Abstract
The destruction of viral-infected and tumor cells is mediated in part via the lysis receptor of natural killer (NK) cells, NKp46. The nature, however, of its lysis ligands expressed on target cells is poorly defined. Recently, we have identified a novel functional interaction between the lysis receptors NKp46 and NKp44 and the hemagglutinin of influenza and hemgglutininneuroaminidase of Sendai viruses. This recognition depends on the sialylation of NKp46 and NKp44 receptors. In this study, we expand the significance of these observations by demonstrating a conserved pattern of NKp46 and NKp44 recognition by various hemagglutinins derived from different viral strains. We further establish that this recognition is direct and mainly mediated via α2,6-linked sialic acid carried by NKp46. In addition, we demonstrate that the ability of NKp46 to recognize target cells is confined to the membrane proximal domain, and largely relies on the highly conserved sugar-carrying residue, Thr 225. This residue plays a critical dual role in NKp46 interactions with both viral hemagglutinins and the unknown tumor ligands via different mechanisms. These results may explain the ability of NK cells to kill such a broad spectrum of viral-infected and tumor cells.
Introduction
Natural killer (NK) cells are bone marrow–derived lymphocytes that constitute a key frontline defense against a range of hazardous conditions, including viral infection and tumor transformation.1 Although NK cells can kill target cells spontaneously without prior stimulation, a delicate balance between inhibitory and activating signals tightly regulates their activation.1-9
A significant breakthrough in the understanding of the specific activation of NK cells was achieved following the recent identification of 3 novel NK-specific triggering receptors collectively termed natural cytotoxic receptors (NCRs). The NCRs, which include NKp46, NKp44, and NKp30, belong to the immunoglobulin (Ig) super family but share no homology with each other and only low identity with other known human molecules.10-12
The most distinctive role of the NCRs in NK cell activity has been attributed to their involvement in recognition and killing of tumor cells. This has become evident by the ability of anti-NCR monoclonal antibodies to block NK-mediated killing of most tumor lines10-14 and by the strict correlation that exists between the density of the expression of NCRs on NK cells and the ability of NCRs to kill tumor targets.13 More recently, the importance of NCRs in vivo was illustrated in patients with acute myeloid leukemia (AML) expressing an insufficient amount of either NCR or NCR ligands, thereby rendering the leukemia cells resistant to NK cytotoxicity.15 However, in spite of the accumulating evidence of the important role played by NCRs in antitumor defense mechanisms, little is known about the nature and distribution of the NCR ligands. The identification of such ligands is currently a subject of intense investigation.
Although NCRs have been implicated most conclusively in immunity against transformed cells, there is evidence showing that they also contribute to the defense against other pathogens.1,16,17 This notion is supported by the fact that NK-deficient individuals suffer from a range of recurrent diseases, especially viral infections.5 Recently, our group has demonstrated that soluble Ig fusion proteins NKp46 and NKp44, but not NKp30, specifically bind to hemagglutinin (HA) of influenza virus (IV) and hemagglutininneuroaminidase of Sendai virus (SV).18,19 We demonstrated that these interactions are functional and are largely mediated via sialylated residues of NKp46 and NKp44.18,19
The aim of the current work was to explore the mechanisms of NKp46 recognition. We demonstrate that the ability of HA to recognize NKp46 and NKp44 receptors is direct and represents a general feature conserved among HA molecules derived from various viral strains. We further characterize the NKp46 recognition by viral HA and demonstrate that this interaction is primarily mediated via α2,6-terminal sialic acid expressed on NKp46. In addition, we show that the ability of NKp46 to bind tumor as well as viral ligands is restricted to the membrane proximal domain of the receptor. Finally, we identify the sugar-carrying residue, Thr225, as a central amino acid critically involved in the recognition of viral HAs and the unknown cellular ligands expressed on tumor cells.
Materials and methods
Cells and viruses
The following cell lines were used: melanoma cell lines (1106mel, LBmelA1, 1259mel, 1076mel), prostate carcinoma cell lines (PC3, LnCap), Epstein-Barr virus (EBV)–transformed B cells (DAUDI, 721.221), and the mouse P815 line.
The SV was purchased from Spafas (Preston City, CT). The influenza A/Beijing (A/Beijing/262/95-like [H1N1]), A/Sydney (A/Sydney/5/97-like [H3N2]), and A/Moscow (A/Moscow/10/99-like [H3N2]) viruses were propagated as previously described.20 The cells were infected by incubating one million cells overnight in 3 mL complete medium (RPMI 1640 plus 10% fetal calf serum [FCS]) at 37°C and 5% CO2 with 100 μL of different virus strains.
Monoclonal antibodies and antivirus sera
The anti-CD99 monoclonal antibody (mAb) 12E7 was a kind gift from A. Bernard (Hospital de L'Archet, Nice, France). The generation of the 135.7 mAb directed against SV-HA was previously described.18
Direct binding assay
Rosettes of HA from X31 (A/Aichi/2/68xpr/8/34) H3N2 influenza virus were prepared as previously described.23 Briefly, purified HA obtained either from Dr Skehel (National Institute for Medical Research, London, United Kingdom) or from Protein Sciences Corp (Meriden, CO), was dialyzed overnight at 4°C against 0.1 M citric acid in phosphate-buffered saline (PBS) (pH = 5). For the enzyme-linked immunosorbent assay (ELISA), plates were coated with 0.1 μg HA rosettes overnight at 4°C. After 2 hours of blocking on ice with 1% bovine serum albumin (BSA) in PBS, plates were washed (1% BSA in PBS/0.05%/Tween 20) and incubated overnight at 4°C with 0.1 μg, 0.2 μg, or 0.5 μg various fusion proteins or BSA (final volume of 100 μL of 1% BSA in PBS/0.05%/Tween 20). The detection of the amount of bound protein was performed using alkaline phosphatase (AP)–conjugated secondary mAb following standard ELISA protocol.
Production and purification of fusion proteins
The generation of NKp46-Ig, NKp44-Ig, NKp30-Ig, KIR2DL1-Ig, and CD99-Ig in Simian kidney COS-7 (COS) cells was previously described.18,19,24 Truncated fusion proteins of NKp46D1-Ig (including the leader peptide 1-21 and residues 1-100) and NKp46D2-Ig (residues 101-235) were generated by polymerase chain reaction (PCR) amplification and cloned into a mammalian expression vector containing the Fc portion of human IgG1 as previously described.25 In order to allow expression of NKp46D2-Ig, which lacks its original leader peptide sequence, we added a methionine start codon and cloned the PCR-amplified fragment of NKp46D2 in frame with the leader peptide of CD5. Sequencing of the constructs revealed that all cDNAs were in frame with the human Fc genomic DNA and were identical to the reported sequences. The production of the fusion proteins in COS cells was previously described.25
For the production of NKp46D2 in Chinese hamster ovary (CHO) cells, the NKp46D2-Ig fragment was cloned into the pcDNA 3.1 vector. After recloning, the highest protein-producing clone was adapted for special serum-free medium (CHO-SFM II; Gibco, Grand Island, NY), followed by optimization for growth in large-scale cultures. Supernatants were collected and purified on protein-G columns using fast-protein liquid chromatography (FPLC).
Treatment of fusion proteins with neuraminidase
Various fusion proteins were incubated with 0.015 U insoluble neuraminidase (NA) attached to beaded agarose (N-5254; Sigma, St Louis, MO), or with PBS (control) for 1.5 hours at 17 °C on a roller. SV- and IV-infected, or noninfected, cells were washed and stained either with NA-treated or mock-treated Ig-fusion proteins, followed by phycoerythrin (PE)–conjugated antihuman Fc. The integrity of the treated fusion proteins was assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel analysis.
The generation of NKp46-Ig point mutations
The point mutations in the NKp46 proteins T125A and N216A were generated by using a PCR-based, site-directed mutagenesis approach as previously described.24 The T225A and T225N mutations were generated by GenScript (Scotch Plains, NJ). All products were cloned in frame with human IgG1 as above. Both production and staining with various fusion proteins were previously described.25 All fusion proteins were routinely tested for degradation on SDS-PAGE gels.
Cytotoxicity assays
Generation of mouse lymphoma BW cells (BWs) transfectants and interleukin-2 (IL-2) release assays
Generation of the stable transfectants BW-NKp46-Zeta and BW-CD99-Zeta was previously described.18 The NKp46T225A-Zeta construct was generated by GenScript and the existence of the mutation was verified by sequencing. cDNA was stably transfected into BW cells as previously described.18 For IL-2 release assay, 1106mel cells infected or uninfected with human influenza virus (Sydney strain) were irradiated (6000 rad [60 Gy]), washed, and plated in 96 flat-wells (50 000 cells/well). Various BW cells (50 000 cells/well) were then added and coincubated at 37°C for 48 hours. The presence of IL-2 was measured by ELISA kit (PharMingen, San Diego, CA).
Results
Viral HA is directly recognized by NKp46 and NKp44
In our previous reports we have shown, by using the NKp46 and NKp44 receptors attached to human IgG1, that these proteins interact with the viral HA of IV and SV presented on infected cells and that this interaction leads to an enhanced killing of infected cells.18,19 Our findings also revealed that sialic acids expressed on the NKp46 and NKp44 receptors are dominantly involved in these interactions.18,19,27 However, as HA is known to interact with other surface molecules, it was crucial to determine whether the HA recognition by NKp46 and NKp44 is direct or if this interaction is only an initial step that mediates the binding of another, perhaps more specific ligand.
To address this question, a direct binding assay was employed using rosettes of HA, prepared as previously described.23 These rosettes, which contain from 6 to 10 trimers of HA, do not change the affinity of HA to sialic acid, as has been previously demonstrated,28,29 but allow multivalent interaction in vitro, similarly to the binding of HA to sialic acid receptors in vivo. We used these rosettes to detect NKp46 and NKp44 interactions with HA in an ELISA.
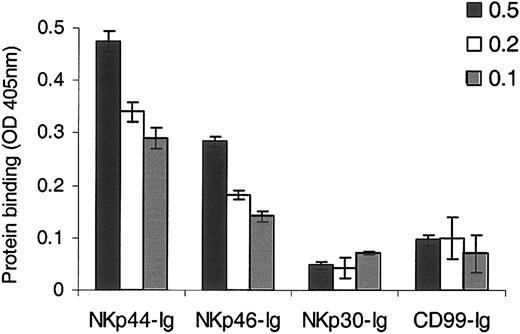
As shown in Figure 1, a clear and specific binding of NKp46-Ig and NKp44-Ig to the HA rosettes was detected, while little or no binding was observed in the case of other Ig fusion proteins tested, such as NKp30-Ig and CD99-Ig. This specific binding was observed when we used various concentrations of fusion protein (Figure 1) or HA (Figure 4B). These results indicate that the HA recognition of NKp46 and NKp44 is direct and does not require the presence of other accessory or mediating molecules.
Direct binding of NKp46-Ig and NKp44-Ig to HA. ELISA plates were coated with 0.1 μg HA rosettes followed by incubation with 0.5 μg, 0.2 μg, and 0.1 μg of the relevant fusion protein, as indicated. Bound proteins were detected using AP-conjugated secondary mAb. The data in the figure represent optical density (OD) absorbance (405 nm). Figure shows one representative experiment of 3 performed. Error bars indicate SD.
Direct binding of NKp46-Ig and NKp44-Ig to HA. ELISA plates were coated with 0.1 μg HA rosettes followed by incubation with 0.5 μg, 0.2 μg, and 0.1 μg of the relevant fusion protein, as indicated. Bound proteins were detected using AP-conjugated secondary mAb. The data in the figure represent optical density (OD) absorbance (405 nm). Figure shows one representative experiment of 3 performed. Error bars indicate SD.
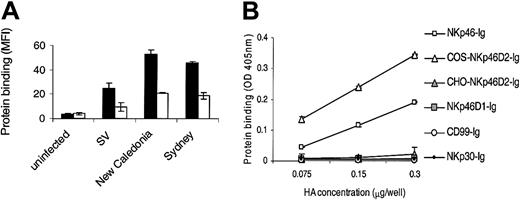
Impaired sialylation of NKp46D2-Ig in CHO cells significantly reduces HA binding. (A) 721.221 cells were infected with SV, human influenza A virus H1N1 (New Caledonia), and H3N2 (Sydney). Infected cells were stained with NKp46D2-Ig produced in either COS (▪) or CHO (□) cells followed by PE-conjugated goat antihuman Fc. Figure shows one representative experiment of 3 performed. (B) ELISA plates were coated with various concentrations of HA rosettes ranging from 0.075 μg/well to 0.3 μg/well as indicated, followed by incubation with 0.2 μg of the relevant fusion protein. Bound proteins were detected using AP-conjugated second mAb. Shown is one representative experiment of 2 performed.
Impaired sialylation of NKp46D2-Ig in CHO cells significantly reduces HA binding. (A) 721.221 cells were infected with SV, human influenza A virus H1N1 (New Caledonia), and H3N2 (Sydney). Infected cells were stained with NKp46D2-Ig produced in either COS (▪) or CHO (□) cells followed by PE-conjugated goat antihuman Fc. Figure shows one representative experiment of 3 performed. (B) ELISA plates were coated with various concentrations of HA rosettes ranging from 0.075 μg/well to 0.3 μg/well as indicated, followed by incubation with 0.2 μg of the relevant fusion protein. Bound proteins were detected using AP-conjugated second mAb. Shown is one representative experiment of 2 performed.
The primary binding site of HA on NKp46 is located in the second domain
The extracellular region of NKp46 comprises 2 C2-type Ig-like domains.11 We therefore prepared smaller versions of NKp46-Ig corresponding to the single domains fused to the Fc portion of human IgG1. The membrane distal domain was named NKp46D1 and the membrane proximal domain (including a stretch of amino acids that probably forms a stem connecting the ectodomain with the transmembrane region) was named NKp46D2. The fusion proteins were produced in COS cells and binding was analyzed to virally infected and uninfected 721.221 and 1106mel cells.
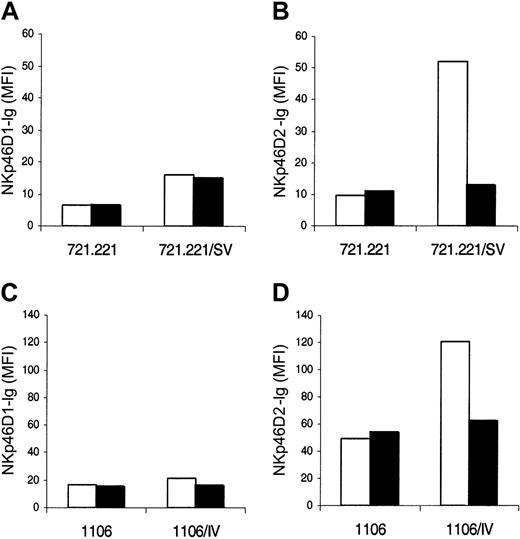
As previously described,18,19 the binding of NKp46-Ig, but not NKp30-Ig, to 721.221 cells was considerably enhanced following SV infection and this binding was negated by preincubation with anti-HA mAb 135.7 (Figure 2). Remarkably, a similar pattern of increased binding to infected cells was also observed in the case of the truncated NKp46D2-Ig. Furthermore, this increased binding of NKp46D2-Ig was significantly blocked by mAb directed against HA 135.7, but not by the control mAb TC-9A1, directed against the other glycoprotein of SV (the fusion protein) or by 12E7, directed against CD99 (Figure 2). In contrast, the binding of NKp46D1-Ig to 721.221 was very weak and not affected by infection or incubation with the various mAbs. Similar results were obtained with 1106mel cells infected with IV (data not shown). These observations indicate that the binding site of HA on NKp46 is located within the second domain of the receptor and that this domain is sufficient to mediate HA binding in a specific and efficient manner.
The recognition of HA by NKp46 is mediated through the membrane proximal domain (D2). Uninfected and SV-infected 721.221 cells were incubated with or without mAb against HA (135.7) or control mAbs (12E7 and TC-9A1) and stained with the indicated various fusion proteins followed by PE-conjugated goat antihuman antibodies. Figure shows one representative experiment of 20 performed. MFI indicates median fluorescence intensity. (A) NKp30-Ig. (B) NKp46-Ig. (C) NKp46D2-Ig. (D) NKp46D1-Ig. Error bars indicate SD.
The recognition of HA by NKp46 is mediated through the membrane proximal domain (D2). Uninfected and SV-infected 721.221 cells were incubated with or without mAb against HA (135.7) or control mAbs (12E7 and TC-9A1) and stained with the indicated various fusion proteins followed by PE-conjugated goat antihuman antibodies. Figure shows one representative experiment of 20 performed. MFI indicates median fluorescence intensity. (A) NKp30-Ig. (B) NKp46-Ig. (C) NKp46D2-Ig. (D) NKp46D1-Ig. Error bars indicate SD.
To extend the implications of these results, we tested the binding of NKp46-Ig, NKp46D2-Ig, and NKp46D1-Ig to 721.221 cells infected with different influenza viruses that differ in their HA type (Table 1).
The membrane proximal domain of NKp46 recognizes the HA of different viruses
Virus . | HA type . | NKp44-Ig . | NKp46-Ig . | NKp46D2-Ig . | NKp46D1-Ig . | NKp30-Ig . | CD99-Ig . |
|---|---|---|---|---|---|---|---|
| Uninfected | — | 7 | 3 | 4 | 2 | 2 | 0 |
| SV | HA-NA | 350 | 77 | 95 | 8 | 7 | 0.5 |
| Beijing | H1N1 | 128 | 44 | 77 | 7 | 4 | 0 |
| X-127 | H1N1 | 220 | 107 | 160 | 5 | 6 | 1 |
| New Caledonia | H1N1 | 518 | 315 | 420 | 5 | 10 | 0 |
| Sydney | H3N2 | 120 | 58 | 100 | 10 | 8 | 1 |
| Moscow | H3N2 | 24 | 20 | 50 | 8 | 8 | 0 |
| Yamanashi | B strain | 117 | 37 | 51 | 8 | 12 | 0 |
Virus . | HA type . | NKp44-Ig . | NKp46-Ig . | NKp46D2-Ig . | NKp46D1-Ig . | NKp30-Ig . | CD99-Ig . |
|---|---|---|---|---|---|---|---|
| Uninfected | — | 7 | 3 | 4 | 2 | 2 | 0 |
| SV | HA-NA | 350 | 77 | 95 | 8 | 7 | 0.5 |
| Beijing | H1N1 | 128 | 44 | 77 | 7 | 4 | 0 |
| X-127 | H1N1 | 220 | 107 | 160 | 5 | 6 | 1 |
| New Caledonia | H1N1 | 518 | 315 | 420 | 5 | 10 | 0 |
| Sydney | H3N2 | 120 | 58 | 100 | 10 | 8 | 1 |
| Moscow | H3N2 | 24 | 20 | 50 | 8 | 8 | 0 |
| Yamanashi | B strain | 117 | 37 | 51 | 8 | 12 | 0 |
721.221 cells were incubated overnight with or without influenza viruses that express different types of HA. Cells (50 000/well) were washed and stained with the various Ig fusion proteins (5 μg/well) followed by PE-conjugated goat antihuman Fc. The level of infection was determined by staining with human sera derived from individuals immunized with a trivalent influenza vaccine. Median fluorescence intensity (MFI) numbers were rounded to the nearest whole number after subtracting background staining (of PE-conjugated goat antihuman Fc). These results represent one experiment of 15 performed. — indicates no infection.
As previously reported,18,19 the binding of all lysis receptors to uninfected 721.221 cells was very low. This might be either because of low-affinity interactions of the various NCRs to their ligands or because the ligands for the various NCRs are expressed in low levels on 721.221 cells. The highest binding to all infected cells was observed with NKp44-Ig. This might be because the NKp44 receptor carries more sugar residues compared with NKp46.11,30 Importantly, in all the cases examined, the infection resulted in an increased parallel binding of both NKp46-Ig and NKp46D2-Ig, but did not effect the binding of NKp46D1-Ig or NKp30-Ig and CD99-Ig (Table 1). The binding of NKp46D2 was consistently stronger than that of NKp46, probably because the smaller NKp46D2 protein is more stable. The differences in the intensity of the enhanced fusion protein binding correlated with infection efficiency determined by staining of infected cells with sera derived from individuals immunized with a trivalent influenza vaccine (see “Material and methods”; data not shown). It is therefore likely that these differences are due to different levels of HA expression. Results of infection of 1106mel cells with these viruses were similar (data not shown).
These findings demonstrate that the recognition of NKp46 and NKp44 by HA is a general phenomenon and that in the case of NKp46, the binding is primarily mediated through the membrane proximal domain.
The sialylation of the second domain of NKp46 is essential for HA recognition
The common feature of all HAs is their ability to bind to oligosacharides containing terminal sialic acid residues. We have previously shown that the recognition of viral HA by the lysis receptors NKp46 and NKp44 depends on the sialylation of these receptors.18,19 NKp46 presumably displays 2 putative O-linked glycosylation sites at Thr125 and Thr225, and one N-linked glycosylation site at Asn216,11 all located within the second domain. Since our results clearly identified the second domain of NKp46 as containing the primary binding site of influenza HAs, we next tested whether the sialic acid–dependent nature of the interactions is also confined to the second domain.
To answer this question, we treated both NKp46D1-Ig and NKp46D2-Ig with bacterial NA and tested for binding to uninfected, and IV- or SV-infected, 1106mel and 721.221 cells, respectively. Indeed, treatment with NA abolished the enhanced binding of NKp46D2-Ig to SV-infected 721.221 cells (Figure 3B) and IV-infected 1106mel (Figure 3D) cells. Importantly however, the NA treatment did not affect the binding of NKp46D2-Ig to the uninfected cells, suggesting that in contrast to the binding to the viral HA, the binding to the cellular tumor ligand is not sialic acid dependent. As expected, 721.221 and 1106mel cells were only minimally recognized by NKp46D1 (Figure 3A,C). These results suggest that the sialylation of the second domain of NKp46 is indeed necessary for binding to the viral HA, but is not involved in the recognition of tumor cells.
The binding of the membrane proximal domain to viral HA is sialic acid dependent. NKp46D1-Ig (A,C) or NKp46D2-Ig (B,D) cells were treated with NA (▪) or with PBS (as control; □). SV-(A-B) or IV-(C-D) infected and uninfected 721.221 or 1106mel cells, respectively, were washed and stained with either the PBS-treated or NA-treated fusion proteins followed by PE-conjugated goat antihuman Fc antibodies. Figure shows one representative experiment of 3 performed.
The binding of the membrane proximal domain to viral HA is sialic acid dependent. NKp46D1-Ig (A,C) or NKp46D2-Ig (B,D) cells were treated with NA (▪) or with PBS (as control; □). SV-(A-B) or IV-(C-D) infected and uninfected 721.221 or 1106mel cells, respectively, were washed and stained with either the PBS-treated or NA-treated fusion proteins followed by PE-conjugated goat antihuman Fc antibodies. Figure shows one representative experiment of 3 performed.
The 2 major linkages between sialic acid and the galactose residues of carbohydrate side chains are Neu5Ac α(2,3)-Gal and Neu5Ac α(2,6)-Gal. Different HAs present different specificities for these linkages and in particular human influenza viruses preferentially bind the Neu5Ac α(2,6)-Gal linkage.28 To further explore the nature of the sialic acids of NKp46 that participate in the HA recognition, we produced NKp46D2-Ig fusion protein in CHO-D2-Ig that express an impaired glycosylation pattern. CHO cells are widely employed to produce glycosylated glycoproteins due to the high similarity of their glycosylation machinery to the human system. Notably however, CHO cells differ from human cells in the sialylation process as they lack a functional copy of the gene-encoding α(2,6)-sialyltransferase.31 As a result, CHO glycoproteins carry only α(2,3)-terminal sialic acids.
We therefore compared the binding of the NKp46D2-Ig proteins produced in CHO or in COS cells to 721.221 cells infected with mouse (SV) and various human influenza viruses. As shown in Figure 4A, the binding of CHO-D2-Ig to 721.221 infected cells was significantly weaker than the binding of the COS-produced NKp46D2-Ig protein. This observation concurs with the binding specificity of HAs of human viruses and suggests that α(2,6)-terminal sialic acid modifications of NKp46 are important for its interaction with viral HAs. Similar results were obtained with other viruses and with 1106mel cells infected with the different viruses (data not shown). Importantly, the integrity of the CHO-D2-Ig fusion protein was confirmed by SDS-PAGE analysis and was supported by the fact that the binding of the CHO-D2-Ig to various tumor cell lines was comparable to the binding of the COS-produced NKp46D2-Ig form (data not shown). Similarly to the variability in the staining intensities observed with the full NKp46-Ig to cells infected with various viruses (see Table 1), differences in the staining by the CHO- and COS-produced NKp46D2-Ig proteins also correlated with infection efficiency (data not shown).
Finally, in order to establish conclusively the sialylated NKp46D2 domain as the major binding subunit of the NKp46 receptor to viral HA, we tested its direct binding to HA rosettes. As shown in Figure 4B, a clear binding of both NKp46-Ig and COS-NKp46D2-Ig, but not NKp46D1-Ig, to HA rosettes was observed. In accordance with the binding pattern to infected cells, a stronger signal was detected in the case of the truncated NKp46D2-Ig compared with the full NKp46-Ig protein. The specificity of this binding was confirmed by the dose-dependent manner of the interactions and by the fact that none of the other control Ig fusion proteins, including CD99-Ig and NKp30-Ig, were bound to the HA rosettes. Importantly, no detectable binding was obtained using the CHO-produced NKp46D2-Ig fusion protein, thus supporting the important role of α(2,6)-terminal sialic acid modifications of NKp46 in recognizing viral HAs. Similar results were obtained using various Ig fusion protein concentrations (data not shown).
Monoclonal antibody specifically directed against the distal membrane domain of NKp46 does not block NK-mediated lysis
The above results suggest that the distal domain of NKp46 (D1) does not contain binding sites to HAs or tumor ligands. However, these observations only relate to the binding properties of the domain and reveal nothing of other possible functional roles it may play in NKp46 activity. In addition, since all the binding assays were based on a truncated fusion protein form of the receptor, it was still possible that the lack of any binding observed was a result of loss of proper folding or weak interactions that could not be detected in immunostaining assays. Therefore, in order to study the possible involvement of the membrane distal domain of NKp46 in its function, we prepared a mAb directed specifically against the membrane distal domain of NKp46.
Using ELISA, we screened spleen-derived B-cell hybridoma supernatants from mice immunized with NKp46-Ig, for increased binding to NKp46-Ig as compared with NKp30-Ig. One hybridoma, 461-G1, recognized BW cells transfected with NKp4618 and NK cells, but not parental BW cells (Figure 5A) or any other non-NK cells tested (data not shown). To further analyze the NKp46 domain to which 461-G1 binds, we performed ELISAs comparing its binding to NKp46D1-Ig, NKp46D2-Ig, and NKp46-Ig, and other Ig fusion proteins (Figure 5B). Results showed that 461-G1 specifically recognized NKp46-D1-Ig and NKp46-Ig, whereas only background recognition was observed with NKp46-D2-Ig or the other Ig fusion proteins tested. Western blot analysis performed with the various Ig-fusion proteins showed similar results (Figure 5C). Thus, 461-G1 specifically recognizes the membrane distal domain of NKp46. As previously reported with regard to other anti-NKp46 mAb,11 ligation of NKp46 on the surface of NK cells by 461-G1 mAb activated the redirected killing assay of the FcγR+ P815 murine target cells, in a manner similar to that observed with the anti-CD16 mAb (Figure 5D). Unfortunately, despite several attempts, we could not obtain a specific anti-NKp46 mAb directed against the NKp46D2.
A specific mAb directed against the membrane distal domain (D1) of NKp46 does not block NK-mediated lysis of target cells. (A) NK cells (NK line), BW, and BW transfected with NKp46 (BW/NKp46) were incubated with or without 461-G1 mAb (thick black line) or control 12E7 mAb (gray histogram). Shown is one representative experiment of 3 performed. (B) ELISA plates were coated with 2 μg/mL of fusion proteins NKp30-Ig, NKp46D2-Ig, NKp46D1-Ig, NKp46-Ig, KIR2DL1-Ig (KIR-I-Ig), and KIR2DL2-Ig (KIR-II-Ig). Fusion proteins were incubated with no antibody or with 461-G1 for 1 hour on ice. HRP-conjugated rabbit antimouse IgG diluted 1:2500 was used as secondary mAb. PBS was used as negative control. Shown is one representative experiment of 3 performed. (C) The indicated proteins (10 μg), NKp30-Ig (1), NKp46D2-Ig (2), NKp46D1-Ig (4), NKp46-Ig (5), KIR2DL1-Ig (6), and the control low-protein medium (LPM [3]) were analyzed on SDS-PAGE. The marker is indicated by M. Fusion proteins were analyzed with the 461-G1 mAb and HRP-conjugated goat antimouse Ig antibodies. Shown is one representative experiment of 2 performed. (D) 35S-labeled P815 cells were incubated either with no mAb (□), anti-CD16 mAb (B73.1.1; ♦), anti-NKp46 mAb (461-G1; ○), or anti-CD99 mAb (12E7; ▴) for 1 hour on ice. NK cells were next added in effector to target ratio (E/T) of 3:1. Shown is one representative experiment of 7 performed. (E) NK cells were preincubated with the indicated mAb for 1 hour on ice, washed and incubated with either uninfected 1106mel or 1106mel infected with various influenzas as indicated. This experiment was preformed in E/T ratio of 20:1 and represents one of 4 performed.
A specific mAb directed against the membrane distal domain (D1) of NKp46 does not block NK-mediated lysis of target cells. (A) NK cells (NK line), BW, and BW transfected with NKp46 (BW/NKp46) were incubated with or without 461-G1 mAb (thick black line) or control 12E7 mAb (gray histogram). Shown is one representative experiment of 3 performed. (B) ELISA plates were coated with 2 μg/mL of fusion proteins NKp30-Ig, NKp46D2-Ig, NKp46D1-Ig, NKp46-Ig, KIR2DL1-Ig (KIR-I-Ig), and KIR2DL2-Ig (KIR-II-Ig). Fusion proteins were incubated with no antibody or with 461-G1 for 1 hour on ice. HRP-conjugated rabbit antimouse IgG diluted 1:2500 was used as secondary mAb. PBS was used as negative control. Shown is one representative experiment of 3 performed. (C) The indicated proteins (10 μg), NKp30-Ig (1), NKp46D2-Ig (2), NKp46D1-Ig (4), NKp46-Ig (5), KIR2DL1-Ig (6), and the control low-protein medium (LPM [3]) were analyzed on SDS-PAGE. The marker is indicated by M. Fusion proteins were analyzed with the 461-G1 mAb and HRP-conjugated goat antimouse Ig antibodies. Shown is one representative experiment of 2 performed. (D) 35S-labeled P815 cells were incubated either with no mAb (□), anti-CD16 mAb (B73.1.1; ♦), anti-NKp46 mAb (461-G1; ○), or anti-CD99 mAb (12E7; ▴) for 1 hour on ice. NK cells were next added in effector to target ratio (E/T) of 3:1. Shown is one representative experiment of 7 performed. (E) NK cells were preincubated with the indicated mAb for 1 hour on ice, washed and incubated with either uninfected 1106mel or 1106mel infected with various influenzas as indicated. This experiment was preformed in E/T ratio of 20:1 and represents one of 4 performed.
We have previously demonstrated an efficient blocking of NK cell cytotoxicity against influenza-infected cells using anti-NKp46 serum.18 In addition, other anti-NKp46 mAbs were shown to block the killing of various tumor targets.10-13 In contrast, preincubation of NK cells with 461-G1 mAb did not block the lysis of the melanoma cell line 1106mel before or after infection with various influenza viruses (Figure 5E). Similarly, this mAb did not block the killing of other tumor lines tested, including 721.221 or Hela cells (data not shown). Thus, the membrane distal domain of NKp46 is probably not involved in the recognition of tumor- and virus-infected cells.
The sugar modification of Thr225 of NKp46 is important for HA recognition
The specific nature of the interactions between NKp46 and viral HAs is still not fully understood. The difficulty rises from the fact that although we demonstrate a crucial role of sialic acid moieties in the recognition of NKp46 by HA, other NK receptors that are heavily glycosylated, such as NKp30-Ig, KIR2DL1-Ig, or CD99-Ig, are not recognized by HAs18,19 (Table 1; Figure 1; data not shown).
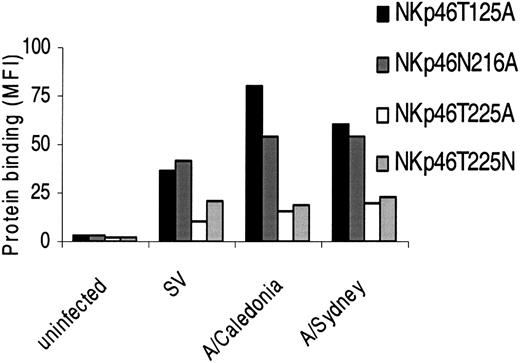
To determine the specific sugar-carrying residue that is important for HA recognition, we performed site-directed mutagenesis in the 3 residues predicted to carry the glycan modifications, produced the fusion proteins in COS cells as above, and tested their binding to 721.221 cells infected with different influenza viruses. Substitution of Thr125 or Asn216 with Ala (NKp46T125A and NKp46N216A, respectively) had no significant effect on the increased binding to infected cells (Figure 6). Importantly however, when Thr225 was substituted with Ala, a sharp decrease in the enhanced binding to the infected cells was observed (Figure 6). Similar results were obtained when 1106mel cells infected with the various influenzas were used (data not shown). Thus, Thr225 of NKp46 is crucial for the binding to HA.
The sialylation of Thr225 of NKp46 is important for HA recognition. 721.221 cells infected with the various influenza viruses were stained with the indicated Ig fusion proteins followed by PE-conjugated goat antihuman Fc. Figure shows one representative experiment of 9 performed.
The sialylation of Thr225 of NKp46 is important for HA recognition. 721.221 cells infected with the various influenza viruses were stained with the indicated Ig fusion proteins followed by PE-conjugated goat antihuman Fc. Figure shows one representative experiment of 9 performed.
These results raised 2 possibilities: either that the substitution of Thr225 with Ala abolished an important glycosylation that is dominantly involved in the HA recognition, or that the absence of Thr225 itself hindered the overall interactions with HA in a direct or an indirect manner. To distinguish between these 2 possibilities, we replaced Thr225 with Asn (NKp46T225N), which like Thr (but not Ala) is classified as a neutral hydrophilic amino acid.32 The reason why we preferred to replace Thr with Asn and not with Ser, which shares greater resemblance to Thr, is that Ser can potentially carry O-glycosylations in this location, whereas Asn can not.32 As shown in Figure 6, expression of Asn instead of Thr at position 225 did not restore the enhanced binding to infected cells, but instead was similar to the binding of NKp46T225A.
It is clear that the interaction between NKp46 and HA is not restricted to one amino acid only and that other amino acid residues also contribute to the enhanced recognition of cells following infection. Indeed, the reduced binding to infected cells resulting from Thr to Ala or Asn substitution was incomplete (Figure 6). Unfortunately, NKp46 mutant in which all the predicted glycosilated residues were replaced by Ala could not be expressed.
In light of these observations, it is unlikely that Thr225 is directly involved in the interaction with viral HA. Instead, this residue probably carries a critical glycosylation that is crucial, although not exclusive, for the recognition by HA.
The identification of the binding site of NKp46 to tumor ligands
The NKp46 receptor, which is constitutively expressed on NK cells, is thought to be the major lysis receptor involved in the killing of transformed cells.15,33,34 However, since no specific tumor ligand for any of the lysis receptors has been identified yet, very little is known about the interactions between NKp46 and its tumor antigen ligand.
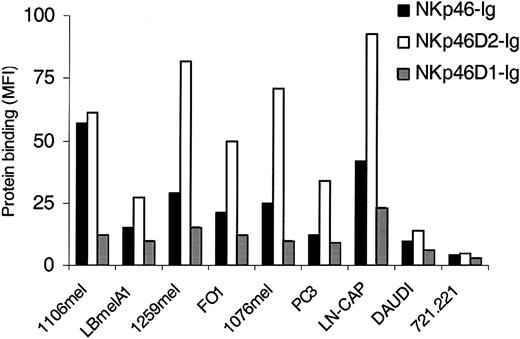
To gain a better understanding of tumor recognition by NKp46, we immunostained a number of different tumor lines, including melanomas, carcinomas, and EBV-transformed B cells with the intact NKp46 protein and the fragmented NKp46D2-Ig and NKp46D1-Ig fusion proteins. NKp46 and NKp46D2 recognized the various tumor lines to varying degrees (Figure 7), suggesting that the tumor ligands for the NKp46 receptors are expressed on various tumors in different levels. In general, and as reported above, the highest binding was observed with the NKp46D2-Ig fusion protein. Importantly, all the different tumors tested were stained by the NKp46D2-Ig in correlation with the staining by the full NKp46-Ig, whereas relatively weak or no staining was observed by the NKp46D1-Ig protein (Figure 7). Thus, similarly to the binding to viral HA and in agreement with the inability of 461-G1 to block tumor cell killing (Figure 5), NKp46 recognizes different tumor cells via the second domain.
The recognition of tumor cells by NKp46 is confined to the membrane proximal domain. Melanomas (1106mel, LBmelA1,1259mel, 1076mel), prostate carcinomas (PC3, LN-CAP), and EBV-transformed B cells (DAUDI, 721.221) were stained with NKp46-Ig, NKp46D1-Ig, or NKp46D2-Ig fusion proteins followed by PE-conjugated goat antihuman Fc. Figure shows one representative experiment of more than 30 performed.
The recognition of tumor cells by NKp46 is confined to the membrane proximal domain. Melanomas (1106mel, LBmelA1,1259mel, 1076mel), prostate carcinomas (PC3, LN-CAP), and EBV-transformed B cells (DAUDI, 721.221) were stained with NKp46-Ig, NKp46D1-Ig, or NKp46D2-Ig fusion proteins followed by PE-conjugated goat antihuman Fc. Figure shows one representative experiment of more than 30 performed.
As mentioned above, impairment of the expression of sialic acid residues in NKp46, either by NA treatment or by the production in CHO, did not affect its binding to tumor cells, suggesting no involvement of carbohydrate moieties of NKp46 in tumor cell recognition. To support this conclusion, we next tested the binding of the various glycosylation mutants to tumor cells. To our surprise, NKp46-Ig expressing the point mutation of Thr225 to Ala completely lost its binding to 1106mel (Figure 8) and to other tumors tested (data not shown). Remarkably, binding was fully restored when Thr225 was replaced with Asn. Mutations in the other 2 glycosylation sites had no effect on the binding (Figure 8). These results support the conclusion that unlike the binding to influenza-infected cells, tumor recognition by NKp46 is not mediated via sialic acid interactions. However, the sialylated amino acid Thr225 of NKp46 seems to play a crucial role in the ability of the lysis receptor to interact with various targets, including virally infected as well as tumor cells.
Thr225 of NKp46 is crucial for the recognition of tumor cells by NKp46. 1106mel cells were stained with the wild-type NKp46-Ig or with the sugar-mutated forms NKp46T125A, NKp46N216A, NKp46T225A, or NKp46T225N, followed by PE-conjugated goat antihuman Fc. Gray histograms represent the background staining of PE-conjugated goat antihuman Fc. Open histograms represent fusion protein staining. Figure shows one representative experiment of 4 performed.
Thr225 of NKp46 is crucial for the recognition of tumor cells by NKp46. 1106mel cells were stained with the wild-type NKp46-Ig or with the sugar-mutated forms NKp46T125A, NKp46N216A, NKp46T225A, or NKp46T225N, followed by PE-conjugated goat antihuman Fc. Gray histograms represent the background staining of PE-conjugated goat antihuman Fc. Open histograms represent fusion protein staining. Figure shows one representative experiment of 4 performed.
Thr225 is important for NKp46 functional effect
So far, our findings place Thr225 in the category of a central amino acid, important to the recognition of both viral HA as well as tumor ligand by NKp46. It was therefore interesting to test whether the dramatic influence that this amino acid confers on the binding properties of NKp46 also has implications on its function.
For this purpose, we stably transfected BW cells with constructs encoding chimeric proteins in which the transmembrane and tail of CD3ξ chain is fused to the extracellular portion of various NK receptors, including a control receptor CD99 (BW/CD99Z), a wild-type NKp46 (BW/NKp46Z), and a mutated NKp46 that expresses Ala in position 225 instead of Thr (BW/NKp46T225AZ). Crosslinking of the chimeric receptor by its ligand will induce specific IL-2 secretion, should a functional interaction have occurred.18
As shown in Figure 9A, we used wild-type and mutated NKp46-Zeta transfectants that were identically stained by anti-NKp46 mAb, indicating the same levels of NKp46 expression. This staining was low but specific, as it was not observed in the parental BW cells or CD99-Zeta transfectants. Higher levels of NKp46 expression could not be obtained. Importantly however, this level of expression is similar to the physiologic expression of NKp46 on NK cells (Figure 5).
Thr225 of NKp46 is important for NKp46 activation. (A) Parental BW cells, or stably transfected BW cells expressing the chimeric receptors CD99-Zeta, NKp46-Zeta, or NKp46T225A-Zeta, were stained with anti-NKp46 mAb (461-G1), followed by FITC-conjugated goat antimouse Fc. Gray histograms represent the background staining of FITC-conjugated goat antimouse Fc. The MFI values of the anti-NKp46 mAb staining are indicated in the figure. Figure shows one representative experiment of 3 performed. (B) 1106mel cells were incubated overnight with or without human influenza virus (Sydney strain). Next, cells (50 000/well) were irradiated (6000 rad [60 Gy]), washed, and incubated with 50 000 cells/well of BW cells expressing CD99-Zeta (BW/CD99Z), wild-type NKp46-Zeta (BW/NKp46Z wild type), mutated NKp46T225A-Zeta (BW/NKp46T225AZ), or parental BW cells. After 48 hours, IL-2 levels in the supernatants were measured by ELISA. Optical density (OD) absorbance (650 nm) was translated into amount of IL-2 (pg/well) according to a standard curve. Results represent one experiment of 2 performed.
Thr225 of NKp46 is important for NKp46 activation. (A) Parental BW cells, or stably transfected BW cells expressing the chimeric receptors CD99-Zeta, NKp46-Zeta, or NKp46T225A-Zeta, were stained with anti-NKp46 mAb (461-G1), followed by FITC-conjugated goat antimouse Fc. Gray histograms represent the background staining of FITC-conjugated goat antimouse Fc. The MFI values of the anti-NKp46 mAb staining are indicated in the figure. Figure shows one representative experiment of 3 performed. (B) 1106mel cells were incubated overnight with or without human influenza virus (Sydney strain). Next, cells (50 000/well) were irradiated (6000 rad [60 Gy]), washed, and incubated with 50 000 cells/well of BW cells expressing CD99-Zeta (BW/CD99Z), wild-type NKp46-Zeta (BW/NKp46Z wild type), mutated NKp46T225A-Zeta (BW/NKp46T225AZ), or parental BW cells. After 48 hours, IL-2 levels in the supernatants were measured by ELISA. Optical density (OD) absorbance (650 nm) was translated into amount of IL-2 (pg/well) according to a standard curve. Results represent one experiment of 2 performed.
In correlation with the binding properties of NKp46, a clear dependency was observed between the expression of Thr225 of NKp46 and the functional activity of the receptor (Figure 9B). As previously reported,18 stimulation with virally infected 1106mel cells led to a dramatic enhanced IL-2 secretion from BW-NKp46 transfectants. However, this enhancement was significantly lower in the case of BW cells expressing the mutated receptor. Importantly, the IL-2 release was specific, since neither the parental nor the CD99-Zeta BW cells showed any enhanced IL-2 secretion.
With regard to stimulation with uninfected 1106mel cells, less dramatic but still specific increased secretion of IL-2 was observed from both wild-type and mutated NKp46-Zeta transfectants (Figure 9B). In agreement with the binding results (Figure 8), the IL-2 secretion from the mutated NKp46 was markedly lower compared with the secretion from the wild-type receptor (Figure 9B). In summary, these findings reveal that indeed Thr225 is critically involved in the interactions of NKp46 with tumor- and virally infected cells.
Discussion
NK cells are experts in the recognition and killing of hazardous cells. This ability is largely dependent on the ligation of the NCRs by their specific activating ligands expressed on target cells.1,4,9 Recently, we reported that the HAs of IV and SV viruses recognize 2 NK lysis receptors, NKp46 and NKp44.18,19 Here we demonstrate that this recognition is a conserved pattern shared by HAs derived from different influenza strains and may therefore represent a general mechanism by which NK cells recognize and destroy an impressive range of viruses.
Although the HA molecule is subjected to a considerable antigenic drift, its ability to bind sialic acid–containing receptors is well preserved throughout the antigenic variation.28,35,36 Our data suggest that NK cells have outsmarted the influenza virus by exploiting this viral HA binding characteristic to their advantage. Thus, NK cells have developed a killing machinery that is based on the natural conserved tendency of HA to bind sialic acids, and manipulated it to directly and specifically activate 2 sialylated lysis receptors, NKp46 and NKp44. We also suggest that this ability of NKp46 and NKp44 to recognize such a wide range of influenza HAs could be used as an analytical research tool to assess influenza infection in addition to specific anti-HA mAbs, which only have limited HA strain specificity.
In agreement with the human influenza viruses preferential binding to Neu5Ac α(2,6)-Gal linkage over Neu5Ac α(2,3)-Gal linkage, the binding of HAs derived from different influenzas was strongly, though not exclusively, dependent on α(2,6)-terminal sialic acids expressed on NKp46. As this dependency was not complete, it suggested that NKp46 recognition by HA may also rely on other elements in the NKp46 protein. We next mutated all 3 predicted glycosylation sites on NKp46. Our findings pointed to the O-linked glycosylation modification carried on Thr225 of NKp46 as the dominant factor involved in HA recognition. This is further supported by the fact that this amino acid is required for proper activation of NKp46. Importantly however, elimination of Thr225, or any of the glycosylation sites, did not completely abolish the ability of NKp46 to bind or respond to virally infected or transformed cells. Therefore, it is clear that either other amino acids or another unknown glycosylation site are important factors in NKp46 interactions.
NKp46 displays 2 Ig-C2–type domains.11 Here we demonstrate that by itself the membrane proximal domain is sufficient for binding to both the viral HAs and the unknown tumor ligands on various tumor lines. The significance of the distal domain of NKp46 remains unclear, as we could not detect binding of a soluble Ig fusion protein of this domain to HAs or any of the cell lines tested, nor did we observe any interference in NK-mediated killing against various target cells using a mAb specifically directed against this domain. Interestingly, data bank survey reveals that the NKp46 isoforms c (accession number CAA06873) and d (accession number CAA06874) almost completely lack the distal domain of the receptor.
NK cells are essential for efficient elimination of tumors and also probably for their prevention.1,33,34 In vitro, NK cells spontaneously lyse different tumor cell lines largely via the ligation of the lysis receptors.34 This suggests that transformation of cells leads to the up-regulation of specific antigens or to the formation of surface modifications common to many tumor cells recognized by the NCRs. The identity or nature of such ligands is unknown. It is worth noting however, that in spite of the sometimes relatively low binding of the NCRs to tumor cells, this binding is yet sufficient to activate the lysis function of NK cells. Indeed, low levels of NKp46-Zeta expression on BW cells that resemble the physiologic expression of NKp46 on NK cells was sufficient to induce IL-2 secretion in response to tumor cells. Furthermore, previous reports have demonstrated that mAbs directed against the NCRs block NK-mediated killing of various tumor targets.10-14 Also, the correlation that exists between the density of the NCRs and the ability of NK cells to lyse tumor, but not normal cells, contributes to the understanding of the important part NCRs play in tumor elimination.13
Here we demonstrate that not only is the recognition of both viral and cellular ligands by NKp46 mediated via the same domain of the receptor, but that it also depends on the same amino acid (Thr225). Importantly however, and in contrast to viral HA recognition, the role of Thr225 in tumor recognition does not involve its sialylation. Hence, the amino acid Thr at position 225 of NKp46 seems to play a dramatic role in the recognition of various targets by NK cells via different mechanisms. This remarkable feature should be examined in light of the inflection of O-glycosylation on the structure of proteins. Recently, Tagashira et al41 showed that changes in the O-glycosylation pattern of a biologically active peptide had dramatic implications on its activity that were attributed to structural changes. Thus, while the O-glycans carried on Thr225 of NKp46 are dominantly involved in its recognition by HA, Thr225 itself is unlikely to be a direct contact residue of the unknown tumor ligand, but rather is more likely to be crucial to the proper folding of the binding site of NKp46. This may explain why the nonsialylated amino acid Asn, which is closer in its chemical characteristics to Thr than Ala, efficiently restored binding to tumor cells but not to HAs. Interestingly, Thr225 is preserved in NKp46 of human, murine, and primates with the exception of cattle that expresses Val in this position.37-40 This evolutionary preservation may also explain the fact that human NKp46 recognizes murine tumor lines with considerable efficiency.10-13,19
NK cells have developed mechanisms to attack virus-infected and tumor cells via a relatively limited panel of receptors. In this study, we identified a crucial amino acid involved in this recognition. However, the specific tumor ligand or ligands that are recognized by NKp46 are currently unknown. The identification of these ligands will not only enable us to better understand the ways by which NK cells recognize and kill tumor cells, but it also might result in the development of new medicines for the treatment of a broad spectrum of tumors.
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-05-1716.
Supported by research grants to O.M. and A.P. from the CapCure and the CRI foundations, and by research grants to O.M. from the European Commission (QLK2-CT-2002-011112) and the Israel Science Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


![Figure 5. A specific mAb directed against the membrane distal domain (D1) of NKp46 does not block NK-mediated lysis of target cells. (A) NK cells (NK line), BW, and BW transfected with NKp46 (BW/NKp46) were incubated with or without 461-G1 mAb (thick black line) or control 12E7 mAb (gray histogram). Shown is one representative experiment of 3 performed. (B) ELISA plates were coated with 2 μg/mL of fusion proteins NKp30-Ig, NKp46D2-Ig, NKp46D1-Ig, NKp46-Ig, KIR2DL1-Ig (KIR-I-Ig), and KIR2DL2-Ig (KIR-II-Ig). Fusion proteins were incubated with no antibody or with 461-G1 for 1 hour on ice. HRP-conjugated rabbit antimouse IgG diluted 1:2500 was used as secondary mAb. PBS was used as negative control. Shown is one representative experiment of 3 performed. (C) The indicated proteins (10 μg), NKp30-Ig (1), NKp46D2-Ig (2), NKp46D1-Ig (4), NKp46-Ig (5), KIR2DL1-Ig (6), and the control low-protein medium (LPM [3]) were analyzed on SDS-PAGE. The marker is indicated by M. Fusion proteins were analyzed with the 461-G1 mAb and HRP-conjugated goat antimouse Ig antibodies. Shown is one representative experiment of 2 performed. (D) 35S-labeled P815 cells were incubated either with no mAb (□), anti-CD16 mAb (B73.1.1; ♦), anti-NKp46 mAb (461-G1; ○), or anti-CD99 mAb (12E7; ▴) for 1 hour on ice. NK cells were next added in effector to target ratio (E/T) of 3:1. Shown is one representative experiment of 7 performed. (E) NK cells were preincubated with the indicated mAb for 1 hour on ice, washed and incubated with either uninfected 1106mel or 1106mel infected with various influenzas as indicated. This experiment was preformed in E/T ratio of 20:1 and represents one of 4 performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-05-1716/6/m_h80245510005.jpeg?Expires=1769081588&Signature=Gnm2QSOSrY48jxSw70qJZwN1GaD9RG8rMTnybzxi977eMRCakzCKqh39iJm0KTxRgpTlQtXiJZ4~nT9riGtmsn5WNZemVJZGBsMrtry65PAX8mgerWt~YRsd1SWpEmdVSY-7FOgRL-XMnc~RXV70QigCP5i4byx~gD5iTwyDReRVvRUNHyEHG9x44SuRF~9YGxhMWCrBWEIAfc1DUEsK4nBoyLC3x8KWKohRkjfkAs-TYxi-iQ-zzTKBe06bvQW65Eibme7sBa1HzzZHAU3u2RkxS4Z0wrSr7SmQmi3lH4DNWgH~5hPAVZP4dDfyngvgcsb7dBq8VNQ7t3vk9d2xBA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 9. Thr225 of NKp46 is important for NKp46 activation. (A) Parental BW cells, or stably transfected BW cells expressing the chimeric receptors CD99-Zeta, NKp46-Zeta, or NKp46T225A-Zeta, were stained with anti-NKp46 mAb (461-G1), followed by FITC-conjugated goat antimouse Fc. Gray histograms represent the background staining of FITC-conjugated goat antimouse Fc. The MFI values of the anti-NKp46 mAb staining are indicated in the figure. Figure shows one representative experiment of 3 performed. (B) 1106mel cells were incubated overnight with or without human influenza virus (Sydney strain). Next, cells (50 000/well) were irradiated (6000 rad [60 Gy]), washed, and incubated with 50 000 cells/well of BW cells expressing CD99-Zeta (BW/CD99Z), wild-type NKp46-Zeta (BW/NKp46Z wild type), mutated NKp46T225A-Zeta (BW/NKp46T225AZ), or parental BW cells. After 48 hours, IL-2 levels in the supernatants were measured by ELISA. Optical density (OD) absorbance (650 nm) was translated into amount of IL-2 (pg/well) according to a standard curve. Results represent one experiment of 2 performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-05-1716/6/m_h80245510009.jpeg?Expires=1769081588&Signature=QtGUD6lXkeTOmTJUenPuofMH08brjQ7aVWcOPs-MO7cNHer511mXipgob-LGSJtjY825z8T9jrK~MrqyI8u9HbIo7FIMWSfzGPHTzHqsR3eSl0GmyVmUlkTqI~R008Fp3o4Gc8p8tJF6R~lJCuN1l1IonKkFnrpUJKIZcyl8zC94blIeSCjTMtgmcBpzIT8mgSP0TyxxrJ9VXBm0rRn1tcyVxFEHDUtRRA4o0TIRkXz2euLtuSJSkpKn1j7M5KOLMaZ5qPWUylV1MOq65VqZexbxZS6edMQivJnaLL-fMBNTnJhXccV1XFGONPwORMwNI2Jmq2IP-bu15C8kFzH7qg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal