Abstract
Point mutations of D835/I836 of the FLT3 gene have been reported in adult acute myeloid leukemia (AML), but not in pediatric AML or acute lymphoblastic leukemia (ALL). FLT3-D835/I836 mutations were found in 6 (5.4%) of 112 children with ALL older than 1 year and in 8 (16.0%) of 50 infants with ALL. Missense mutations were found in 11 patients, 3-base pair deletions in 2 patients, and a deletion/insertion in 1 patient. Remarkably, FLT3-D835/I836 mutations were found in 8 (18.2%) of 44 infants with ALL with MLL rearrangements and in 4 (21.5%) of 19 patients with hyperdiploid ALL, but they were not found in any patients older than 1 year who had TEL-AML1 (n = 11), E2APBX1 (n = 4), or BCR-ABL (n = 6) fusion genes. Although infant ALL patients with mutations had poorer prognoses than did those without mutations, pediatric ALL patients with mutations who were older than 1 year had good prognoses. We also found FLT3-D835 mutations in 2 of 11 leukemic cell lines with MLL rearrangements. FLT3 was highly phosphorylated in these cell lines with FLT3-D835 mutations, leading to constitutive activation of downstream targets such as signal transducer and activator of transcription 5 (STAT5) without FLT3 ligand stimulation. These results suggested that FLT3-D835/I836 mutations are one of the second genetic events in infant ALL with MLL rearrangements or pediatric ALL with hyperdiploidy. (Blood. 2004;103:1085-1088)
Introduction
Tyrosine kinases (TKs) function as the control of cellular signal transmission. Some TKs are closely associated with normal hematopoietic regulation and cell function.1 Of TKs involved in hematopoiesis, some genes related to TKs fuse to functionally important genes, such as ABL or ALK-related fusion genes, and others have mutations, such as the FMS or c-KIT genes.1 Constitutive phosphorylation of TK induced by these gene aberrations leads to a strong proliferative activity in hematopoietic cells, and often it is involved in the development of leukemia/lymphoma.1,2 In these TKs, the FLT3 gene is a receptor TK that has a transmembrane domain and plays an essential role in hematopoiesis.2 Internal tandem duplication (ITD) of the juxtamembrane (JM) domain of the FLT3 gene was identified in acute myeloid leukemia (AML).3 The frequency of FLT3-ITDs is 17% to 27% of de novo adult AML, 5% to 17% of childhood AML, 3% to 5% of myelodysplastic syndrome (MDS), and 3% of acute lymphoblastic leukemia (ALL).3-9 FLT3-ITD leads to constitutive activation and is recognized as a significant prognostic factor in adult3-6 and pediatric7-9 AML. FLT3-ITD activates the signal transducer and activator of transcription 5 and MAP kinase pathway.10 In a murine bone marrow transplant model, the FLT3-ITD mutant induces a myeloproliferative disorder.11
Recently, point mutations of D835/I836 in the activation loop of the second TK domain of FLT3 were found in adult AML, MDS, and ALL.5,12,13 The incidence of FLT3-D835/I836 mutations was 7% of de novo adult AML, 3% of adult MDS, and 2.8% of ALL.5,12,13 The D835/I836-mutant FLT3 induces the constitutive tyrosine-phosphorylation and interleukin-3 (IL-3)-independent proliferation of 32Dcl3 cells.12 AML patients with FLT3-D835/I836 mutations tend to have poor prognoses,5,12,13 suggesting that the FLT3-activating mutations of ITDs and D835/I836 mutations play important roles in AML. However, these FLT3-ITDs have rarely been found in pediatric ALL patients carrying myeloid markers and have not been associated with a poor prognosis.8 In pediatric ALL, FLT3-D835/I836 mutations have not been reported previously. To clarify the relationship between the clinical features of ALL and FLT3-D835/I836 mutations, we analyzed FLT3-D835/I836 mutations in pediatric ALL, including infant ALL, and found the mutations to be relatively frequent in infant ALL with MLL rearrangements and pediatric ALL with hyperdiploidy.
Study design
Patient samples
We analyzed 162 patients with pediatric ALL—50 infants younger than 11 months and 112 children older than 1 year (93 B-cell precursors, 19 T-cell phenotypes)—in addition to 81 healthy donors. ALL was diagnosed in these patients according to the French-American-British (FAB) classification. Chromosomal analysis was performed using G-banding, as previously reported,14 and karyotyping was successful in 136 (84.0%) patients. MLL rearrangements were examined in 50 infants with ALL with the restriction enzymes EcoRI and HindIII using Southern blotting, as previously reported.14 DNA indexing was performed in 112 ALL patients 1 year and older.15 A DNA index greater than 1.14 indicated hyperdiploidy. Pediatric ALL patients (age range, 1-15 years; median, 6 years) were mainly treated according to the Tokyo Children's Cancer Study Group (TCCSG) L95-14 protocol,15 and infant ALL patients (age range, 0-11 months; median, 5 months) were mainly treated with the Japan Infant Leukemia Study MLL96 protocol.16 Informed consent was obtained from the patients and/or the patients' parents and the healthy donors.
Leukemia cell lines
FLT3-D835/I836 mutations and ITDs were examined in 44 leukemia cell lines as follows8,17 : 20 B-precursor ALL cell lines (LC4-1, NALM-26, NALM-17, UTP-L5, REH, UTP-L10, UTP-2, NALM-20, NALM-24, BV173, OM9;22, SCMC-L10, KOCL-33, KOCL-44, KOCL-45, KOCL-58, KOCL-69, HAL-01, KOPN-41, KOPN-1), 3 B-ALL cell lines (BALM-6, DAUDI, BAL-KH), 3 T-ALL cell lines (ALL-SIL, CCRF-HSB-2, KCMC-T), 6 AML cell lines (YNH-1, KASUMI-3, KG-1, SN-1, NB4, HEL), 7 acute monocytic leukemia (AMOL) cell lines (MV4;11, THP-1, CTS, P31/FUJ, MOLM-13, KOCL-48, IMS/M1), 2 acute megakaryoblastic leukemia (AMKL) cell lines (CMS, CMY), and 3 chronic myelogenous leukemia (CML) cell lines (MOLM-1, MOLM-7, TS9;22).
RFLP-mediated PCR for the detection of FLT3-D835/I836 mutations and PCR for the detection of FLT3-ITDs
High-molecular-weight DNA or total RNA was extracted from bone marrow or peripheral blood samples from the patients using standard methods.8,17 Total RNA (4 μg) was reverse transcribed to cDNA with a cDNA Synthesis Kit (Amersham Pharmacia Biotech, Buckinghamshire, England). FLT3-D835/I836 mutations were examined by restriction fragment length polymorphism (RFLP)-polymerase chain reaction (PCR) or reverse transcription-PCR (RT-PCR). The sense primer used for PCR and RT-PCR was 17F,12 and the antisense primer for PCR or RT-PCR was FLT3-TK-R1 5′-AGTAAGCAGACTGCTGTGAG-3′) or FLT3-TK-R2 5′-GTAGAAGTTAGCATCAACCGG-3′), respectively. PCR procedure has been reported previously.8,17 Five microliters of the PCR product were digested with 5 U EcoRV for 1 hour at 37°C, then electrophoresed on 3% agarose gel. Undigested PCR products were directly sequenced by the fluorometric method. The FLT3-ITD was analyzed by PCR or RT-PCR, as previously reported.8
Antibodies and Western blot analysis
Monoclonal antibodies against phosphotyrosine (clone 4G10), signal transducer and activator of transcription 5 (STAT5) (clone 89), and α-tubulin (clone TU-01) were obtained from Upstate Biotechnology (Lake Placid, NY), Transduction Laboratories (Lexington, KY), and Sanbio (Uden, Netherlands), respectively. Rabbit polyclonal antibodies against FLT3 (C-20) and phospho-STAT5, which reacts to the Tyr694-phosphorylated active form of STAT5a and STAT5b, were from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling Technology (Beverly, MA), respectively. Western blot analysis was performed as reported previously.18 In brief, lysates of leukemia cells were separated, transferred, and incubated with the primary antibody, followed by incubation with horseradish peroxidase-conjugated second antibody. In the experiment for FLT3 phosphorylation, FLT3 was immunoprecipitated by anti-FLT3 antibody and protein A beads and then immunoblotted by antiphosphotyrosine antibody. The detection of bands was performed using an enhanced chemiluminescence kit (Amersham Japan, Tokyo).
Results and discussion
We detected FLT3-D835/I836 mutations in 6 (5.4%) of 112 children older than 1 with ALL and in 8 (16.0%) of 50 infants with ALL (Table 1). Missense mutations were found in 11 patients—Asp835Tyr in 3, Asp835His in 2, Asp835Glu in 2, and Ile836Met in 4. Deletions of 3 base pairs (Ile836del) were found in 2 patients. One patient had a deletion and an insertion mutation consisting of a 4-amino acid (Asp835-Ser838) deletion in 1 allele and a 3-amino acid (Ala835, Leu836, and Gly837) insertion in another allele. All the mutations were heterozygous, and the open reading frame was conserved. The Asp835 mutation probably was an activating mutation because the Asp835Tyr, His, and Glu found in this study were constitutively tyrosine phosphorylated.12 Ile836del has also been shown to have strong autophosphorylation,19 suggesting an association with the proliferation of leukemic cells because these mutations were detected in adult AML and tended to reduce disease-free survival.5,12,13
Clinical features of infant and pediatric ALL patients with FLT3-D835/I836 mutations
Patient . | Age . | Sex . | WBCs × 104/μL . | DNA index . | Relapse . | Prognosis . | FLT3-D835/I836 mutations . |
|---|---|---|---|---|---|---|---|
| Infants | |||||||
| 1* | 4 mo | M | 67.6 | ND | + | Dead | I836M |
| 2* | 3 mo | M | 14.6 | ND | − | Alive | D835E |
| 3† | 3 mo | F | 2.5 | ND | + | Dead | D835Y |
| 4† | 6 mo | F | 12.3 | ND | + | Dead | D835Y |
| 5† | 6 mo | M | 99.3 | ND | + | Dead | D835H |
| 6† | 4 mo | M | 53.7 | ND | − | Dead | I836del |
| 7* | 2 mo | F | 95.3 | ND | + | Dead | D835Y |
| 8* | 2 mo | F | 50.0 | ND | − | Alive | I836del |
| Children | |||||||
| 9 | 3 y | M | 7.4 | 1.19‡ | − | Alive | I836M |
| 10 | 2 y | F | 18.5 | 1.15‡ | − | Alive | I836M |
| 11 | 4 y | F | 6.9 | 1.2‡ | − | Alive | D835H |
| 12 | 2 y | F | 5.6 | 1.19§ | − | Alive | I836M |
| 13 | 3 y | M | 5.8 | 0.99‡ | − | Alive | D835E |
| 14 | 7 y | M | 102.7 | 1.0§ | − | Alive | 9-bp del + 6-bp ins∥ |
Patient . | Age . | Sex . | WBCs × 104/μL . | DNA index . | Relapse . | Prognosis . | FLT3-D835/I836 mutations . |
|---|---|---|---|---|---|---|---|
| Infants | |||||||
| 1* | 4 mo | M | 67.6 | ND | + | Dead | I836M |
| 2* | 3 mo | M | 14.6 | ND | − | Alive | D835E |
| 3† | 3 mo | F | 2.5 | ND | + | Dead | D835Y |
| 4† | 6 mo | F | 12.3 | ND | + | Dead | D835Y |
| 5† | 6 mo | M | 99.3 | ND | + | Dead | D835H |
| 6† | 4 mo | M | 53.7 | ND | − | Dead | I836del |
| 7* | 2 mo | F | 95.3 | ND | + | Dead | D835Y |
| 8* | 2 mo | F | 50.0 | ND | − | Alive | I836del |
| Children | |||||||
| 9 | 3 y | M | 7.4 | 1.19‡ | − | Alive | I836M |
| 10 | 2 y | F | 18.5 | 1.15‡ | − | Alive | I836M |
| 11 | 4 y | F | 6.9 | 1.2‡ | − | Alive | D835H |
| 12 | 2 y | F | 5.6 | 1.19§ | − | Alive | I836M |
| 13 | 3 y | M | 5.8 | 0.99‡ | − | Alive | D835E |
| 14 | 7 y | M | 102.7 | 1.0§ | − | Alive | 9-bp del + 6-bp ins∥ |
MLL rearrangements were found in all infants. WBCs refers to white blood cells.
t (4;11).
t (11;19).
Normal karyotype.
Not available.
Deletion (D835-S838) + insertion (A835, L836, G837).
No FLT3-ITDs were found in this study. FLT3-ITDs are rarely found in pediatric and adult ALL,8,20 although they are frequently found in AML.3-8 FLT3-ITDs may not be involved in growth advantage in ALL because ALL carrying FLT3-ITDs were not associated with poor prognoses.8 Neither ITDs nor D835/I836 mutations were detected in the healthy donors in this study, as previously reported.12,13
FLT3-D835/I836 mutations were frequently found in infants with ALL with MLL rearrangements and children with ALL with hyperdiploidy
Of 50 infants with ALL, 44 patients had CD10- early pre-B cell ALL with MLL rearrangements, 5 had CD10+ B-cell precursor ALL, and 1 had T-cell ALL. Interestingly, FLT3-D835/I836 mutations were found in 8 (18.2%) of 44 infants with ALL with MLL rearrangements—t(4;11)(q21;q23)/MLL-AF4 in 4 patients and t(11;19)(q23;p13)/MLL-ENL in 4 patients—but not in the remaining 6 patients without MLL rearrangements. However, this difference was statistically not significant. Six of 8 patients with the mutations died, and the overall survival rate in the patients with the mutations was lower than that in the patients with wild-type FLT3, though the difference was not significant (P = .42).
B-cell precursor ALL (CD10+, CD19+, CD3-, CD13-, CD33-) was diagnosed in 6 children with the FLT3-D835/I836 mutations. Interestingly, FLT3-D835/I836 mutations were found in 4 (21.5%) of 19 children with ALL with hyperdiploidy (all were alive). Frequencies of FLT3-D835/I836 mutations were significantly higher in pediatric ALL with hyperdiploidy than in the other patients (P = .0074). No mutations were found in patients with TEL-AML1 (n = 11), E2A-PBX1 (n = 4), or BCR-ABL (n = 6) fusion genes. FLT3-D835/I836 mutations were not associated with sex, age, white blood cell (WBC) or platelet counts, hepatosplenomegaly, or involvement of the central nervous system at onset in either pediatric or infant ALL (data not shown).
Of 44 leukemia cell lines with the FLT3 expression, FLT3-D835 mutations were found in 2 (4.5%) leukemia cell lines, including 1 ALL (KOCL-33 carrying t(11;19)) and 1 AMOL (KOCL-48 carrying t(4;11)) cell line, which were derived from infant leukemia. FLT3-ITDs were found in 2 (4.5%) AMOL cell lines—MV4;11 carrying t(4;11) and MOLM-13 carrying t(9;11)—which were derived from pediatric or adult AMOL patients. Interestingly, 4 (36.3%) of 11 leukemia cell lines with MLL rearrangements had FLT3-activating mutations. On the other hand, no FLT3-activating mutations were found in any B-ALL, T-ALL, AML, AMKL, or CML cell lines.
FLT3 and STAT5 were highly phosphorylated in lymphoid and myeloid leukemic cell lines with FLT3-D835/I836 mutation
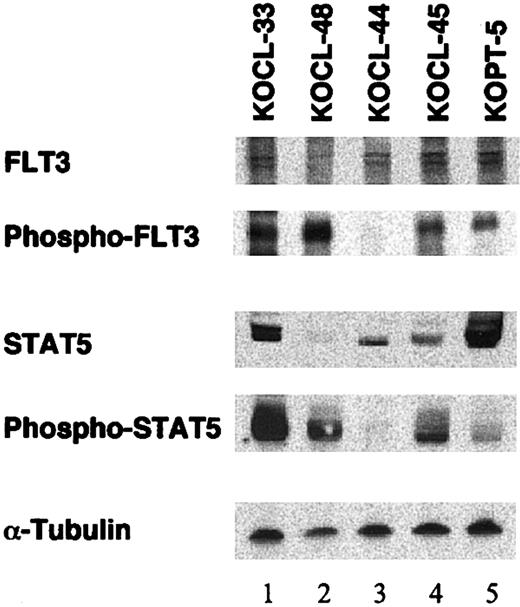
It is known that FLT3 ligand (FL) stimulation of FLT3 results in the activation of STAT5a in murine myeloid Ba/F3 cells through a Janus kinase (JAK)-independent mechanism.21 It is also known that ITDs and D835/I836 mutations in AML induce the constitutive phosphorylation of FLT3 and the activation of its downstream targets.2 However, it remains unknown whether the phosphorylation status of FLT3 and its downstream signal transduction pathways are different between myeloid and lymphoid leukemia cells with FLT3-D835/I836 mutations. To address this point, we examined the phosphorylation of FLT3 and STAT5 on Western blot in 5 leukemia cell lines, including 2 cell lines with D835 mutations. As shown in Figure 1, lymphoid (KOCL-33, lane 1) and monocytoid (KOCL-48, lane 2) cell lines with D835 mutations showed marked tyrosine phosphorylation of FLT3 and STAT5. Of note, the expression of FLT3 and STAT5 in KOCL-48 was lowest among the cell lines examined, but their phosphorylation status was markedly elevated. Thus, FLT3 in lymphoid and myeloid MLL rearrangement-positive leukemias with FLT3-D835/I836 mutations might be constitutively and highly phosphorylated, leading to the constitutive activation of downstream targets such as STAT5 without FLT3 ligand stimulation.
Western blot analysis on tyrosine phosphorylation of FLT3 and STAT5 in leukemia cell lines with or without D835 mutations. Lysates of leukemia cell lines with (lanes 1-2) or without (lanes 3-5) FLT3-D835 mutations were separated, blotted, and stained with antibodies against FLT3, phosphotyrosine (after immunoprecipitation by anti-FLT3), STAT5, phospho-STAT5, and α-tubulin, respectively. Lanes 1, 3, and 4 were B-precursor cell lines with MLL rearrangement; lane 2 was a monocytoid cell line with MLL rearrangement; lane 5 was a T-lymphoid cell line.
Western blot analysis on tyrosine phosphorylation of FLT3 and STAT5 in leukemia cell lines with or without D835 mutations. Lysates of leukemia cell lines with (lanes 1-2) or without (lanes 3-5) FLT3-D835 mutations were separated, blotted, and stained with antibodies against FLT3, phosphotyrosine (after immunoprecipitation by anti-FLT3), STAT5, phospho-STAT5, and α-tubulin, respectively. Lanes 1, 3, and 4 were B-precursor cell lines with MLL rearrangement; lane 2 was a monocytoid cell line with MLL rearrangement; lane 5 was a T-lymphoid cell line.
FLT3-D835/I836 mutations are the second genetic events in infant ALL with MLL rearrangements and ALL with hyperdiploidy
Frequent additional alterations of proliferation-related genes or tumor-suppressor genes as the second genetic events in ALL with hyperdiploidy22,23 or MLL rearrangements have not yet been reported.18,24 Thus, the FLT3-D835/I836 mutations found in this study are considered to be the second genetic events in ALL. As for prognoses, our results indicated that FLT3-D835/I836 mutations affected the poorer prognoses of infants with ALL and MLL rearrangements. In contrast, patients with hyperdiploid ALL and the mutations had good clinical outcomes, suggesting that the mutations may not affect the growth advantage of hyperdiploid ALL cells. Further larger prospective studies are needed. Recently, gene expression profiling by microarray showed that FLT3 expression was higher in acute leukemia with MLL rearrangements25 and in ALL with hyperdiplody.26 These studies suggest that FLT3 high-expression or constitutive tyrosine phosphorylation possibly caused by D835/I836 mutations in ALL with MLL rearrangements or hyperdiploidy might contribute to the pathogenesis of ALL with MLL rearrangements or hyperdiploidy and that a molecularly targeted drug against activating FLT3 should be considered in the future.
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-02-0418.
Supported by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor and Welfare of Japan, a Grant-in-Aid for Scientific Research on Priority Areas, and a Grant-in-Aid for Scientific Research (B) and (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Supported also by the Nagao Medical Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Professor Seiji Yamaguchi (Department of Pediatrics, Shimane Medical University) for critical comments and Shoko Sohma, Hisae Soga, and Yumiko Taketani for their excellent technical assistance. We thank Dr Takeyuki Sato (Department of Pediatrics, Chiba University School of Medicine) for providing AMKL (CMS, CMY) cell lines; Dr Yoshinobu Matsuo (Hayashibara Biochemical Laboratories, Inc, Fujisaki Cell Center) for providing varieties of ALL cell lines; and Dr Kazuma Ohyashiki (First Department of Internal Medicine, Tokyo Medical University) for providing Philadelphia chromosome ALL and CML cell lines.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal