Abstract
The ability of donor lymphocyte infusions (DLIs) to induce complete responses (CRs) in patients with relapsed myeloma after allogeneic bone marrow transplantation (BMT) provides clear evidence of an effective graft-versus-myeloma (GVM) response. To identify target antigens of the GVM response, we screened a myeloma cDNA expression library with post-DLI serum from 4 patients with myeloma who achieved CR after DLI and 1 patient who was in CR before DLI. We identified a panel of 13 gene products reactive with post-DLI serum but negative with pre-DLI and pre-BMT serum. Antibodies to these proteins were not detected in the sera of 10 patients who underwent allogeneic BMT without DLI and 5 patients with acute graft-versus-host disease (GVHD). Minimal reactivity with these proteins was detected in the sera of 20 healthy donors and 20 patients with chronic GVHD. In contrast, 5 of these proteins were recognized by more than 1 myeloma DLI responder. Testing of serial serum samples showed an association between antibody response and time of best response after DLI. The expression of these genes was evaluated in primary myeloma cells and in normal plasma cells. This study demonstrates that the GVM response is associated with antibody responses to highly expressed myeloma-associated antigens.
Introduction
Previous studies have demonstrated that allogeneic bone marrow transplantation (BMT) can provide effective therapy for patients with multiple myeloma.1-5 Although the benefit of BMT is in part provided by the administration of high-dose chemotherapy and total body irradiation, donor immune cells also play an important role in the curative effects of allogeneic BMT.4,6-8 One of the clearest demonstrations of the effectiveness of graft-versus-myeloma (GVM) immunity is the clinical response achieved after donor lymphocyte infusion (DLI). Several studies have shown clinically significant responses to DLI in 40% to 50% of patients with myeloma in the absence of any other therapy.9,10 Some of these patients only achieve partial responses after DLI, but 20% to 30% of patients with relapsed myeloma achieve long-lasting complete remission after a single infusion of donor lymphocytes. Although GVM effects after DLI are often associated with the development of GVHD,10-12 previous clinical reports have also documented GVM activity in the absence of GVHD. The distinction between GVM and GVHD is also supported by laboratory studies that have identified different T-cell populations that mediate GVM and GVHD.13 These laboratory studies also suggest that at least some of the target antigens of the GVM response are distinct from those that also cause GVHD.14 Taken together, these clinical and laboratory studies demonstrate that the immune response directed against myeloma tumor cells provides an example of effective tumor immunity, and further characterization of this immune response may help elucidate the immune mechanisms that mediate tumor rejection in vivo.
Although T cells clearly play a critical role in the GVM response, several lines of evidence suggest that tumor rejection in vivo represents a coordinated immune response involving B and natural killer (NK) cells as well as T cells. In animal models, interactions between antibody responses and both CD4+ and CD8+ T cells facilitate the elimination of tumor cells in vivo.15,16 In patients who respond to tumor vaccines, histologic examination of vaccination sites and regressing tumors demonstrates the presence of a diverse inflammatory response and of polyclonal B cells and T cells.17-19 Additional characterization of B-cell responses in patients with widely varying tumors has demonstrated the presence of high-titer polyclonal antibody responses to a diversity of tumor-associated antigens. Using patient serum to directly clone these target antigens from a cDNA expression library, investigators have identified a large number of novel tumor-associated antigens. 20-24 Several antigens (eg, NY-ESO-1) initially identified by this method (serological identification of antigens by recombinant expression cloning, termed SEREX) have subsequently been shown to be targets of specific T-cell responses in vivo.25-27 Similarly, antigens (eg, MAGE-1 and WT1) first identified as targets of tumor-specific T cells28 have also been identified using the SEREX approach.20,21,29 These observations support the hypothesis that effective tumor rejection in vivo results from a complex and coordinated immune response involving different classes of effector cells targeting a variety of tumor-associated antigens.
In a previous study in patients with myeloma who underwent allogeneic BMT, we found that prophylactic infusions of CD4+ donor lymphocytes successfully induced GVM responses in most patients who received DLIs.30 Infusions of CD4+ donor T cells also resulted in the significant expansion of peripheral B cells, in contrast to outcomes in patients who underwent allogeneic BMT without DLI.31 We hypothesized that the expansion of peripheral B cells at the time of clinical response after DLI reflected a strong antibody response directed against myeloma-associated antigens. To test this hypothesis, we used sera from patients who responded to DLI to screen a myeloma cDNA expression library. These experiments identified a panel of 13 myeloma-associated antigens recognized by antibodies that developed after DLI but that were not detectable in patient sera before BMT or before DLI. Although these antigens represent a diverse group of nuclear, cytoplasmic, and membrane proteins, antibody responses to these antigens appeared to correlate with tumor rejection in vivo. This approach thus provides a useful method for identifying myeloma-associated tumor antigens. Additional studies can be undertaken to characterize the coordination of B and T responses to these antigens during tumor rejection.
Patients, materials, and methods
Patient samples and treatment
Serum samples were obtained after informed consent from healthy donors and patients enrolled in clinical trials of allogeneic stem cell transplantation. All patients received myeloablative therapy followed by infusions of marrow stem cells from HLA-identical sibling donors. The 5 patients with multiple myeloma used to screen the cDNA expression library received CD6+ T-cell–depleted marrow followed by prophylactic infusions of CD8-depleted lymphocytes from the same donor 6 months after marrow transplantation, as previously described.30,31 RNA for gene expression analysis was obtained from purified CD138+ plasma cells from 33 patients with myeloma, 5 patients with monoclonal gammopathy of unknown significance (MGUS), and 5 healthy donors. Cell lines for Northern blot analysis were purchased from American Type Culture Collection (Manassas, VA). Clinical protocols were approved by the institutional review board of the Dana-Farber/Harvard Cancer Center, and informed consent was obtained from each patient.
Complete remission (CR) was defined as the absence of clonal plasma cells in bone marrow and the lack of detectable monoclonal protein in serum or urine by immunofixation. Partial response (PR) was defined as a 50% or greater reduction in monoclonal immunoglobulin or light chain; decreases of less than 25% were defined as no response. A 25% or more increase in monoclonal protein concentration indicated progressive disease (PD).
Myeloma cDNA library
Total RNA was isolated from CD138+ myeloma cells purified from the bone marrow of a patient with immunoglobulin G (IgG) myeloma. Poly (A)+ RNA was prepared with a messenger (mRNA) isolation kit (Stratagene, La Jolla, CA). The cDNA expression library was constructed using 5 μg poly (A)+ RNA. First-strand synthesis was performed using an oligo (dt) primer with an internal XhoI site and 5-methyl-CTP. The cDNA was ligated to an EcoRI adapter and digested with XhoI. Fragments were cloned directionally into the bacteriophage expression vector 1ZAP II (Stratagene), packaged into phage particles, and transfected into XLI-blue Escherichia coli. The library was amplified and used for antibody screening.
Myeloma cDNA library screening
Serum samples from 5 patients with myeloma who underwent CD6–T-cell–depleted BMT and CD4+ donor lymphocyte infusions 6 months after BMT30 were used to screen the cDNA expression library. Primary screening of the library was performed as previously described,32 with minor modification. Briefly, 5 × 104 recombinant phages per dish were cultured at 42°C for 4 hours. Expression of recombinant protein was induced by incubation with isopropyl β-D-thiogalactoside (IPTG)–treated nitrocellulose membranes for 3.5 hours at 37°C. To exclude IgG clones in the primary screening, an identical second set of IPTG-treated nitrocellulose membranes was incubated for another 3.5 hours at 37°C. After washing and blocking with 2% nonfat dry milk in Tris-buffered saline (TBS), one set of membranes was incubated overnight with post-DLI (1:500 dilution) patient sera followed by incubation with alkaline-phosphatase–conjugated goat antihuman IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted at 1:2000 in TBS/Tween 20 (TBST). The second set of membranes was only incubated with the secondary antibody. Antigen-antibody complexes were visualized by staining with 5-bromo-4-chloro-3-indolylphosphatase and nitroblue tetrazolium (BCIP/NBT) (Promega, Madison, WI). Individual clones identified in both sets of membranes were presumed to express human IgG and were discarded. All other clones were plated for secondary and tertiary screenings until single plaques were isolated. The cDNA inserts of selected clones were excised and sequenced using T3 and T7 primers.
Phage-plate assay
Purified positive clones were mixed with nonreactive phages of the cDNA library as internal negative controls at a 1:10 ratio. Two hundred microliters XL1-blue E coli bacteria was transfected with this mix and was plated in NZY agar plates for 4 hours at 42°C. Recombinant protein expression was induced by incubation with IPTG-treated nitrocellulose membranes for an additional 3.5 hours at 37°C. After washing in TBST and blocking overnight with 2% nonfat dry milk in TBS, filters were incubated overnight with test serum diluted 1:200; this was followed by incubation with alkaline phosphatase-conjugated goat antihuman IgG antibody diluted at 1:2000 in TBST. Antigen-antibody complexes were visualized by staining with BCIP/NBT. The intensity of positive staining was visually graded on a scale of 1 to 4.
Microarray gene expression
Total RNA from primary myeloma cells, plasma cells from patients with MGUS, or plasma cells from healthy donors was extracted using Trizol reagent (Gibco BRL, Grand Island, NY) and was purified using the SV Total RNA Isolation System (Promega) according to the manufacturer's protocol, with minor modifications. The detailed protocol for sample preparation and microarray processing is available from Affymetrix (Santa Clara, CA) (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Briefly, first-strand cDNA was synthesized from 5 μg total RNA using a T7-(dT)24 primer (Genset Corporation, San Diego, CA) and was reverse transcribed with the SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen Life Technologies, Carlsbad, CA). After synthesis of second-strand cDNA, the product was used for in vitro transcription (bioarray kit; Enzo Diagnostics, Farmingdale, NY) to generate biotinylated complementary RNA (cRNA). Fragmented cRNA (15 μg/sample) was hybridized for 16 hours to the Affymetrix U95Av2 GeneChip, which contains 12 600 sequences and is derived from the GenBank database. After hybridization, each microarray was washed, stained, and scanned with an argon-ion confocal laser, with excitation at 488 nm and detection at 570 nm. The CEL files were imported into d-Chip (http://www.dchip.org),33 and normalized values were obtained. Expression of specific genes of interest was evaluated in each set of plasma cell samples.
Statistical methods
To compare the gene expression in plasma cells among healthy donors, patients with MGUS, and patients with myeloma, the Wilcoxon rank-sum test was used for all 2-way comparisons. No adjustment was made for multiple comparisons.
Northern blot
Total RNA from myeloma and leukemia cell lines, peripheral blood leukocytes (PBLs), and BM from healthy donors was extracted using Trizol reagent (Gibco) and was purified using the SV total RNA isolation system (Promega). Fifteen micrograms RNA was loaded in each lane in a formaldehyde agarose gel and was run for 3.5 hours. Gels were blotted into nylon membranes (Hybond XL; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) and were ultraviolet light (UV) cross-linked in a UV light box (Stratalinker; Stratagene, La Jolla, CA). Hybridizations were conducted with the purified clones used as a probe labeled with 32P in QuikHyb hybridization solution (Stratagene) at 68°C for 1 hour according to the manufacturer's protocol. The same blots were then stripped and hybridized with 32P-labeled human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe as control.
Results
Myeloma cDNA library screening
To determine whether patients who respond to DLI develop antibody responses to myeloma-associated antigens, we constructed a cDNA expression library from CD138+ myeloma cells obtained from a patient with IgG myeloma. The library was screened with post-DLI serum (at 1:500 dilution) from 5 patients who underwent allogeneic BMT for relapsed myeloma. As shown in Table 1, all 5 patients had IgG-secreting myeloma. Except for patient 3, who achieved CR after BMT, patients did not achieve CR until after DLI. The time of the best response after DLI varied. Early responses (3-4 months) were noted in 2 patients, whereas CR was not achieved until much later (1-2.4 years) in 2 other patients. The myeloma cDNA library was therefore screened with early (4 months) and late (1 and 2.6 years) post-DLI serum from each patient.
Patient characteristics
Patient . | Myeloma type . | Disease status after BMT . | Response to DLI . | Time of best response after DLI . |
|---|---|---|---|---|
| 1 | IgG | PD | CR | 4 mo |
| 2 | IgG | PR | CR | 4 mo |
| 3 | IgG | CR | — | — |
| 4 | IgG | PR | CR | 2.4 y |
| 5 | IgG | PD | CR | 1 y |
Patient . | Myeloma type . | Disease status after BMT . | Response to DLI . | Time of best response after DLI . |
|---|---|---|---|---|
| 1 | IgG | PD | CR | 4 mo |
| 2 | IgG | PR | CR | 4 mo |
| 3 | IgG | CR | — | — |
| 4 | IgG | PR | CR | 2.4 y |
| 5 | IgG | PD | CR | 1 y |
— indicates that the patient remained in CR.
Library screening with early post-DLI serum resulted in the identification of 12 clones, and late post-DLI serum identified 14 clones. Each of these clones was only reactive with post-DLI serum and was negative with pre-BMT and pre-DLI serum. After restriction enzyme digestion and DNA sequence analysis, these clones were found to represent 13 unique gene products (Table 2). Five genes were identified with early serum, 5 genes were identified with late serum, and 3 genes were identified with early and late serum samples (Table 2). Interestingly, no clones were identified with early or late serum from patient 3, who was in CR before DLI. As summarized in Table 2, though 4 of the genes have unknown functions, 9 genes have been previously described. Among the known genes, 8 are intracellular proteins involved in a variety of functions, including DNA and RNA binding, cell signaling, and apoptosis. One gene (BCMA) is a transmembrane receptor of the tumor necrosis factor (TNF) superfamily, recently shown to be selectively expressed on mature B cells.34
Gene products identified with early and late post-DLI serum
Gene products . | Chromosome localization . | mRNA size, bp . | Serum screening . | Comments . |
|---|---|---|---|---|
| DHA | 11q23.1 | 2583 | E + L | E2 component of human pyruvate dehydrogenase complex (E2-PDC) |
| KIAA0053 | 2p14 | 2739 | E | Unknown function |
| BCMA | 16p13.1 | 995 | E | B-cell maturation antigen |
| FLJ10330 | 1 | 3239 | E | Unknown function |
| FLJ10534 | 17 | 4138 | E | Unknown function |
| SON DNA | 21q22.11 | 8368 | E | DNA-binding protein |
| ROCK-1 | 18 | 4065 | L | Downstream effector protein of Rho |
| Hepatoma-derived growth factor—related protein | 19p13.3 | 2305 | L | Unknown function |
| Homer-3 | 19p13.1 | 982 | E + L | Neuronal immediate early gene |
| Bif-1 | 1p22 | 1561 | E + L | Regulator of Bax |
| Heterogeneous nuclear ribonucleoprotein D—like | 4q13 | 4919 | L | RNA-binding protein |
| SFRS | 7q22 | 3702 | L | Protein kinase |
| AT-rich sequence-binding protein | 3p23 | 2907 | L | DNA-binding protein |
Gene products . | Chromosome localization . | mRNA size, bp . | Serum screening . | Comments . |
|---|---|---|---|---|
| DHA | 11q23.1 | 2583 | E + L | E2 component of human pyruvate dehydrogenase complex (E2-PDC) |
| KIAA0053 | 2p14 | 2739 | E | Unknown function |
| BCMA | 16p13.1 | 995 | E | B-cell maturation antigen |
| FLJ10330 | 1 | 3239 | E | Unknown function |
| FLJ10534 | 17 | 4138 | E | Unknown function |
| SON DNA | 21q22.11 | 8368 | E | DNA-binding protein |
| ROCK-1 | 18 | 4065 | L | Downstream effector protein of Rho |
| Hepatoma-derived growth factor—related protein | 19p13.3 | 2305 | L | Unknown function |
| Homer-3 | 19p13.1 | 982 | E + L | Neuronal immediate early gene |
| Bif-1 | 1p22 | 1561 | E + L | Regulator of Bax |
| Heterogeneous nuclear ribonucleoprotein D—like | 4q13 | 4919 | L | RNA-binding protein |
| SFRS | 7q22 | 3702 | L | Protein kinase |
| AT-rich sequence-binding protein | 3p23 | 2907 | L | DNA-binding protein |
DHA indicated dihydrolipoamide acetyltransferase; E, early and L, late.
Testing normal, post-BMT, and post-DLI sera for reactivity with GVM-associated antigens
After identifying 13 gene products that were potential target antigens of the DLI response, we examined whether antibodies against these proteins were also present in healthy donors or in other patients after allogeneic BMT. Serum samples (1:200 dilution) from 20 healthy donors, 5 patients with acute GVHD, 20 patients with chronic GVHD, and 10 patients (5 with myeloma and 5 with other hematologic malignancies) who underwent allogeneic BMT but without DLI were tested for reactivity against each of these gene products using a phage-plate assay. Serum samples from patients who underwent only allogeneic BMT or patients with chronic GVHD were obtained 1 year after BMT. Samples from patients with acute GVHD were obtained at the time of onset of GVHD. All patients were in CR. As shown in Table 3, serum from 1 healthy donor was reactive with 1 of these gene products (AT-rich sequence-binding protein), and sera from 3 patients with chronic GVHD were reactive with 1 of these gene products each. In contrast, 5 of these proteins were recognized by the sera of other myeloma patients who responded to DLI. One of these proteins (ROCK-1, identified in patient 4) was reactive with the sera of 3 additional patients of the total group of 9 myeloma DLI responders. Dihydrolipoamide acetyltransferase (DHA), BCMA, FLJ10330 (identified in patient 2), and Homer-3 (identified in patient 1) were each reactive with the serum of 1 additional patient in the group of 9 DLI responders tested. Antibodies to DHA were also detected in the serum of 1 patient with chronic myelogenous leukemia (CML) who had a complete cytogenetic and molecular response after DLI. We also tested sera from 5 patients with myeloma who did not respond to DLI, but none of these samples had detectable antibodies against this panel of proteins.
Recognition of target antigens with normal, post-BMT, and post-DLI serum
Gene products . | Healthy donors, n = 20 . | Acute GVHD, n = 5 . | Chronic GVHD, n = 20 . | TCD BMT, n = 10 . | Myeloma DLI responders, n = 9 . | Myeloma DLI nonresponders, n = 5 . | CML DLI responders, n = 5 . |
|---|---|---|---|---|---|---|---|
| Dihydrolipoamide acetyltransferase | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| KIAA0053 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| BCMA | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| FLJ10330 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ROCK-1 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| Similar to hepatoma-derived growth factor | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Homer-3 | 0 | 0 | 1 | 0 | 2 | 0 | 0 |
| Bif-1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Heterogeneous nuclear ribonucleoprotein D—like | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| SON DNA | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| FLJ10534 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| SFRS | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| AT-rich sequence-binding protein | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
Gene products . | Healthy donors, n = 20 . | Acute GVHD, n = 5 . | Chronic GVHD, n = 20 . | TCD BMT, n = 10 . | Myeloma DLI responders, n = 9 . | Myeloma DLI nonresponders, n = 5 . | CML DLI responders, n = 5 . |
|---|---|---|---|---|---|---|---|
| Dihydrolipoamide acetyltransferase | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| KIAA0053 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| BCMA | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| FLJ10330 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ROCK-1 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| Similar to hepatoma-derived growth factor | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Homer-3 | 0 | 0 | 1 | 0 | 2 | 0 | 0 |
| Bif-1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Heterogeneous nuclear ribonucleoprotein D—like | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| SON DNA | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| FLJ10534 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| SFRS | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| AT-rich sequence-binding protein | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
Presence of antibodies to GVM-associated antigens at different times after DLI
As noted previously, only 3 proteins were identified in the screening of early and late post-DLI serum. The remaining 10 proteins were only identified with either early or late post-DLI samples. To better characterize the development of antibody responses to this panel of proteins, we used the phage-plate assay to test serum samples obtained at multiple times after DLI for the development of antibodies to these gene products. Serum samples (1:200 dilution) obtained over a 2-year period from each of the patients used to screen the myeloma library were compared with samples obtained before BMT and before DLI. As shown in Table 4, repeat testing of pre-BMT and pre-DLI serum showed no evidence of reactivity against any of these gene products. For the 5 proteins that were initially identified by screening serum samples obtained 4 months after DLI, evidence of antibody reactivity against 3 of these proteins was present in earlier samples obtained 2 months after DLI. Although titers were lower, antibodies against 4 proteins remained detectable 1 year after DLI, and antibodies against 3 proteins persisted for more than 2 years after DLI. Analysis of antibody responses to the 5 proteins initially identified by late post-DLI serum showed a different pattern of reactivity. Although antibody responses to 4 of these proteins were detectable 4 months after DLI, these were weak responses, and antibody titers were higher at later time points. Antibody responses to the 3 proteins identified by early and late post-DLI samples generally showed persistence at relatively high levels throughout this long period of observation.
Serial reactivity of post-DLI serum with myeloma-associated antigens
Gene products . | Screening serum . | Pre-BMT . | Pre-DLI . | 2 mo . | 4 mo . | 6 mo . | 1 y . | 2 to 2.6 y . |
|---|---|---|---|---|---|---|---|---|
| KIAA0053 | E | - | - | ++ | + | + | - | - |
| BCMA | E | - | - | + | ++++ | +++ | ++ | + |
| FLJ10330 | E | - | - | + | +++ | ++ | + | + |
| FLJ10534 | E | - | - | - | + | + | + | - |
| SON DNA | E | - | - | NA | ++ | ++ | + | + |
| DHA | E + L | - | - | ++ | ++++ | +++ | +++ | ++ |
| Homer-3 | E + L | - | - | NA | ++ | ++ | ++ | + |
| Bif-1 | E + L | - | - | + | ++ | ++++ | +++ | ++ |
| ROCK-1 | L | - | - | - | - | - | + | ++++ |
| Similar to hepatoma-derived growth factor | L | - | - | - | + | + | + | ++++ |
| Heterogeneous nuclear ribonucleoprotein D-like | L | - | - | + | + | + | ++ | ++ |
| SFRS | L | - | - | NA | + | + | ++ | ++ |
| AT-rich sequence-binding protein | L | - | - | NA | + | ++ | ++ | ++ |
Gene products . | Screening serum . | Pre-BMT . | Pre-DLI . | 2 mo . | 4 mo . | 6 mo . | 1 y . | 2 to 2.6 y . |
|---|---|---|---|---|---|---|---|---|
| KIAA0053 | E | - | - | ++ | + | + | - | - |
| BCMA | E | - | - | + | ++++ | +++ | ++ | + |
| FLJ10330 | E | - | - | + | +++ | ++ | + | + |
| FLJ10534 | E | - | - | - | + | + | + | - |
| SON DNA | E | - | - | NA | ++ | ++ | + | + |
| DHA | E + L | - | - | ++ | ++++ | +++ | +++ | ++ |
| Homer-3 | E + L | - | - | NA | ++ | ++ | ++ | + |
| Bif-1 | E + L | - | - | + | ++ | ++++ | +++ | ++ |
| ROCK-1 | L | - | - | - | - | - | + | ++++ |
| Similar to hepatoma-derived growth factor | L | - | - | - | + | + | + | ++++ |
| Heterogeneous nuclear ribonucleoprotein D-like | L | - | - | + | + | + | ++ | ++ |
| SFRS | L | - | - | NA | + | + | ++ | ++ |
| AT-rich sequence-binding protein | L | - | - | NA | + | ++ | ++ | ++ |
E indicates early; L, late; NA, sample not available; -, negative. Other table symbols (+, -) are described in the footnote to Table 5.
Association of antibody response with the time of the best response after DLI
To assess whether antibody reactivity correlated with clinical response after DLI, we compared the development of antibody responses to the same target antigens in myeloma patients who responded to DLI (Table 5). For example, patients 2 and 5 developed antibody responses to DHA, BCMA, and FLJ10330. Patient 2 was an early responder, and the strongest response to each of these antigens was found in the early post-DLI samples. In contrast, patient 5 achieved CR at 1 year after DLI, and antibodies to DHA and BCMA were not detectable until 6 months after DLI. The antibody response to FLJ10330 was present 4 months after DLI, but the strongest response was noted at the time of CR, 1 year after DLI. Similarly, patients 1 and 7 developed antibody responses to Homer-3 protein, and the strongest responses to this antigen were found at the time of best clinical response. As shown in Table 5, 4 patients developed antibody responses to ROCK-1. In early and late responders, the development of antibody to ROCK-1 appeared to correlate with the time of best clinical response after DLI. Figure 1 shows results of the phage-plate assay for ROCK-1 with serum samples from patient 2 (early responder) and patient 4 (late responder). Although the phage-plate assay is not quantitative, the intensity of reactivity was clearly stronger at 4 months for patient 2, whereas the strongest reactivity for patient 4 was noted 2.6 years after DLI.
Comparison of antibody responses after DLI
Gene product . | Patient . | Pre-BMT . | Pre-DLI . | 2 mo . | 4 mo . | 6 mo . | 1 y . | 2 to 2.6 y . |
|---|---|---|---|---|---|---|---|---|
| DHA | 2, ER | - | - | ++ | ++++ | +++ | +++ | ++ |
| DHA | 5, LR | - | - | - | - | + | ++ | ++ |
| BCMA | 2, ER | - | - | + | ++++ | +++ | ++ | + |
| BCMA | 5, LR | - | - | - | - | + | + | + |
| FLJ10330 | 2, ER | - | - | + | +++ | ++ | + | + |
| FLJ10330 | 5, LR | - | - | - | + | + | ++ | + |
| Homer-3 | 1, ER | - | - | NA | ++ | ++ | ++ | + |
| Homer-3 | 7, LR | - | - | NA | - | + | ++ | + |
| ROCK-1 | 4, LR | - | - | - | - | - | + | ++++ |
| ROCK-1 | 5, LR | - | - | - | - | + | + | + |
| ROCK-1 | 2, ER | - | - | + | +++ | ++ | + | + |
| ROCK-1 | 9, ER | - | - | + | + | + | NA | NA |
Gene product . | Patient . | Pre-BMT . | Pre-DLI . | 2 mo . | 4 mo . | 6 mo . | 1 y . | 2 to 2.6 y . |
|---|---|---|---|---|---|---|---|---|
| DHA | 2, ER | - | - | ++ | ++++ | +++ | +++ | ++ |
| DHA | 5, LR | - | - | - | - | + | ++ | ++ |
| BCMA | 2, ER | - | - | + | ++++ | +++ | ++ | + |
| BCMA | 5, LR | - | - | - | - | + | + | + |
| FLJ10330 | 2, ER | - | - | + | +++ | ++ | + | + |
| FLJ10330 | 5, LR | - | - | - | + | + | ++ | + |
| Homer-3 | 1, ER | - | - | NA | ++ | ++ | ++ | + |
| Homer-3 | 7, LR | - | - | NA | - | + | ++ | + |
| ROCK-1 | 4, LR | - | - | - | - | - | + | ++++ |
| ROCK-1 | 5, LR | - | - | - | - | + | + | + |
| ROCK-1 | 2, ER | - | - | + | +++ | ++ | + | + |
| ROCK-1 | 9, ER | - | - | + | + | + | NA | NA |
ER indicates early responder; LR, late responder; NA, sample not available; -, negative; +, weak; ++, moderate; +++,strong; and ++++, very strong.
Serial time points analysis of ROCK-1. Assessment of antibody reactivity with ROCK-1 determined by phage-plate assay in serial samples from patient 2 (early responder [ER]) and patient 4 (late responder [LR]). Quantification was based on the intensity of reactivity of the positive plaques.
Serial time points analysis of ROCK-1. Assessment of antibody reactivity with ROCK-1 determined by phage-plate assay in serial samples from patient 2 (early responder [ER]) and patient 4 (late responder [LR]). Quantification was based on the intensity of reactivity of the positive plaques.
Expression of DHA, Homer-3, and ROCK-1 in tumor cell lines, primary myeloma, MGUS, and normal plasma cells
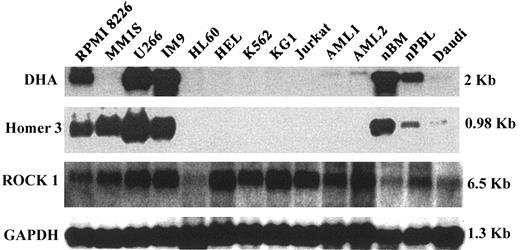
We next examined the expression of 3 of these genes in a series of tumor cell lines by Northern blot. These particular genes were selected from the larger panel because antibodies to these proteins had been detected in several patients. As shown in Figure 2, DHA was highly expressed in 3 of 4 myeloma tumor cell lines but not in several other myeloid (HL-60, K562, KG1) or lymphoid (Jurkat, Daudi) tumor cell lines. DHA was expressed at a lower level in normal bone marrow and peripheral blood mononuclear cells but not in primary acute myelogenous leukemia (AML) cells from 2 patients. The pattern of expression of the HOMER 3 gene was similar except that high-level expression was seen in all 4 myeloma cell lines. In contrast, ROCK-1 was expressed in all cells tested; expression was lowest in HL60 (promyelocytic leukemia cell line) and in normal bone marrow.
Northern blot analysis of DHA, ROCK-1, and Homer-3. mRNA expression was analyzed in 4 myeloma cell lines (RPMI 8226, MM1S, U266, and IM9), 5 leukemia cell lines, HL60 (acute promyelocytic leukemia), HEL (erythroleukemia), K562 (chronic myelogenous leukemia), KG1 (acute myelogenous leukemia), Jurkat (acute T-cell leukemia), 1 Burkitt lymphoma cell line (Daudi), 2 primary AML cells, and PBL and BM from healthy donors.
Northern blot analysis of DHA, ROCK-1, and Homer-3. mRNA expression was analyzed in 4 myeloma cell lines (RPMI 8226, MM1S, U266, and IM9), 5 leukemia cell lines, HL60 (acute promyelocytic leukemia), HEL (erythroleukemia), K562 (chronic myelogenous leukemia), KG1 (acute myelogenous leukemia), Jurkat (acute T-cell leukemia), 1 Burkitt lymphoma cell line (Daudi), 2 primary AML cells, and PBL and BM from healthy donors.
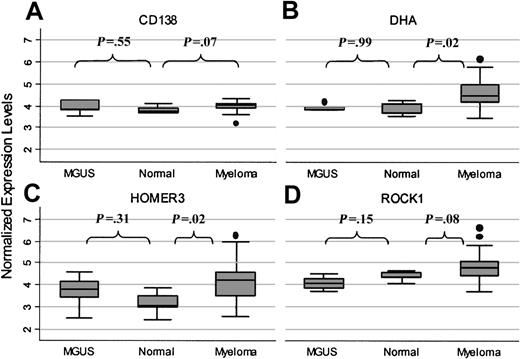
Expression of these 3 genes in primary myeloma tumor cells was also evaluated using oligonucleotide microarrays (Affymetrix Hu95av2). As shown in Figure 3, expression of these genes in 33 primary myeloma tumor cells was compared with expression in purified plasma cells from 5 patients with MGUS and purified plasma cells from 5 healthy donors. For each of these genes, the range of expression values was much broader in myeloma tumor cells than in the other plasma cells. Expression of DHA (Figure 3B) was significantly higher in myeloma than in MGUS plasma cells (P = .01) or normal plasma cells (P = .02). Expression of the HOMER 3 gene (Figure 3C) was not significantly higher in myeloma than in MGUS (P = .38), but HOMER 3 gene expression was significantly higher in myeloma than in normal plasma cells (P = .02). ROCK-1 gene expression (Figure 3D) was also highest in primary myeloma cells, but expression values were not significantly higher than in normal plasma cells (P = .08). For comparison, CD138, an antigen known to be highly expressed in myeloma and in normal plasma cells (Figure 3A), was uniformly expressed in all 3 populations.
Quantitative expression in primary plasma cells. Quantitative expression of CD138 (A), DHA (B), HOMER 3 (C), and ROCK-1 (D) in primary plasma cells measured by Affymetrix U95Av2 microarray. Gene expression was examined in myeloma (n = 33), MGUS (n = 5), and normal BM plasma cells (n = 5). Box plots define the median values, 25% to 75% of values around the median, and the range of values.
Quantitative expression in primary plasma cells. Quantitative expression of CD138 (A), DHA (B), HOMER 3 (C), and ROCK-1 (D) in primary plasma cells measured by Affymetrix U95Av2 microarray. Gene expression was examined in myeloma (n = 33), MGUS (n = 5), and normal BM plasma cells (n = 5). Box plots define the median values, 25% to 75% of values around the median, and the range of values.
Discussion
Although clinical responses to DLI in patients with multiple myeloma provide convincing examples of effective tumor immunity, the immunologic mechanisms and the target antigens of this response have not been well characterized. Importantly, previous studies in patients with CML demonstrated the presence of a coordinated immune response involving T and B cells at the time of tumor rejection.16,26,32 Similarly, patients with myeloma who responded to DLI were found to have increased numbers of circulating B cells.31 The present study was, therefore, undertaken to determine whether patients with myeloma who respond to DLI also develop antibody responses to myeloma-associated antigens and to directly identify the antigenic targets of this response. Using serum from 5 patients who received DLI, these experiments resulted in the identification of a panel of 13 distinct antigens representing a diverse set of cellular proteins. Antibodies to this panel of 13 antigens could not be detected before BMT or before DLI and only developed after DLI. In those patients who responded soon after DLI, a strong antibody response developed 2 to 4 months after DLI. In those patients who did not achieve CR until 1 to 2 years after a single infusion of donor cells, the strongest antibody response did not occur until 1 to 2 years after DLI. In 1 patient with myeloma who had achieved CR after allogeneic transplantation, before the prophylactic infusion of donor cells, no antibodies specific for myeloma-associated antigens could be detected in any post-DLI serum. Although additional studies to define the role of these 13 antigens in the GVM response after DLI are necessary, our experiments show that antibody responses to this set of antigens were temporally related to the immunologic elimination of myeloma cells in vivo and suggest that this panel of proteins represents potential targets of this immune response. Given that 12 of these antigens are intracellular proteins, it is unlikely that antibody responses alone were responsible for the elimination of myeloma cells in vivo. In responding patients, the long-term persistence of high-titer IgG antibodies also suggests that donor T cells specific for these antigens developed after DLI. These T cells likely play a primary effector role in the GVM response. Further studies will be conducted to identify T-cell responses to these antigens to better characterize the immune responses that mediate rejection of myeloma cells after DLI.
Considering that this panel of 13 antigens was targeted after the infusion of allogeneic donor lymphocytes, it is necessary to consider the possibility that genetic polymorphisms that distinguish recipient from donor proteins contribute to the immunogenicity of these antigens. Single nucleotide polymorphisms (SNPs) are the most common manifestation of human genetic diversity and often lead to amino acid polymorphisms. When these polymorphisms affect antigen processing or peptide binding to major histocompatibility complex proteins on the cell surface, these protein disparities result in the generation of minor histocompatibility antigens (mHAs) that can be recognized by donor T cells after allogeneic transplantation.35 Minor histocompatibility antigens have traditionally been defined as targets of donor T cells, but recent studies from our laboratory have demonstrated that mHAs can also elicit high-titer antibody responses after allogeneic transplant.54 Nevertheless, it is unlikely that the immune response to most of the myeloma-associated antigens identified in our panel was caused by genetic disparities between donor and recipient. For 6 of the 13 antigens (DHA, KIAA0053, BCMA, ROCK-1, Homer-3, Bax-interacting factor 1 [Bif-1]), the DNA sequence of the immunogenic clone was 100% identical to the GenBank sequence. This suggests that SNPs do not contribute to the immunogenicity of these proteins. Moreover, little antibody reactivity against this panel of antigens was found in patients with acute or chronic GVHD. Nevertheless, further comparison of the DNA sequence of these genes in patients and their donors will be needed to definitively exclude the possibility that the immunogenicity of these proteins is attributed to genetic disparities between donor and recipient.
Although several of the antigens we identified were only immunogenic in individual responders, extending our analysis to include additional patients with myeloma who responded to DLI resulted in the identification of 5 antigens that were recognized in more than 1 patient. Antibodies to DHA, BCMA, FLJ10330, and Homer-3 were each detected in 2 myeloma DLI responders, and antibodies to ROCK-1 were present in 4 responders. In these instances, peak antibody responses also occurred when patients achieved CR. Antibodies to this set of proteins were not detected in 5 patients with myeloma who underwent allogeneic BMT or in 5 patients with myeloma who did not respond to DLI. Antibody to only 1 antigen (DHA) was found in 1 of 5 patients with CML who responded to DLI. These antigens were also not generally immunogenic after allogeneic BMT or in patients with acute or chronic GVHD. Taken together, these observations suggest that the antibody response after DLI appears to be directed against a selected set of highly immunogenic antigens. Many of these proteins have not previously been identified as potential targets of antitumor immunity. However, antibodies specific for 4 of these proteins (FLJ10330, Heterogeneous nuclear ribonucleoprotein D–like, ROCK-1, and SFRS) have previously been identified in patients with other tumors (http://www.licr.org/SEREX). For example, antibodies against ROCK1 have been found in patients with breast cancer, renal cancer, and fibrosarcoma, and antibodies to SFRS family proteins have been identified in patients with colorectal and ovarian cancer. These observations suggest that at least some of the antigens targeted after DLI may be immunogenic in other cancers and might be appropriate targets for immunotherapy in other malignancies.32,36
To develop a better understanding of this diverse set of antigens, we began to examine the expression of several of the antigens that were recognized by more than 1 DLI responder. Of the antigens identified in our panel, BCMA, a member of the TNF-receptor family, is the only antigen in our panel that is expressed on the cell membrane.34,37 The expression of BCMA is known to be restricted to the B-cell lineage and is preferentially expressed by plasma cells.38,39 However, all the other known genes are expressed in nonhematopoietic tissues and other hematopoietic cells. For example, ROCK-1 was highly expressed in a variety of myeloid and lymphoid leukemia cell lines and in lymphoma and myeloma cell lines. ROCK-1 kinase is a downstream effector of Rho, a GTPase of the Ras superfamily.40,41 The Rho pathway is known to regulate actin cytoskeleton function and cell proliferation.42 ROCK-1 activation contributes to the stabilization of filamentous actin, myosin ATPase activity and to the development of apoptotic morphology.43 Several recent reports have suggested that Rho GTPases and ROCK-1 are highly expressed in various tumors and that they play a role in tumor invasion and metastasis.44,45 Our studies demonstrated that ROCK-1 has highly variable expression in primary myeloma tumors, but the average level of expression was not significantly higher than in normal plasma cells.
Dihydrolipoamide acetyltransferase (DHA) is the E2 component of the multienzyme pyruvate dehydrogenate complex (PDC), which plays an important role in carbohydrate metabolism. DHA is known to be the most common antigen recognized by autoantibodies in patients with primary biliary cirrhosis (PBC). Several studies have identified the epitope of human PDC-E2 that is targeted by autoantibodies in PBC.46-48 Moreover, recent studies have also identified an HLA-A2–restricted peptide epitope of DHA.49 This peptide (aa 159-167 of PDC-E2) induced specific major histocompatibility complex (MHC) class 1–restricted CD8+ CTL lines from 10 of 12 HLA-A2 patients with PBC. Thus far, we have identified 2 myeloma and 1 CML DLI responders who developed strong antibody responses specific for DHA. These patients did not have any evidence of chronic liver disease. Moreover, antibodies to DHA were not present in serum from 20 patients with chronic GVHD or 5 patients with acute GVHD, in whom hepatic toxicity is a common feature. Northern blots demonstrated that the DHA gene was highly expressed in 3 of 4 myeloma cell lines but was not expressed in other hematopoietic tumor cell lines. DHA was also expressed in normal bone marrow and peripheral blood, but expression of this gene in primary myeloma tumor cells was significantly greater than in normal plasma cells (P = .02).
Two of 9 DLI responders and 1 of 20 patients with chronic GVHD developed antibodies to Homer-3 protein. Through the differential expression of various isoforms, 3 members of the Homer gene family encode a large number of related proteins. The function of Homer proteins has been studied extensively in neural cells, where they are expressed at high levels in postsynaptic densities. In neural cells, Homer proteins form complexes with metabotropic glutamate receptors (mGluR) and regulate their coupling to ion channels and intracellular Ca2+ pools.50-52 Homer proteins thus regulate synaptic signaling in neural cells,53 but the expression of these proteins in hematopoietic cells has not previously been demonstrated, and their function in other cell types has not been studied. In our experiments, Northern blots demonstrated high-level expression of Homer-3 in all 4 myeloma cell lines and low-level expression in Daudi, a Burkitt lymphoma cell line. Homer-3 was not expressed in myeloid cell lines or primary AML but was expressed in normal bone marrow and peripheral blood. Microarrays demonstrated a significantly higher expression in primary myeloma cells compared with normal plasma cells (P = .02). The expression of this gene in myeloma and in normal plasma cells suggests that the Homer proteins may also have important functions in B lymphocytes and possibly other hematopoietic cells.
In summary, our studies demonstrate that patients with myeloma who respond to the infusion of allogeneic donor cells develop a sustained high-titer antibody response to a variety of myeloma-associated antigens. Antibodies to this panel of antigen targets are rarely found in other patients undergoing allogeneic BMT or in healthy donors. Although these antigens have diverse cellular functions, the most immunogenic of these antigens appear to have particularly high levels of expression in myeloma tumor cells. In most instances, the expression of these genes in myeloma cells is significantly higher than in normal plasma cells. As a group, these novel antigens may be appropriate targets for further immunologic interventions to enhance the development and effectiveness of tumor immunity in patients with myeloma.
Prepublished online as Blood First Edition Paper, October 16, 2003; DOI 10.1182/blood-2003-07-2559.
Supported by National Institutes of Health grants CA078378 and AI29530 and the Ted and Eileen Pasquarello Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 1. Serial time points analysis of ROCK-1. Assessment of antibody reactivity with ROCK-1 determined by phage-plate assay in serial samples from patient 2 (early responder [ER]) and patient 4 (late responder [LR]). Quantification was based on the intensity of reactivity of the positive plaques.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2559/6/m_h80245539001.jpeg?Expires=1769149862&Signature=wvwY~amDKy4k97wlV9Y9OVEz4yG63YD2JjjWHeq~QoyQEjg4AfUdsDBl6nfpJGJLUEMWSHUu~4I5EfiSZ0Fd4bP8sYzrWlxHCje3koseJyn6~Q1Ck3xVYtBeinWb-347YhR7sxfKYW8h8ea4C6XvAAaxn5VuJcgq1nCwVVJitiFYm1WAVVkV2GVVxoRLI6b2Q30TU2ShTS5hDDy7y9MrX3RVHj7j5Jhpb3K3H75KdyWrBsck~UzYSfjN52SWk3dDPF~GRql9hAgsSu8-8Zff3Cy93aEzBPIsRxmZN6Cy~PLNjt-6L9PZsvoXOcz65Fp-ltBzUPcOZ080lnEXNSWuaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal