median treatment-free interval, mo* . | Median survival, mo* . |
|---|---|---|---|
| 17p- | 7 | 9 | 32 |
| 11q- without 17p- | 17 | 13 | 79 |
| Trisomy 12 without 11q- or 17p- | 14 | 33 | 114 |
| Normal karyotype | 18 | 49 | 111 |
| 13q- as sole abnormality | 36 | 92 | 133 |
| Other abnormalities | 8 | — | — |
— indicates heterogeneous group.
Median follow-up, 70 months.
Others subsequently evaluated the relationship between FISH, IgVH mutation status, and CD38 expression. In the largest reported study mutational status, CD38 status, and FISH detectable chromosome defects were evaluated in 325 consecutive patients.35 On univariate analysis, IgVH mutation status and CD38 status were confirmed as poor prognosis markers. On multivariate analysis, however, nonmutated IgVH genes and abnormalities of chromosome 17 and 11 but not CD38 status remained significant prognostic markers. Patients with nonmutated IgVH genes and 17p abnormalities also had significantly shorter times to progression. The investigators proposed a hierarchical risk profile based on a combination of high-risk chromosomal abnormalities (17p– or 11q–) and IgVH mutational status.35 Other investigators have proposed similar models based on FISH and CD38 status.60
The P53 gene, located on the short arm of chromosome 17, plays an integral role in inducing apoptosis or cell cycle arrest after DNA damage. In CLL, a functional p53 pathway is an important indicator of responsiveness to purine nucleoside analogues, perhaps explaining the prognostic importance of 17p–abnormalities.61,62 Multiple mechanisms by which p53 dysregulation may occur include direct p53 mutation because of abnormalities of chromosome 17p and inactivation by regulator genes, some of which reside on chromosome 11q.58,63 Because abnormal p53 function is present in a larger subset of patients with CLL (26%) than abnormalities of chromosome 17 (7%) by FISH analysis, some have proposed functional assays of p53 may be a better prognostic marker than identifying abnormalities on chromosome 17 or 11.35,40,58
To evaluate the interaction between mutational status, CD38 status, and p53 function, these variables were explored using an assay of p53 function according to the response of CLL B cells to ionizing radiation.40 p53 function was abnormal in roughly 25% of patients, and p53 function, IgVH mutation status, and CD38 status all had prognostic significance on univariate analysis. Of importance, all the patients with p53 dysfunction also had nonmutated IgVH genes.40 Immunohistochemical assessment of p53 protein expression may also have prognostic significance in CLL.28,64
New chromosomal abnormalities can be acquired by the CLL clone during the course of disease, leading to the concept of “clonal evolution.”45,57,60 Thus, as subsets of leukemic cells acquire additional chromosomal abnormalities, subclones with different cytogenetic profiles and gain or loss of function of molecular machinery may develop. These subclones may have a survival advantage(s) that results in a more aggressive disease. The use of interphase FISH for both initial prognostication and subsequent clinical evaluation to assess possible chromosome evolution is reasonable and clinically available. The probe targets of the FISH panel currently used for evaluation of patients with CLL at the Mayo Clinic are presented in Figure 2.
Probe targets of Mayo Clinic CLL FISH panel. This figure displays the chromosome and gene targets of the probes currently used in the CLL FISH Panel at Mayo Clinic (see Dewald et al60 ). CEN indicates centromere; $, abnormalities of this chromosome associated with more indolent clinical course; *, abnormalities of this chromosome associated with more aggressive clinical course. Produced with the permission of Gordon Dewald, Mayo Clinic, Rochester, MN (unpublished artwork, November 2003).
Probe targets of Mayo Clinic CLL FISH panel. This figure displays the chromosome and gene targets of the probes currently used in the CLL FISH Panel at Mayo Clinic (see Dewald et al60 ). CEN indicates centromere; $, abnormalities of this chromosome associated with more indolent clinical course; *, abnormalities of this chromosome associated with more aggressive clinical course. Produced with the permission of Gordon Dewald, Mayo Clinic, Rochester, MN (unpublished artwork, November 2003).
Serum thymidine kinase
Thymidine kinase (TK) is a cellular enzyme involved in a salvage pathway for DNA synthesis. The predominant form of TK is present in dividing cells and absent in nondividing cells; thus, this enzyme is a potentially useful marker of proliferative activity. Early studies reported that elevated TK levels correlated with advanced Rai stage and progressive disease,65 and, on retrospective analysis, untreated patients with indolent disease could be separated into 2 prognostic groups on the basis of their initial serum TK. In a more recent study of 113 patients with CLL, TK was an independent prognostic variable on multivariate analysis; however, this study did not include assessment of IgVH mutation status, CD38, or chromosome analysis by FISH.17 Later, these same investigators reported on the significance of TK in 122 patients with untreated CLL.66 When patients with Binet stage A were stratified by TK level, median progression-free survival was 9 months (range, 5-13) for patients with high serum TK levels (> 7.1 U/L) compared with 49 months (range, 24-74) for patients with low serum TK levels (P < .001).
Another recent study reported on the relationship of TK and mutation status and found that a TK level more than 15 U/L was a strong predictor of nonmutated IgVH genes (odds ratio, 31.7) with a positive predictive value of 86%.23 Others have reported a relationship between TK levels and LDT, markers of disease burden (ALC), and markers of cell turnover (lactate dehydrogenase [LDH], β-2 microglobulin).67,68 In one retrospective review of 188 patients, serum TK was a significant predictor of both survival and response to treatment.68 Assays for TK are feasible and commercially available, making it a potentially widely available and useful prognostic marker.
Other candidates for prognostic markers
Gene expression profile analysis: roadmap to other prognostic markers
Several recent studies have used gene expression profiling (GEP) analysis for the simultaneous evaluation of the expression of multiple genes.69,70 The rationale for the use of genomics-based technology is to render a more detailed and comprehensive molecular view of human cancers. Reviews of analytic methods and clinically relevant advances in hematologic malignancies using this technique have recently been published.71
Several investigators have performed unsupervised analysis using DNA microarrays in CLL. These studies found a gene signature common to all CLL cells independent of the IgVH mutation status, implying that there is a common cell of origin and/or mechanism leading to malignant transformation.70,72-74 Supervised analysis of GEP by IgVH status, however, revealed that certain restricted sets of genes were able to discriminate between the nonmutated and mutated type clones.72,73 These genes may reveal the molecular map/pathways that lead to the evolution of the CLL B cell.69,72,73
By using immunoblot assays to confirm results from GEP, investigators found the gene encoding zeta-associated protein 70 (ZAP-70), a tyrosine kinase required for T-cell receptor (TCR) signaling normally expressed in T cells, was expressed in 100% of patients with nonmutated IgVH genes but only 10% of patients with mutated IgVH genes.75 Other studies using supervised analysis of GEP reported that ZAP-70 expression is also found to be elevated in CD38+ patients70 and that ZAP-70 may play a role in CD38 signaling.76 A recent report of GEP in 107 patients with CLL confirmed that, of the genes studied, ZAP-70 was the single gene best able to distinguish IgVH mutation status and correctly predicted mutation status in 93% of patients.69 The ability of ZAP-70 to predict shorter time to treatment was also similar to IgVH gene mutation status.
In a retrospective series, ZAP-70 expression as determined by flow cytometry was evaluated as a surrogate for IgVH mutation status.77 By using an arbitrary cutoff classifying patients with 20% or more of CLL cells expressing ZAP-70 as positive, ZAP-70 expression correctly identified 32 (91%) of 35 patients with nonmutated IgVH genes. No patient with mutated IgVH genes was ZAP-70+, and, on multivariate regression, ZAP-70 was found to correlate with IgVH gene mutation status.77 ZAP-70 expression did not change on sequential analysis, and it was an independent prognostic marker for overall survival.77 The estimated median survival for patients with Binet A who were ZAP-70+ was 90 months, whereas median survival had not been reached in ZAP-70– patients (median follow-up, 63 months).
Although ZAP-70 is a promising prognostic marker, a number of questions remain. As a surrogate for mutation status, the test characteristics of the flow cytometry ZAP-70 assay (sensitivity, 91%; specificity, 100%) are nearly identical to the original reports of CD38 flow cytometry (sensitivity, 85%; specificity, 100%) which also reported no variation on sequential analysis.7,77 One small series of sequential ZAP-70 analysis by flow cytometry found 2 of 16 patients had a change in ZAP-70 status on serial testing by using the 20% threshold and 2 other patients had large changes in the percentage of CLL B cells expressing ZAP-70.78 The 20% cutoff to classify cells ZAP-70+ is arbitrary, and the optimal cutoff remains to be determined.77 ZAP-70 detection by flow cytometry is a technically difficult assay for an intracellular antigen that requires the cell membrane be permeabilized.77 If ZAP-70 is confirmed as an important prognostic variable for patients with CLL, the stability and optimal method of detection will need to be determined.69,77,78 Table 5 reports the sensitivity and specificity of markers proposed as surrogates for IgVH mutation status.
Sensitivity and specificity of proposed surrogates for nonmutated IgVH gene mutation status
Surrogate for IgVH . | Sensitivity, % . | Specificity, % . | References . |
|---|---|---|---|
| CD38 using 30% cutoff* | 45-85 | 71-100 | Damle et al,7 † Hamblin et al,38 ‡ Krober et al,35 ‡ Crespo et al77 † |
| CD38 using 7% cutoff* | 63-66 | 63-68 | Jelinek et al,36 † Krober et al35 ‡ |
| CD38 using presence of clone* | 80 | 85 | Ghia et al50 † |
| Thymidine kinase* | 76 | 91 | Magnac et al23 ‡ |
| ZAP-70*§ | 89-91 | 95-100 | Wiestner et al,69 ‡ Crespo et al77 † |
Surrogate for IgVH . | Sensitivity, % . | Specificity, % . | References . |
|---|---|---|---|
| CD38 using 30% cutoff* | 45-85 | 71-100 | Damle et al,7 † Hamblin et al,38 ‡ Krober et al,35 ‡ Crespo et al77 † |
| CD38 using 7% cutoff* | 63-66 | 63-68 | Jelinek et al,36 † Krober et al35 ‡ |
| CD38 using presence of clone* | 80 | 85 | Ghia et al50 † |
| Thymidine kinase* | 76 | 91 | Magnac et al23 ‡ |
| ZAP-70*§ | 89-91 | 95-100 | Wiestner et al,69 ‡ Crespo et al77 † |
These markers have all been shown to have prognostic significance independent of their relationship to IgVH gene mutation status.
Sensitivity and specificity as reported in reference text.
Sensitivity and specificity calculated from published data.
The results from Wiestner et al69 are based on ZAP-70 mRNA expression by gene expression profile analysis; results from Crespo et al77 are based on flow cytometry using 20% threshold to classify the patient's expression of ZAP-70. See “Gene expression profile analysis: roadmap to other prognostic markers.”
Markers of angiogenesis
Angiogenesis, the growth of new capillaries from preexisting blood vessels, has increasingly been recognized as an important process in the growth of solid tumors,79 and similar observations have linked angiogenesis with tumor growth in various hematologic malignancies.80 Early studies in CLL revealed bone marrow microvessel density correlates with disease stage,81 and subsequent work found a high bone marrow microvessel count identifies patients with a shorter progression-free survival.82 Other reports suggest serum and urine levels of proangiogenic factors are increased in CLL.82,83
Several preliminary investigations have explored the relationship between vascular factors and survival in CLL. Vascular endothelial growth factor (VEGF) is a stimulator of angiogenesis and is believed to be a significant mediator of tumor angiogenesis.84 Serum VEGF levels in 68 patients with CLL and 31 control patients were found to be similar. Patients with Binet stage A or Rai stages I-II who had serum VEGF levels greater than the median, however, had significantly shorter progression-free survival.84 In aggregate these results suggest that angiogenesis may be an important process in the pathogenesis of CLL and could provide important early clues about patients destined to have more aggressive disease. If validated, the measurement of serum VEGF could help discriminate patients with high-risk early stage CLL.
Other
Although beyond the scope of this review, several other cytokines, enzymes, and molecular markers, including interleukin 6 (IL6),27,85 IL10,27 tumor necrosis factor (TNF),86 intracellular Bcl-2,87 and soluble intracellular adhesion molecule 1 (ICAM1),88 have shown promise as potential prognostic markers in CLL and deserve further investigation. Serum levels of soluble CD23 are particularly interesting, although optimal thresholds for prognostication are unknown.6,44,89,90
Integration of multiple prognostic variables into clinical practice: use of statistical modeling
The increasing array of biologic markers provides the challenge and opportunity to develop more exact algorithms that integrate combinations of markers and tools to guide counseling and treatment decisions for individual patients. Several modeling techniques can be used to evaluate the prognostic significance of multiple factors used in combination. By using time-to-event endpoints of overall survival and time to progression, it is appropriate to focus on Cox proportional hazards modeling. The greatest challenge using this approach is determination of which risk factors belong in the final prognostic model. Relying on P values alone to identify significant prognostic factors is inadequate when a prediction model based only on statistical significance does not take into account the clinical significance of the marker. To be included in the prognostic model a marker must provide additional prognostic information over existing tools to justify its cost in the care of the patient. For CLL, a new marker would need to improve our ability to predict the patient's course over Rai or Binet stage. Although measures of disease burden (bone marrow histologic pattern) may be statistically significant prognostic markers, they measure risk already captured in staging criteria and are unlikely to markedly improve prognostication.
Multiple methods to determine the value of information added by a new marker to existing prognostic tools have been proposed.91-93 The concordance index (ie, c-index or Harrell c statistic) used in conjunction with Cox proportional hazards modeling can quantify the marginal utility of a marker to predict survival and time to progression. Guidelines to ensure the validity of these methods and the predictive model are available.91,94 A prediction model built from such analysis needs to be validated in some additional manner, whereby bootstrap validation is particularly helpful to correct for potential overfitting.
An important caveat to these approaches is that a valid model requires a very large data set. Such models have been constructed for prostate cancer and soft tissue sarcoma, with inclusion of more than 1000 and 2000 patients, respectively.95,96 To facilitate the development of a prognostic model for CLL, we propose a multiple institution effort that pools patients involved in clinical CLL research to provide sufficient numbers of patients to generate a generalizable risk stratification model. This approach is under way in the multiple myeloma community, whereby data from international groups are being pooled to develop an International Staging System (ISS).97 Furthermore, the National Cancer Institute's (NCI) mandate to use common data elements in trials and databases should facilitate such modeling endeavors. Applying these statistical techniques to a large, multi-institution data set with information on age, sex, Rai stage, IgVH mutational status, FISH analysis, B2M, CD38 status, and other recently identified prognostic measures (eg, TK, ZAP-70, sCD23) should allow optimization of a clinically useful prognostic index. Factors that contribute to the model in a statistically significant fashion and independently improve the ability to identify patients with shorter survival by at least 5% should be considered for inclusion in the prognostic index.
Recommendations for clinicians
Consensus guidelines for treatment of patients with CLL have been proposed by the NCI Working Group.98 Indications for treatment include constitutional symptoms (weight loss > 10% body weight in < 6 months, fever in absence of infection, night sweats, extreme fatigue), marrow failure (progressive anemia or thrombocytopenia), massive splenomegaly (> 6 cm below costal margin), massive lymphadenopathy (> 10 cm in greatest dimension), more than 50% increase in ALC in less than 2 months (or estimated LDT < 6 months), or autoimmune hemolytic anemia or immune thrombocy topenic purpura (ITP) refractory to other therapies. Importantly, treatment of unselected patients with early stage has been shown to increase toxicity without improving overall survival.99,100 Thus, patients in early stage who do not meet the NCI Working Group treatment criteria should not be treated outside of clinical trials at the present time.
Unfortunately, the NCI treatment criteria do not identify patients in early stage with biologically aggressive disease. As discussed, the identification of high-risk, early stage patients is increasingly feasible. Pending large scale statistical modeling to determine a prognostic index, current use of prognostic tools for patients with newly diagnosed CLL should include staging, LDT, B2M, CD38 testing, CLL-relevant FISH panel, and, if available, IgVH gene mutation status (Table 6). Patients with a good Eastern Cooperative Oncology Group (ECOG) performance status and unfavorable prognostic features (nonmutated IgVH genes, CD38+, unfavorable FISH results [17p–, 11q–], elevated B2M, or short LDT) should be offered participation in clinical trials. The ability of these markers to predict survival in previously treated patients is not well defined and needs further investigation.
Recommended studies to perform at diagnosis for patients with CLL to identify high-risk patients
. | Clinical practice* . | Clinical trials† . |
|---|---|---|
| Staging (Rai or Binet) | Yes | Yes |
| Lymphocyte doubling time | Yes | Yes |
| β-2 Microglobulin | Yes | Yes |
| Cytogenetics by FISH | Yes | Yes |
| CD38 status | Yes | Yes |
| IgVh mutation status | If available | Yes |
| ZAP-70 | Not available | Yes |
| Thymidine kinase | No | Consider |
| Markers of angiogenesis‡ | No | Consider |
| Statistical modeling | Not available | Yes |
. | Clinical practice* . | Clinical trials† . |
|---|---|---|
| Staging (Rai or Binet) | Yes | Yes |
| Lymphocyte doubling time | Yes | Yes |
| β-2 Microglobulin | Yes | Yes |
| Cytogenetics by FISH | Yes | Yes |
| CD38 status | Yes | Yes |
| IgVh mutation status | If available | Yes |
| ZAP-70 | Not available | Yes |
| Thymidine kinase | No | Consider |
| Markers of angiogenesis‡ | No | Consider |
| Statistical modeling | Not available | Yes |
Recommendations for clinicians in “routine” evaluation of their patients with CLL.
Recommendations for risk stratification parameters that should be measured in clinical trials in which formal biostatistical analysis can be performed on well-characterized groups of patients with CLL.
Including both marrow angiogenesis levels as well as serum levels of proangiogenic and antiangiogenic factors.
Conclusion
Future studies of CLL using molecular biologic techniques will continue to refine our understanding of the biology and clinical behavior of this disease and identify novel targets for treatment (Figure 3). Correlation of molecular abnormalities with clinical course is likely to refine risk stratification for individual patients in a wise and timely fashion. Most important, they will need to identify individuals who may benefit from an early or more aggressive treatment strategy. Integration of multiple markers and tools through use of statistical modeling should identify which markers or groups of markers incrementally improve the ability to identify high-risk patients who may benefit from tailored treatment strategies.
Novel risk stratification: nonmutated versus mutated CLL B-cell clones. Figure presents a schematic of some of the biologic differences of CLL B cells on the basis of IgVH gene mutation status. NM indicates CLL B cells with nonmutated IgVH genes; M, CLL B cells with mutated IgVH genes.
Novel risk stratification: nonmutated versus mutated CLL B-cell clones. Figure presents a schematic of some of the biologic differences of CLL B cells on the basis of IgVH gene mutation status. NM indicates CLL B cells with nonmutated IgVH genes; M, CLL B cells with mutated IgVH genes.
Prepublished online as Blood First Edition Paper, October 23, 2003; DOI 10.1182/blood-2003-07-2281.
We thank Morie Gertz for inspiration and Clive Zent, MD, Joseph Colgan, MD, and Gordon DeWald, PhD, for their critical review of this manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal