Abstract
In a series of 153 children with T-cell malignancies enrolled in 2 consecutive European Organization for Research and Treatment of Cancer (EORTC) trials, we assessed the HOX11L2 expression and/or the presence of a t(5;14)(q35;q32). Additionally, in 138 of these patients, HOX11 expression and SIL-TAL rearrangement were also assessed. These alterations were mutually exclusive, and their frequency was 23% (n = 35), 7% (n = 10), and 12% (n = 17), respectively. HOX11L2/t(5;14) positivity was more frequent in acute lymphoblastic leukemia (ALL) with cortical T immunophenotype and in children aged between 6 and 9 years. In contrast with previously reported data, patients positive and negative for HOX11L2/t(5;14) were comparable with regard to clinical outcome as well as to the response to a 7-day prephase treatment or to residual disease at completion of induction therapy. The 3-year event-free survival (EFS) rate (± SE percentage) for patients positive and negative for HOX11L2/t(5;14) was 75.5% (± 8.1%) and 68.3% (± 5.0%), respectively; the hazard ratio was 0.84 (95% confidence interval, 0.40-1.80). Patients with HOX11-high expression and those with SIL-TAL fusion had low levels of residual disease at the end of induction and a favorable prognosis: the 3-year EFS rate was 83.3% (± 8.5%) and 75.3% (± 12.6%), respectively. The results obtained in HOX11L2/t(5;14) patients in this study do not confirm the unfavorable prognosis reported in previous studies.
Introduction
Childhood T-cell malignancies include T-cell acute lymphoblastic leukemia (T-ALL), which accounts for approximately 15% of ALL, and T-cell lymphoblastic lymphoma, which represents about one third of non-Hodgkin lymphoma. As compared with precursor B-cell ALL, T-cell malignancies are often associated with unfavorable features. The use of more intensive treatments and risk-adapted therapy have significantly improved the outcome of patients with T-ALL as well as of patients with T-cell lymphoblastic lymphoma, and event-free survival rates of 60% to 70% are now reported in children.1,2 Whether classical risk factors such as age or white blood cell (WBC) count are relevant in patients with T-ALL is a matter of debate. A study of the Pediatric Oncology Group suggested that the prognostic significance of initial features such as age, leukocyte count, involvement of central nervous system (CNS) is reduced in T-lineage ALL as compared with B-lineage ALL.3 Some genetic abnormalities present in the leukemic clone of B-lineage ALL are of strong prognostic importance. However, there are only few data concerning the possible prognostic significance of genetic abnormalities in T-lineage malignancies.
Some nonrandom translocations that are specific to T-lineage malignancies have been identified. They involve genes that code for transcriptional regulators and are transcriptionally deregulated in malignancies.
Alteration of the TAL-1 (SCL; TCL-5) locus located on chromosome 1p32 is considered as the most common nonrandom genetic defect in childhood T-ALL. In 1% to 3% of childhood T-ALL, TAL-1 disruption is associated with a t(1;14)(p33;q11).4,5 In another 9% to 26% of T-ALL, TAL-1 defect occurs via a nonrandom submicroscopic interstitial deletion between a locus called SIL and the 5′ untranslated region (UTR) of TAL-1, giving rise to an SIL-TAL fusion transcipt.6,7 Both alterations disrupt the coding potential of TAL-1 in a similar manner, leading to its ectopic expression in T cells.8,9
Two other translocations, t(10;14)(q24;q11) and its variant t(7; 10)(q35;q24), were identified in T-ALL. Either of these is present in about 5% of cases. They both lead to the transcriptional activation of an homeobox gene HOX11 (TLX1; TCL3) which is not expressed in healthy T cells,10,11 by bringing the HOX11 coding sequence under the transcriptional control of regulatory sequences of the T-cell receptor gene.10,12-14 Overexpression of HOX11 has also been demonstrated in the absence of a 10q24 rearrangement, suggesting that other mechanisms can lead to this aberrant gene expression.11,15
Recently, a new cryptic translocation, t(5;14)(q35;q32), was found in T-ALL by fluorescence in situ hybridization (FISH).16,17 This translocation leads to the ectopic expression of another homeobox gene, HOX11L2 (RNX; TLX3), possibly by bringing it under the influence of regulatory elements of CTIP2, a gene highly expressed during T-lymphoid differentiation.16 Ectopic expression of HOX11L2 has also been found in a subset of T-ALL by quantitative reverse transcription–polymerase chain reaction (RT-PCR).15 HOX11 and HOX11L2 are called orphan homeobox genes because they are located outside the 4 mammalian HOX clusters. They are members of a class of homeobox genes that also includes the mammalian HOX11L1 (ENX; TLX2) gene.18 Preliminary studies indicate that t(5;14) is restricted to T-lineage ALL and is more frequent in children than in adults. It could represent a frequent specific genetic alteration in childhood T-ALL.19,20 Moreover, t(5;14) and/or HOX11L2 ectopic expression have been reported as being associated with a very poor outcome in children with T-ALL.15,19
The aim of our study was to retrospectively evaluate the frequency and prognostic value of the newly described t(5;14) and/or HOX11L2 expression in childhood T-cell malignancies. For this purpose, we studied blastic samples from 153 children with newly diagnosed T-cell malignancies enrolled in 2 consecutive European Organization for Research and Treatment of Cancer Children's Leukemia Group (EORTC-CLG) ALL studies. We also reassessed the frequency of 2 additional alterations specifically found in T-ALL which have not been extensively investigated in children, HOX11 overexpression and SIL-TAL fusion, as well as their possible association with a distinct leukemic phenotype and/or outcome.
Patients and methods
Patients
A total of 153 children, aged 3 months to 17 years, with newly diagnosed T-cell malignancies (141 with T-ALL and 12 with T-cell lymphoblastic lymphoma) were included in the study. Patients were selected retrospectively on the basis of the availability of frozen material at diagnosis and on the successful assessment of the presence or absence of either HOX11L2 expression by RT-PCR and/or t(5;14) by FISH. Informed consent was provided according to the Declaration of Helsinki. All patients were treated according to the protocols of the EORTC 58881 (75 patients) or 58951 (78 patients), between August 1989 and September 2002.21 These protocols have been accepted by the EORTC Protocol Review Committee and by the Ethics Committee of each participating center. Main characteristics of the patients studied are described in Table 1. The median follow-up was 3.1 years. Eleven of these children were previously included in the study of Mauvieux et al.20
Characteristics of patients with T-malignancies according to genetic subgroups
. | Total population studied . | Subgroup . | . | . | . | All HOX11L2 negative† . | |||
|---|---|---|---|---|---|---|---|---|---|
. | . | HOX11L2 positive . | HOX11-high . | SIL-TAL . | Other* . | . | |||
| Total, N (%) | 138 (100) | 35 (100) | 10 (100) | 17 (100) | 76 (100) | 118 (100) | |||
| Disease, n | |||||||||
| ALL | 127 (92.0) | 34 (97.1) | 9 (90) | 15 (88.2) | 69 (90.8) | 107 (90.7) | |||
| Lymphoblastic lymphoma | 11 (8.0) | 1 (2.9) | 1 (10) | 2 (11.8) | 7 (9.2) | 11 (9.3) | |||
| Sex, n | |||||||||
| Male | 102 (73.9) | 28 (80) | 9 (90) | 13 (76.5) | 52 (68.4) | 87 (73.7) | |||
| Female | 36 (26.1) | 7 (21) | 1 (10) | 4 (23.5) | 24 (31.5) | 31 (26.3) | |||
| Age at diagnosis, y | |||||||||
| Median | 8 | 6 | 5 | 11 | 8.5 | 8 | |||
| Range | 0-17 | 3-13 | 0-14 | 1-14 | 1-17 | 1-17 | |||
| WBC count | |||||||||
| Less than 50 × 109/L, n | 69 (50.0) | 22 (62.9) | 6 (60) | 4 (23.5) | 37 (48.7) | 54 (45.8) | |||
| 50-99 × 109/L, n | 18 (13.0) | 5 (14.3) | 0 (0) | 2 (11.8) | 11 (14.5) | 19 (16.1) | |||
| At least 100 × 109/L or greater, n | 51 (37.0) | 8 (22.9) | 4 (40) | 11 (64.7) | 28 (36.8) | 45 (38.1) | |||
| Median, × 109/L | 49 | 34 | 29 | 124 | 51 | 58 | |||
| Range, × 109/L | 1.9-999.8 | 3.4-288 | 9-346 | 3.6-646 | 1.9-998 | 1.9-998 | |||
| Mediastinal involvement, n | |||||||||
| No | 14 (10.1) | 4 (11.4) | 0 (0) | 3 (17.6) | 7 (9.2) | 15 (12.7) | |||
| Yes | 124 (89.9) | 31 (88.6) | 10 (100) | 14 (82.4) | 69 (90.8) | 103 (87.3) | |||
| Immunophenotype, n‡ | |||||||||
| T-lineage (no further specified) | 38 (27.5) | 11 (31.4%) | 1 (10) | 5 (29.4) | 21 (27.6) | 36 (30.5) | |||
| Early-T | 24 [24.0] | 2 [8.3] | 1 [11.1] | 3 [25.0] | 18 [32.7] | 23 [28.0] | |||
| Cortical-T | 44 [44.0] | 16 [66.7] | 6 [66.7] | 5 [41.7] | 17 [30.9] | 32 [39.0] | |||
| Mature-T | 32 [32.0] | 6 [25.0] | 2 [22.2] | 4 [33.3] | 20 [36.4] | 27 [32.9] | |||
| Cytogenetics in ALL patients, n‡ | |||||||||
| Not done or failure | 16 (12.6) | 1 (2.9) | 0 (0.0) | 2 (13.3) | 13 (18.8) | 16 (15.0) | |||
| Normal | 48 [43.2] | 21 [63.6] | 5 [55.6] | 4 [30.8] | 18 [32.1] | 33 [36.3] | |||
| Abnormal | 63 [56.8] | 12 [36.4] | 4 [44.4] | 9 [69.2] | 38 [67.9] | 58 [63.7] | |||
. | Total population studied . | Subgroup . | . | . | . | All HOX11L2 negative† . | |||
|---|---|---|---|---|---|---|---|---|---|
. | . | HOX11L2 positive . | HOX11-high . | SIL-TAL . | Other* . | . | |||
| Total, N (%) | 138 (100) | 35 (100) | 10 (100) | 17 (100) | 76 (100) | 118 (100) | |||
| Disease, n | |||||||||
| ALL | 127 (92.0) | 34 (97.1) | 9 (90) | 15 (88.2) | 69 (90.8) | 107 (90.7) | |||
| Lymphoblastic lymphoma | 11 (8.0) | 1 (2.9) | 1 (10) | 2 (11.8) | 7 (9.2) | 11 (9.3) | |||
| Sex, n | |||||||||
| Male | 102 (73.9) | 28 (80) | 9 (90) | 13 (76.5) | 52 (68.4) | 87 (73.7) | |||
| Female | 36 (26.1) | 7 (21) | 1 (10) | 4 (23.5) | 24 (31.5) | 31 (26.3) | |||
| Age at diagnosis, y | |||||||||
| Median | 8 | 6 | 5 | 11 | 8.5 | 8 | |||
| Range | 0-17 | 3-13 | 0-14 | 1-14 | 1-17 | 1-17 | |||
| WBC count | |||||||||
| Less than 50 × 109/L, n | 69 (50.0) | 22 (62.9) | 6 (60) | 4 (23.5) | 37 (48.7) | 54 (45.8) | |||
| 50-99 × 109/L, n | 18 (13.0) | 5 (14.3) | 0 (0) | 2 (11.8) | 11 (14.5) | 19 (16.1) | |||
| At least 100 × 109/L or greater, n | 51 (37.0) | 8 (22.9) | 4 (40) | 11 (64.7) | 28 (36.8) | 45 (38.1) | |||
| Median, × 109/L | 49 | 34 | 29 | 124 | 51 | 58 | |||
| Range, × 109/L | 1.9-999.8 | 3.4-288 | 9-346 | 3.6-646 | 1.9-998 | 1.9-998 | |||
| Mediastinal involvement, n | |||||||||
| No | 14 (10.1) | 4 (11.4) | 0 (0) | 3 (17.6) | 7 (9.2) | 15 (12.7) | |||
| Yes | 124 (89.9) | 31 (88.6) | 10 (100) | 14 (82.4) | 69 (90.8) | 103 (87.3) | |||
| Immunophenotype, n‡ | |||||||||
| T-lineage (no further specified) | 38 (27.5) | 11 (31.4%) | 1 (10) | 5 (29.4) | 21 (27.6) | 36 (30.5) | |||
| Early-T | 24 [24.0] | 2 [8.3] | 1 [11.1] | 3 [25.0] | 18 [32.7] | 23 [28.0] | |||
| Cortical-T | 44 [44.0] | 16 [66.7] | 6 [66.7] | 5 [41.7] | 17 [30.9] | 32 [39.0] | |||
| Mature-T | 32 [32.0] | 6 [25.0] | 2 [22.2] | 4 [33.3] | 20 [36.4] | 27 [32.9] | |||
| Cytogenetics in ALL patients, n‡ | |||||||||
| Not done or failure | 16 (12.6) | 1 (2.9) | 0 (0.0) | 2 (13.3) | 13 (18.8) | 16 (15.0) | |||
| Normal | 48 [43.2] | 21 [63.6] | 5 [55.6] | 4 [30.8] | 18 [32.1] | 33 [36.3] | |||
| Abnormal | 63 [56.8] | 12 [36.4] | 4 [44.4] | 9 [69.2] | 38 [67.9] | 58 [63.7] | |||
Numbers in parentheses and brackets are percentages.
Negative for 3 genetic markers.
This group included 118 patients: 10 patients with HOX11-high, 17 patients positive for SIL-TAL, 76 patients with other, and 15 additional patients who tested negative for HOX11L2 and not tested for HOX11 and SIL-TAL status.
Percentages between [] were calculated taking into account only documented cases
T-cell lineage was defined by the presence of the T-cell antigen CD3 either on the cell surface (sCD3) or in the cytoplasm (cCD3). At least 2 of the B-cell markers tested (CD19, s/cCD22, cCD79a) and at least 3 of the myeloid markers tested (CD13, CD33, CD117, myeloperoxidase [MPO]) had to be negative. Subclassification of T-cell lineage was performed as follows. T-cell malignancies positive for cCD3, CD7, ±CD2, ±CD5, ±CD8 and negative for sCD3 and CD1a were considered “early-T.” When positive for CD1a, T-cell malignancies were considered “cortical-T,” and when positive for CD3 but negative for CD1a they were considered “mature-T.”
Karyotyping of malignant clone at diagnosis was centrally reviewed. For patients with T-ALL, the hematologic status after the prephase (one intrathecal methotrexate injection on day 1 and 7 days of glucocorticoid treatment), conventionally called “response” to the prephase, was assessed on day 8 by the number of leukemic blasts: patients with more than 1000 blasts/mm3 of blood after completion of this treatment were considered poor responders and classified in a very high risk (VHR) group. Minimal residual disease (MRD) was assessed at day 35, after completion of a 1-month induction therapy, in 59 patients. The technique used for MRD monitoring was based on the quantification by competitive PCR of T-cell delta and gamma receptor gene rearrangements. After PCR, specific rearrangements were detected either by clono-specific hybridization22 or by fluorescent genescan23 for more recent patients. Results were expressed as a ratio of leukemia cells in mononucleated marrow cells. Patients with T-cell lymphoblastic lymphoma received the same treatment but were not assessed for response to the prephase or for MRD.
Detection of the t(5;14)(q35;q32) by FISH
All samples analyzed by FISH were collected from bone marrow (BM) or lymph node samples with major blastic involvement. FISH analyses for detection of the t(5;14) were carried out on cytogenetic pellets and interpreted as positive when metaphases with t(5;14) were present.
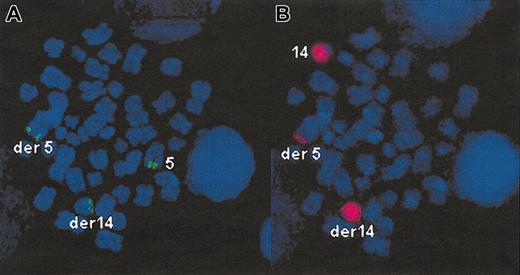
The t(5;14)(q35;q32) was detected by using the combination of signals obtained with a chromosome 14 painting (wcp14) and with the 885A6 yeast artificial chromosome (YAC) assigned at 5q35. The rhodamine direct-labeled wcp14 was purchased from Oncor (CP5614-RW; Appligene Oncor, Gaithersburg, MD) and used according to manufacturer's instructions. The 885A6 YAC was obtained from the CEPH library (Fondation Jean Dausset-CEPH, Paris, France). The YAC and the wcp14 were either cohybridized or sequentially hybridized on the same slide, and hybridizations were carried out according to methods previously described.24 Metaphases with a t(5;14) were characterized by a splitting of one YAC signal, one part translocating onto the telomeric end of a chromosome 14 (green signals in Figure 1). The wcp14 (red signal in Figure 1) pointed at the derivative 14 as well as at the derivative 5, characterized by a small translocated fragment from chromosome 14 at its telomeric part.
FISH detection of t(5;14)(q35;q32). Sequential hybridization of the 885A6 YAC (A), then of wcp14 (B) on the same metaphase. (A) The normal chromosome 55 and the derivative 5 (der 5) are identified by green signals (885A6 YAC) at 5q35. (B) The derivative 5 is characterized by a small red signal (wcp14) at 5q35, corresponding to the translocated fragment from chromosome 14. (A) The derivative 14 (der 14) is identified by a green signal at its telomeric end (14q32), corresponding to the translocated part of the YAC. (B) The derivative 14 and the normal chromosome 14 are identified by wcp14 (red signal). Original magnification, × 100.
FISH detection of t(5;14)(q35;q32). Sequential hybridization of the 885A6 YAC (A), then of wcp14 (B) on the same metaphase. (A) The normal chromosome 55 and the derivative 5 (der 5) are identified by green signals (885A6 YAC) at 5q35. (B) The derivative 5 is characterized by a small red signal (wcp14) at 5q35, corresponding to the translocated fragment from chromosome 14. (A) The derivative 14 (der 14) is identified by a green signal at its telomeric end (14q32), corresponding to the translocated part of the YAC. (B) The derivative 14 and the normal chromosome 14 are identified by wcp14 (red signal). Original magnification, × 100.
Sample preparation for RNA analysis
For each patient, a sample of malignant cells was collected on EDTA (ethylenediaminetetraacetic acid) at diagnosis, according to the EORTC-CLG protocol. It consisted of bone marrow for patients with ALL and of pleural liquid or lymph node punctuate or bone marrow (in stage IV) for patients with lymphoma. In case of bone marrow sampling, mononuclear cells were separated by Ficoll centrifugation, counted, and stored at –80°C. The percentage of blasts after Ficoll separation was evaluated on a cytospin. Total RNA was extracted using RNA Plus (Appligene, Illkirch, France), according to the instructions of the manufacturer. Determination of the quality and approximate concentration of total RNA was performed by electrophoresis on a 0.8% agarose gel stained with ethidium bromide.
SIL-TALRT-PCR
HOX11andHOX11L2real-time PCR assays
Because HOX11 and HOX11L2 are normal transcripts, they are potentially expressed by some subsets of healthy cells present in the blastic samples. Thus, we used quantitative real-time RT-PCR assays to reliably discriminate true overexpression from background. RT-PCR products were detected by the fluorescent TaqMan methodology. The transcript coding for the TATA box-binding protein (TBP) was studied in parallel and used as the endogenous RNA control.
TBP, HOX11L2, and HOX11 were amplified by using primers TBP-HS1/TBP-HS2 with probe TBP,28 primers RQ-HOX11L2-3/RQ-HOX11L2-4 with probe HOX11L2,19 and primers HOX11-S/HOX11-AS with probe HOX11 (Asnafi et al, unpublished communication, March 2002), respectively. Reverse-transcription product (2 μL) was added to the PCR reaction mixture containing 1X TaqMan Buffer (Invitrogen, Cergy Pontoise, France), 5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate (dNTP), 300 nM of each primer, 150 nM probe, and 1.25 U AmpliTaq Gold (Applied Biosystems, Courtaboeuf, France) in a total reaction volume of 50 μL. PCR was performed on the ABI PRISM 7700 Sequence detector system (Applied Biosystems). After 10 minutes at 95°C, 50 cycles (15 seconds at 95°C and 15 seconds at 65°C) were performed. All experiments were done in duplicate. The threshold cycle (Ct), defined as the fractional cycle number at which the signal reached a preset threshold, was measured in each sample for the target and control (TBP) cDNA.
Samples displaying a TBP Ct (CtTBP) higher than 30 were considered as degraded and were discarded. When good quality RNA was available, results obtained for HOX11 and HOX11L2 transcripts were compared with those obtained for TBP transcripts, which were used as a reference, by calculating the delta Ct (ΔCtHOX11L2-TBP and ΔCtHOX11-TBP) for each sample.
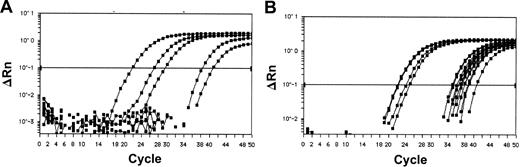
When present, HOX11L2 expression was unambiguous in virtually all cases, with expression levels equal or higher than those obtained for TBP (ΔCtHOX11L2-TBP ranging from –5.5 to 0.6; mean, –1.1; standard deviation, 2.6). A weak expression of HOX11L2, with ΔCtHOX11L2-TBP more than 15, was rarely seen, and these cases were considered as negative (Figure 2). In one case, an intermediate ΔCtHOX11L2-TBP value of 4 was obtained. This case was positive for t(5;14), and blast content was more than 80%. The relatively low HOX11L2 expression observed in this case might be due to the translocation being restricted to a subclone of leukemia cells.
Amplification of HOX11L2 and HOX11 transcripts by real-time RT-PCR. Logarithmic representation of amplification plots obtained for patient samples are shown for HOX11L2 (A) and HOX11 (B). The normalized fluorescence signal (ΔRn) is plotted on the y-axis and the cycle number on the x-axis. Real-time RT-PCR permits to clearly distinguish patients with high HOX11L2 or HOX11 expression (low Ct), from those with low expression (high Ct) which were grouped with those negative for the statistical analysis.
Amplification of HOX11L2 and HOX11 transcripts by real-time RT-PCR. Logarithmic representation of amplification plots obtained for patient samples are shown for HOX11L2 (A) and HOX11 (B). The normalized fluorescence signal (ΔRn) is plotted on the y-axis and the cycle number on the x-axis. Real-time RT-PCR permits to clearly distinguish patients with high HOX11L2 or HOX11 expression (low Ct), from those with low expression (high Ct) which were grouped with those negative for the statistical analysis.
Almost one third of samples expressed very low levels of HOX11 transcript (Figure 2), with ΔCtHOX11-TBP values lower than 15. These samples were grouped with negative ones for further analyses. Another smaller group of samples expressed high levels of HOX11 with ΔCtHOX11-TBP ranging from –5.2 to 5 (mean, –2.12; standard deviation, 3.8). These samples were termed “HOX11-high.”
Statistical analysis
Event-free survival was calculated from the date of complete remission to the date of first relapse or death. For patients who failed to reach complete remission by the end of induction-consolidation, the failure was considered as an event at time 0. All patients alive and in first continuous remission were censored at their last follow-up. The duration of survival was calculated from the date of start of treatment until the date of death; patients still alive were censored at their last follow-up.
Actuarial curves were calculated according to the Kaplan-Meier technique.29 The curves have been cut at 8 years because too few patients remained at risk beyond that time point; one relapse occurring at 9 years has, therefore, not been displayed. The standard errors (SEs) of the estimates were computed by using the Greenwood formula.29 The differences between actuarial curves were tested for statistical significance by using the 2-tailed log-rank test factor. The Cox proportional hazards model has been used to obtain the estimate and the 95% confidence interval of the hazard ratio (HR) of the instantaneous event rate in one group versus the one in another group, as specified by a given variable. The Wald test has been used to determine the prognostic significance.29 The Cox model has also been used to determine the independent prognostic importance of several variables. All analyses were based on the intent-to-treat principle.
The relationship between 2 or more genetic groups and a continuous variable (eg, age, WBC count) has been tested by using the Kruskal-Wallis test. Because of the limited number of patients included, P values should be interpreted with caution.
The cutoff date was April 2003. The SAS 8.2 statistical software (SAS Institute, Cary, NC) has been used.
Results
t(5;14)(q35;q32) and HOX11L2 expression
HOX11L2 expression and/or the presence of a t(5;14) were investigated in 153 patients with T-cell malignancies. Expression of HOX11L2 was measured by real-time RT-PCR in 135 patients. The search of the t(5;14)(q35;q32) was done by FISH analysis in 75 patients. Of 153 patients, 35 (22.9%) displayed a t(5;14) and/or expressed HOX11L2. Fifty-seven patients were tested by both techniques: 38 were negative using the 2 techniques, 16 were positive for HOX11L2 expression and t(5;14), and only 3 (5%) patients showed HOX11L2 expression without evidence of t(5;14).
Frequency of genetic alterations in T-ALL and T-lymphoblastic lymphoma
Expression of HOX11L2 and/or t(5;14) was found in 34 (24.1%) of 141 patients with T-ALL and in one (8.3%) of 12 patients with T-lymphoblastic lymphoma. Of the 153 patients tested for HOX11L2 expression and/or t(5;14), 138 could also be studied for the presence of HOX11 expression or of SIL-TAL fusion. Of the 127 patients with T-ALL, 9 (7.1%) displayed high HOX11 expression and 15 (11.8%) SIL-TAL fusion. Of the 11 patients with lymphoblastic lymphoma, 1 (9.1%) displayed high HOX11 expression and 2 (18.2%) SIL-TAL fusion. In this group of 138 patients, a combination of 2 or 3 of these abnormalities was never observed.
Characteristics of children with T-cell malignancies according to genetic subgroups
Among the 138 patients in whom all 3 genetic alterations could be investigated, 4 subgroups were distinguished: the HOX11L2 group contained patients displaying either a t(5;14) and/or HOX11L2 expression (n = 35), the HOX11-high group contained those patients exhibiting high HOX11 expression (n = 10), the SIL-TAL group contained those patients who were positive for the SIL-TAL fusion (n = 17), and the group “other” contained those patients displaying none of these abnormalities (n = 76). In addition, a fifth group containing all patients with HOX11L2-negative expression, irrespective of their HOX11 or SIL-TAL status, has been analyzed (n = 118).
Characteristics of the patients according to genetic subgroups are described in Table 1. No significant difference in sex ratio or of incidence of mediastinal involvement was observed between these genetic subgroups. The median ages were 6, 5, 11, and 8.5 years in these 4 groups, respectively, which is not significantly different (Kruskal-Wallis P = .13) (Table 1). However, the age distribution was slightly different between the 4 groups (Figure 3). The age of patients with a HOX11L2-positive ALL displayed a symmetric distribution, 48.6% of them being aged 6 to 9 years at diagnosis, whereas in the HOX11-high group, no patient was present in this age category. The frequency of SIL-TAL–positive T-cell malignancies increased regularly with age to reach 53% in children older than 10 years at diagnosis.
Histograms showing the distribution of patients' age according to the 4 genetic subgroups. (A) HOX11L2. (B) HOX11-high. (C) SIL-TAL. (D) Other.
Histograms showing the distribution of patients' age according to the 4 genetic subgroups. (A) HOX11L2. (B) HOX11-high. (C) SIL-TAL. (D) Other.
WBC count at diagnosis was higher (Kruskal-Wallis P = .12) for patients of the SIL-TAL group as compared with the HOX11L2, HOX11-high, and other groups of patients: the medians were 124, 34, 29, and 51 × 109/L, respectively.
Detailed immunophenotyping was available in 100 of 138 patients: 24 were categorized as early, 44 as cortical, and 32 as mature-T lineage. Even though HOX11-high and HOX11L2 malignancies were predominantly associated with a cortical-T phenotype, phenotype was rather heterogeneous in the HOX11L2 group with 25% of the cases displaying a mature-T phenotype. Patients with a SIL-TAL fusion had an immunophenotyping distribution very similar to that of the entire group. Overall, there was no association between the genetic subgroup and immunophenotype maturation (P = .95).
Cytogenetic evaluation was successful in 111 of 127 patients with ALL, the karyotype failure/unknown rate ranged from 0% in patients with HOX11-high to 18.8% in the other group. Forty-eight (43.2%) of these 111 patients had a normal karyotype when using conventional cytogenetic techniques. Thirty (62.5%) of these 48 patients with a normal karyotype had a genetic abnormality: 21 (43.8%) had a HOX11L2, 5 (10.4%) HOX11, and 4 (8.3%) a SIL-TAL alteration. In most of the HOX11L2 and HOX11-high malignancies, no cytogenetic alteration was found, whereas SIL-TAL fusion was more often associated with additional cytogenetic alterations (Kruskal-Wallis P = .02). Surprisingly, none of the 9 cases with a high HOX11 expression and a successful karyotype displayed a t(10;14) or a 10q24 rearrangement.
Prognosis associated with t(5;14) and/or HOX11L2 expression
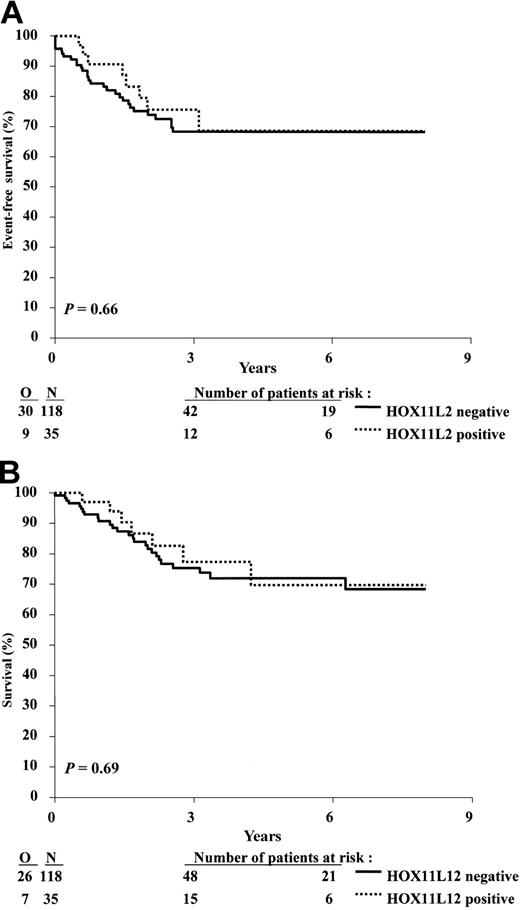
The group of 153 patients included in the study had overall 3-year event-free survival (EFS) and survival rates of 70.0% and 75.8%, respectively. These outcome data are similar to those observed for the whole population of patients with T-cell malignancies included in the 58881 and 58951 trials during the same period of time. The clinical outcome of patients from the HOX11L2 group was similar to that of the patients who did not display this abnormality (Figure 4A-B): the 3-year EFS (± SE) was 75.5% (± 8.1%) and 68.3% (± 5.0%), respectively, and the 3-year overall survival was 77.3% (± 8.4%) and 75.3% (± 4.6%), respectively. The estimated hazard ratios for these 2 analyses are close to 1, and the upper 95% confidence intervals are lower than 2, indicating that the patients positive for HOX11L2 have not a clearly worse outcome than patients negative for HOX11L2. The pattern of events (no CR, type of event) was quite similar in the 2 groups (Tables 2, 3, 4).
Kaplan-Meier estimates of event-free survival, and survival according to the presence or absence of t(5;14) and/or HOX11L2 expression. (A) Event-free survival. (B) Survival. N indicates number of patients; O, observed number of events. P value was given by the log-rank test.
Kaplan-Meier estimates of event-free survival, and survival according to the presence or absence of t(5;14) and/or HOX11L2 expression. (A) Event-free survival. (B) Survival. N indicates number of patients; O, observed number of events. P value was given by the log-rank test.
In vivo response to chemotherapy, distribution of types of event and of survival status of children with T-ALL according to genetic subgroups
. | Total population studied, n . | Subgroup . | . | . | . | All HOX11L2-negative, n† . | |||
|---|---|---|---|---|---|---|---|---|---|
. | . | HOX11L2-positive, n . | HOX11-high, n . | SIL-TAL, n . | Other, n* . | . | |||
| Total, N (%) | 127 (100) | 34 (100) | 9 (100) | 15 (100) | 69 (100) | 117 (100) | |||
| Response to prephase | |||||||||
| Died before evaluation | 1 (0.8) | 0 (0) | 0 (0) | 1 (6.7) | 0 (0) | 1 (0.9) | |||
| Less than 1000 blasts/μL | 79 (62.2) | 24 (70.6) | 9 (100) | 7 (46.7) | 39 (56.5) | 67 (62.6) | |||
| At least 1000 blasts/μL | 47 (37.0) | 10 (29.4) | 0 (0) | 7 (46.7) | 30 (43.5) | 39 (36.4) | |||
| MRD‡ | |||||||||
| ND | 59 (46.5) | 15 (44.1) | 4 (44.4) | 8 (53.3) | 32 (46.4) | 55 (51.4) | |||
| Absence of marker | 12 (9.4) | 1 (2.9) | 1 (11.1) | 0 (0.0) | 10 (14.5) | 11 (10.3) | |||
| Less than 10-2 | 46 [82.1] | 13 [72.2] | 4 [100] | 7 [100] | 22 [81.5] | 35 [85.4] | |||
| At least 10-2 | 10 [17.9] | 5 [27.8] | 0 [0] | 0 [0] | 5 [18.5] | 6 [14.6] | |||
| Type of event | |||||||||
| No event (ie, in CCR) | 94 (74.0) | 25 (73.5) | 8 (88.9) | 12 (80.0) | 49 (71.0) | 25 (73.5) | |||
| No CR | 4 (3.1) | 0 (0) | 0 (0) | 1 (6.7) | 3 (4.3) | 5 (4.7) | |||
| BM only | 12 (9.4) | 4 (11.8) | 0 (0) | 2 (13.3) | 6 (8.7) | 8 (7.5) | |||
| CNS only | 3 (2.4) | 0 (0) | 0 (0) | 0 (0) | 3 (4.3) | 7 (6.5) | |||
| Other isolated relapse | 1 (0.8) | 0 (0) | 0 (0) | 0 (0) | 1 (1.4) | 1 (0.9) | |||
| Combined | 9 (7.1) | 4 (11.8) | 1 (11.1) | 0 (0) | 4 (5.8) | 5 (4.7) | |||
| Death in CR | 4 (3.1) | 1 (2.9) | 0 (0) | 0 (0) | 3 (4.3) | 3 (2.8) | |||
| Survival status | |||||||||
| Alive | 99 (78.0) | 27 (79.4) | 9 (100) | 14 (93.3) | 49 (71.0) | 82 (76.6) | |||
| Dead | 28 (22.0) | 7 (20.6) | 0 (0) | 1 (6.7) | 20 (29.0) | 25 (23.4) | |||
. | Total population studied, n . | Subgroup . | . | . | . | All HOX11L2-negative, n† . | |||
|---|---|---|---|---|---|---|---|---|---|
. | . | HOX11L2-positive, n . | HOX11-high, n . | SIL-TAL, n . | Other, n* . | . | |||
| Total, N (%) | 127 (100) | 34 (100) | 9 (100) | 15 (100) | 69 (100) | 117 (100) | |||
| Response to prephase | |||||||||
| Died before evaluation | 1 (0.8) | 0 (0) | 0 (0) | 1 (6.7) | 0 (0) | 1 (0.9) | |||
| Less than 1000 blasts/μL | 79 (62.2) | 24 (70.6) | 9 (100) | 7 (46.7) | 39 (56.5) | 67 (62.6) | |||
| At least 1000 blasts/μL | 47 (37.0) | 10 (29.4) | 0 (0) | 7 (46.7) | 30 (43.5) | 39 (36.4) | |||
| MRD‡ | |||||||||
| ND | 59 (46.5) | 15 (44.1) | 4 (44.4) | 8 (53.3) | 32 (46.4) | 55 (51.4) | |||
| Absence of marker | 12 (9.4) | 1 (2.9) | 1 (11.1) | 0 (0.0) | 10 (14.5) | 11 (10.3) | |||
| Less than 10-2 | 46 [82.1] | 13 [72.2] | 4 [100] | 7 [100] | 22 [81.5] | 35 [85.4] | |||
| At least 10-2 | 10 [17.9] | 5 [27.8] | 0 [0] | 0 [0] | 5 [18.5] | 6 [14.6] | |||
| Type of event | |||||||||
| No event (ie, in CCR) | 94 (74.0) | 25 (73.5) | 8 (88.9) | 12 (80.0) | 49 (71.0) | 25 (73.5) | |||
| No CR | 4 (3.1) | 0 (0) | 0 (0) | 1 (6.7) | 3 (4.3) | 5 (4.7) | |||
| BM only | 12 (9.4) | 4 (11.8) | 0 (0) | 2 (13.3) | 6 (8.7) | 8 (7.5) | |||
| CNS only | 3 (2.4) | 0 (0) | 0 (0) | 0 (0) | 3 (4.3) | 7 (6.5) | |||
| Other isolated relapse | 1 (0.8) | 0 (0) | 0 (0) | 0 (0) | 1 (1.4) | 1 (0.9) | |||
| Combined | 9 (7.1) | 4 (11.8) | 1 (11.1) | 0 (0) | 4 (5.8) | 5 (4.7) | |||
| Death in CR | 4 (3.1) | 1 (2.9) | 0 (0) | 0 (0) | 3 (4.3) | 3 (2.8) | |||
| Survival status | |||||||||
| Alive | 99 (78.0) | 27 (79.4) | 9 (100) | 14 (93.3) | 49 (71.0) | 82 (76.6) | |||
| Dead | 28 (22.0) | 7 (20.6) | 0 (0) | 1 (6.7) | 20 (29.0) | 25 (23.4) | |||
Numbers in parentheses and brackets are percentages. CCR indicates continuous complete remission; BM, bone marrow relapse; CNS, central nervous system relapse.
Negative for 3 genetic markers.
This group included 107 patients: 9 patients with HOX11-high, 15 patients positive for SIL-TAL, 69 patients with other, and 14 additional patients who tested negative for HOX11L2 and not tested for HOX11 and SIL-TAL status.
Percentages between [] were calculated taking into account only documented cases.
Distribution of types of event and of survival status of children with T-lymphoblastic lymphoma according to genetic subgroups
. | Total population studied, n (%) . | Subgroup . | . | . | . | All HOX11L2-negative, n (%)† . | |||
|---|---|---|---|---|---|---|---|---|---|
. | . | HOX11L2-positive, n (%) . | HOX11-high, n (%) . | SIL-TAL, n (%) . | Other, n (%)* . | . | |||
| Total, N (%) | 11 (100) | 1 (100) | 1 (100) | 2 (100) | 7 (100) | 11 (100) | |||
| Type of event | |||||||||
| No event (ie, in CCR) | 10 (90.9) | 1 (100) | 1 (100) | 1 (100) | 6 (85.7) | 10 (90.9) | |||
| BM only | 1 (9.1) | 0 (0) | 0 (0) | 0 (0) | 1 (14.3) | 1 (9.1) | |||
| Survival status | |||||||||
| Alive | 10 (90.9) | 1 (100) | 1 (100) | 1 (100) | 6 (85.7) | 10 (90.9) | |||
| Dead | 1 (9.1) | 0 (0) | 0 (0) | 0 (0) | 1 (14.3) | 1 (9.1) | |||
. | Total population studied, n (%) . | Subgroup . | . | . | . | All HOX11L2-negative, n (%)† . | |||
|---|---|---|---|---|---|---|---|---|---|
. | . | HOX11L2-positive, n (%) . | HOX11-high, n (%) . | SIL-TAL, n (%) . | Other, n (%)* . | . | |||
| Total, N (%) | 11 (100) | 1 (100) | 1 (100) | 2 (100) | 7 (100) | 11 (100) | |||
| Type of event | |||||||||
| No event (ie, in CCR) | 10 (90.9) | 1 (100) | 1 (100) | 1 (100) | 6 (85.7) | 10 (90.9) | |||
| BM only | 1 (9.1) | 0 (0) | 0 (0) | 0 (0) | 1 (14.3) | 1 (9.1) | |||
| Survival status | |||||||||
| Alive | 10 (90.9) | 1 (100) | 1 (100) | 1 (100) | 6 (85.7) | 10 (90.9) | |||
| Dead | 1 (9.1) | 0 (0) | 0 (0) | 0 (0) | 1 (14.3) | 1 (9.1) | |||
Negative for 3 genetic markers.
This group included 11 patients: 1 patient with HOX11-high, 2 patients positive for SIL-TAL, 7 patients with other, and 1 additional patient negative for HOX11L2 but with unknown HOX11 and SIL-TAL status.
Comparison of the 2 genetic groups, HOX11L2 positive versus HOX11L2 negative, in terms of EFS and survival: 3-year estimate rates and the hazard ratio estimates
. | HOX11L2-positive, N = 35 . | HOX11L2-negative,*N = 118 . |
|---|---|---|
| EFS | ||
| 3-year rate, % (SE %) | 75.5 (8.1) | 68.3 (5.0) |
| Hazard ratio (95% CI) | 0.84 (0.40-1.80) | 1 |
| Survival | ||
| 3-year rate, % (SE %) | 77.3 (8.4) | 75.3 (4.6) |
| Hazard ratio (95% CI) | 0.85 (0.37-1.95) | 1 |
. | HOX11L2-positive, N = 35 . | HOX11L2-negative,*N = 118 . |
|---|---|---|
| EFS | ||
| 3-year rate, % (SE %) | 75.5 (8.1) | 68.3 (5.0) |
| Hazard ratio (95% CI) | 0.84 (0.40-1.80) | 1 |
| Survival | ||
| 3-year rate, % (SE %) | 77.3 (8.4) | 75.3 (4.6) |
| Hazard ratio (95% CI) | 0.85 (0.37-1.95) | 1 |
This group included 118 patients: 10 patients with HOX11-high, 17 patients positive for SIL-TAL, 76 patients with other, and 15 additional patients who tested negative for HOX11L2 and not tested for HOX11 and SIL-TAL status.
In patients with T-ALL, the rate of response to prephase did not differ significantly (Kruskal-Wallis P = .24) between patients positive or negative for HOX11L2 (Table 2): 29.4% of patients positive for HOX11L2 were poor responders to prephase versus 36.4% of the patients negative for HOX11L2. In a limited group of 18 patients positive for HOX11L2 and 41 patients negative for HOX11L2, the level of MRD at the end of induction was available: the rate of high levels of MRD (≥ 10–2) was 27.8% and 14.6% respectively.
The effect of some initial factors on the outcome was limited. The 3-year EFS of patients with a WBC count of more than 50 × 109/L (n = 77) was 64.3% versus 76.3% (P = .07) for patients with a WBC count of 50 × 109/L or less (n = 76), and the estimated hazard ratio was 1.8; the 3-year EFS of patients with a high National Cancer Institute (NCI) risk (n = 104) or standard NCI risk (n = 49) was 71.1% and 68.4%, respectively. However, in patients with T-ALL, those with a good response to prephase (n = 91) had a better outcome than those with a poor response (n = 49): the 3-year EFS rates were 81.9% and 47.5% (P < .001), respectively, the estimated hazard ratio was 4.39, and the 95% confidence interval (CI) was 2.22 to 8.69. The Cox model indicated that the differences between HOX11L2 positive and negative in terms of EFS remained practically unchanged after the adjustment for the initial WBC count (P = .88; HR = .94; 95% CI, 0.44-2.03). Cox model stratified by study (58881 versus 58951) lead to practically the same results.
Prognosis according to genetic subgroups
Restricting the analyses to the subgroup of 138 patients for whom a genetic group could be determined (HOX11L2, HOX11-high, SIL-TAL, or other), the overall results were practically identical to the overall group of 153 patients: the 3-year EFS and survival rates were 70.4% and 75.3%, respectively. The prognostic effect of the genetic group was not significant, either in terms of EFS or survival (Figure 5A-B). The 3-year EFS and survival rates in the 4 groups and the estimates of the hazard ratios, considering the other group as baseline, are given in Table 5. As each EFS curve of genetic subgroups (HOX11L2, HOX11-high, or SIL-TAL) is above the one corresponding to the other group, the estimates of the 3 hazard ratios are below 1. By adding in the Cox model the initial WBC count (< versus ≥ 50 × 109/L), the estimates of the hazard ratios (95% CI) corresponding to the 3 groups were 0.81 (0.37-1.79), 0.29 (0.04-2.14), and 0.53 (0.16-1.79), respectively.
Kaplan-Meier estimates of event-free survival and survival according to the 4 genetic groups. (A) Event-free survival. (B) Survival. N indicates number of patients; O, observed number of events. P value was given by the overall log-rank test.
Kaplan-Meier estimates of event-free survival and survival according to the 4 genetic groups. (A) Event-free survival. (B) Survival. N indicates number of patients; O, observed number of events. P value was given by the overall log-rank test.
Comparison of the 4 genetic groups in terms of EFS and survival: 3-year estimate rates and the hazard ratio estimates by taking other as the baseline group
. | HOX11L2-positive, N = 35 . | HOX11-high, N = 10 . | SIL-TAL, N = 17 . | Other,*N = 76 . |
|---|---|---|---|---|
| EFS | ||||
| 3-year rate, % (SE %) | 75.5 (8.1) | 83.3 (8.5) | 75.3 (12.6) | 65.1 (6.4) |
| Hazard ratio (95% CI) | 0.75 (0.34-1.66) | 0.29 (0.04-2.16) | 0.63 (0.19-2.11) | 1 |
| Survival | ||||
| 3-year rate, % (SE %) | 77.3 (8.4) | 100 (0) | 94.1 (3.0) | 67.4 (6.3) |
| Hazard ratio (95% CI) | 0.62 (0.26-1.46) | —† | 0.21 (0.03-1.56) | 1 |
. | HOX11L2-positive, N = 35 . | HOX11-high, N = 10 . | SIL-TAL, N = 17 . | Other,*N = 76 . |
|---|---|---|---|---|
| EFS | ||||
| 3-year rate, % (SE %) | 75.5 (8.1) | 83.3 (8.5) | 75.3 (12.6) | 65.1 (6.4) |
| Hazard ratio (95% CI) | 0.75 (0.34-1.66) | 0.29 (0.04-2.16) | 0.63 (0.19-2.11) | 1 |
| Survival | ||||
| 3-year rate, % (SE %) | 77.3 (8.4) | 100 (0) | 94.1 (3.0) | 67.4 (6.3) |
| Hazard ratio (95% CI) | 0.62 (0.26-1.46) | —† | 0.21 (0.03-1.56) | 1 |
Negative for 3 genetic markers.
Not assessable, as there were no events in the HOX11-high group.
The clinical outcome of patients from the HOX11-high group was quite good (Figure 4A-B). Only one child had a combined CNS relapse 30 months after the achievement of CR. This good outcome is consistent with the results obtained when studying in vivo sensitivity to the early steps of chemotherapy. None of the children of the HOX11-high group were poor responders to prephase, and none of them had high levels of MRD after completion of induction therapy (Table 2).
Patients positive for SIL-TAL had a particularly high frequency (50%) of poor response to the prephase, but none of them had high levels of MRD 1 month later (Table 2). Of 15 patients positive for SIL-TAL, 2 had a BM relapse, and one had an early toxic death.
In patients with lymphoblastic lymphoma the outcome was extremely favorable regardless of the genetic group (Table 3).
Discussion
Childhood B-lineage ALL harbors different nonrandom chromosomal translocations that identify phenotypically and clinically distinct subgroups of patients. In contrast, T-ALL is infrequently characterized by chromosomal translocations, although several T-cell–specific genetic alterations have been described that correlate with different stages of thymocyte differentiation.
Only recently was t(5;14)(q35;q32) uncovered. It is a cryptic translocation with conventional cytogenetic techniques and seems to be restricted to T-ALL. Analysis of t(5;14)(q35;q32) by FISH and/or analysis by real-time quantitative PCR (RQ-PCR) of its molecular consequence HOX11L2 ectopic expression were performed in 153 cases of childhood T-cell malignancies. Our results show that it represents the most frequent genetic alteration (24%) found in T-cell leukemias so far. This finding confirms, on a larger group of patients, what was previously reported by other groups.15,19,29 HOX11L2 expression in leukemia blasts was almost constantly a consequence of a t(5;14)(q35;q32). The few cases without t(5;14) might result either from another genetic alteration or from a failure to detect a t(5;14) with the FISH technique. Another rare translocation observed in T-ALL, t(5;14)(q33;q11), involves the T-cell receptor α/δ (TCRα/δ) locus on chromosome 14. Its breakpoint on chromosome 5 is located 2 kb upstream of the HOX11L2 coding sequence.30 In this respect it could lead to HOX11L2 transcription deregulation, but this hypothesis has not been tested so far. However, none of our patients displaying high HOX11L2 levels without t(5;14)(q35; q32) displayed a t(5;14)(q33;q11).
Our study suggests that the outcome of the children with HOX11L2/t(5;14)(q35;q32)-positive T-cell malignancies was similar to that observed in children with other T-cell malignancies. The P value was not significant, but with a total of 39 events, the statistical power in detecting a difference between 2 groups in term of EFS rate at 3 years of 20% (50% versus 70%, hazard ratio = 1.94) is only 50%; for a difference of 40% (30% versus 70%, hazard ratio = 3.38) it is approximately 90%. The ectopic expression of HOX11L2 is probably not associated with the poor prognosis previously reported by other studies.15,19 Our study was larger than the latter, the median follow-up was 3.1 years, and the EFS analysis was based on 39 events versus 6 events.19 Therefore, the estimate of the hazard ratio of patients with HOX11L2 positive versus negative is still unreliable, the 95% confidence interval is quite wide (0.40-1.84), but it does not cover 2, which represents the minimum level for a prognostic factor. Actually, the data are compatible with the hypothesis that HOX11L2 may be associated with a good prognosis. Two thirds of the HOX11L2-positive group had a cortical-T immunophenotype (CD1a+) versus 39% in patients negative for HOX11L2. CD1a+ T-ALL has been identified as a distinct subgroup with an excellent prognosis under different treatments, including the Berlin-Frankfurt-Munster (BFM) study.3,31-33 In that study as well, there was a trend in favor of better prognosis for patients with CD1a+. This good prognosis has been shown to be associated with an increased in vitro sensitivity to dexamethasone and doxorubicin.34 In contrast, the most immature T-ALL (CD1a–/CD3–), as well as mature T cells (CD1a–/CD3+), are associated with a worse outcome.3,31-33,35 The presence of a high proportion of HOX11L2-expressing ALL within the immunologic subset associated with a favorable prognosis in several treatment protocols provides an indirect argument against the association of HOX11L2 expression with a very unfavorable outcome in most of the current treatment protocols for T-ALL.
In addition to HOX11L2, SIL-TAL fusion and HOX11 expression were investigated in 138 children. A combination of any of these 3 abnormalities was never observed, suggesting that they are mutually exclusive. This observation, as well as the fact that HOX11L2 and HOX11 transcriptional deregulations are preferentially observed in patients with normal karyotypes, is in favor of the involvement of these genes in the initiation of leukemogenesis.
The frequency of SIL-TAL fusion is usually considered to be around 25% in children, but reported frequencies vary widely, ranging from 9% to 30% depending on the study. The 12% frequency we observed for SIL-TAL fusion is in the lower range of this spectrum. However, if we make a compilation of results obtained in studies gathering more than 70 patients, including ours, we obtain a mean estimation of only 17% for SIL-TAL frequency in childhood T-ALL, suggesting this frequency has probably been overestimated in the past.6,36-39 A possible explanation is that, as shown in our and other studies, patients positive for SIL-TAL usually display a high WBC count and could be preferentially selected in retrospective studies, depending on the availability of stored material. The regular increase in SIL-TAL frequency with age that we observed is surprising because this defect is rarely seen in adult malignancies.6,39,40 In this respect, it may be interesting to know whether the few cases found in adults are preferentially found in younger patients. A recent study, which reports SIL-TAL frequencies of 16% in adults aged between 16 and 30 years, and 5% for adults older than 30, suggests that it could be the case.39 Variations of SIL-TAL frequencies with age may contribute to the differences in frequencies observed from one pediatric study to another because some included children aged up to 15 years and others aged up to 20 years.
Very few data are available concerning the prognostic implication of SIL-TAL fusion in childhood T neoplasms. A trend toward improved EFS for patients displaying SIL-TAL fusion was reported in 2 studies,38,41 including 48 and 9 positive patients, respectively. No data are available for patients positive for SIL-TAL treated according to more recent protocols, in which higher overall EFS rates are obtained for patients with T-cell ALL. Although our study did not show a higher EFS rate for patients positive for SIL-TAL, there was a trend toward a better survival rate. There was a discrepancy between the high frequency of poor response to the prephase and the good MRD results after induction in these patients. Actually, the discrepancy was only apparent, because the blast count after prephase in the peripheral blood, conventionally but inappropriately designated as response, is not a true measure of blast sensitivity to the prephase but also depends on the initial blast count. As patients positive for SIL-TAL tend to display higher initial WBC count, even with equal sensitivity to the prephase, a higher percentage of so called poor responders should be expected.
Translocation (10;14) is a nonrandom alteration observed in both T-ALL and T-cell lymphoblastic lymphomas.42 It leads to high HOX11 expression. There is some evidence that HOX11 may play an important role in leukemogenesis. In particular, it has been shown that constitutive expression of HOX11 favors expansion and, in some instances, immortalization of murine hematopoietic progenitors in vitro.43,44 t(10;14) has been found with a frequency of 1.7% to 3.5% in childhood T-ALL.5 Salvati et al11 reported overexpression of HOX11 in 4 of 12 pediatric T-ALL cases and showed one case of HOX11 overexpression lacking t(10;14). More recently, Ferrando et al,15 using a microarray approach, observed high levels of HOX11 transcript in 8 (13%) of 59 pediatric patients with T-ALL, and half of them only displayed a 10q24 rearrangement. In our study, none of the children expressing high levels of HOX11 had a 10q24 rearrangement. These data strongly suggest that in many T-ALLs another genetic abnormality is responsible for deregulation of HOX11 transcription. Although mutation of negative regulatory elements was never found, epigenetic reactivation by specific demethylation of HOX11 promoter has been shown in ALL expressing HOX11 in the absence of 10q24 rearrangements.45
A large study of childhood T-ALL demonstrated the association of t(10;14) with a better survival in univariate analysis as compared with other chromosomal alterations.5 A favorable outcome was also observed in the study by Ferrando et al15 in which the criteria was HOX11 expression instead of the presence of the t(10;14). t(10;14) is more frequent in adults than in children, and frequencies of 14% have been reported,46 whereas preliminary data on HOX11 expression show 30% of positivity.47 In adults also, this translocation as well as HOX11 overexpression seems to be associated with a favorable outcome. In our series of children with T-ALL, we also found a clear trend toward a better outcome for children displaying a high HOX11 expression, in keeping with a good in vivo response to the early steps of chemotherapy. Because of the rarity of this genetic alteration, all studies, including ours, lacked the statistical power that would have allowed any definitive conclusion about its prognostic significance. Further data are required to establish whether the seemingly better outcome associated with t(10;14) and/or HOX11 overexpression is indeed real.
We studied 12 cases of T-cell lymphoblastic lymphoma. Childhood lymphoma remains relatively unexplored at the cytogenetic and molecular levels. Lymphoblastic lymphoma can in some cases evolve into a leukemic phase morphologically indistinguishable from T-ALL, and, in children, malignant blasts present in lymphoblastic leukemia and in ALL share common phenotypic features. During the period covering our study,ALL and lymphoblastic lymphomas were treated according to the same protocols.2 However, whether ALL and lymphoblastic lymphoma do or do not represent the same disease is still unclear. Kikuchi et al41 having failed to find TAL-1 disruption in 18 cases of T-cell lymphoblastic lymphoma concluded that this alteration was restricted to T-ALL. In our study, the 3 T-cell–specific alterations investigated that were observed in ALL were also found in lymphoblastic lymphoma, including SIL-TAL. This finding is consistent with ALL and lymphoma representing different clinical presentations of the same disease. However, HOX11L2 positivity was found in only one case of T-cell lymphoblastic lymphoma. Although the small number of cases studied precludes any definitive conclusion, HOX11L2 overexpression could be less frequent in T-cell lymphoblastic lymphomas than in T-ALL, the more so as T-cell lymphoblastic lymphomas, including those we studied, generally have a cortical thymocyte phenotype that is preferentially associated with HOX11L2 transcriptional deregulation in ALL.
In conclusion, our study provides additional data in favor of a good prognosis associated with SIL-TAL fusion and with high HOX11 expression in childhood T-ALL. This conclusion is consistent with MRD results obtained in these patients and in line with the high predictive power of MRD in T-ALL that was observed in this study (data not shown) and others.48 In contrast with previous reports, which were based on much smaller number of patients and shorter follow-up, we did not find that children with HOX11L2 expressing ALL had a worse outcome than others. However, as prognosis associated with genetic alterations may vary according to the treatment regimen, more extensive studies are clearly required before definitive conclusions can be drawn.
Appendix
The following members of the EORTC-CLG participated in this study: X. Rialland (Angers); F. Maes, E. Michiels (Antwerpen); E. Racadot, E. Plouvier (Besançon); J. Otten, B. Cantineaux (Brussels); S. Suciu, I. Vande Velde (Brussels); M. Malet, E. Lebrun, P. Boutard (Caen); B. Poppe, F. Speleman, Y. Benoit (Ghent); P. Mossuz, D. Plantaz (La Tronche); A. Hagemeijer, A. Uyttebroeck (Leuven); B. Hoyoux, N. Schaaf-Lafontaine (Liège); C. Preudhomme, N. Grardel, M. Fournier, F. Mazingue, B. Nelken (Lille); M.-P. Pages, E. Homolle, Y. Bertrand (Lyon); M. Dupont, J. F. Eliaou, G. Margueritte (Montpellier); F. Méchinaud, R. Garand (Nantes); H. Cavé, G. Brunie, G. Sterkers, K. Yacouben, E. Vilmer (Paris); F. Millot (Poitiers); C. Béhar, M. Munzer (Reims); C. Bastard (Rouen); L. Mauvieux, M.-P. Gaub, A. Falkenrodt, M. Lessard, P. Lutz (Strasbourg); and N. Dastugue, A. Robert (Toulouse).
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-05-1495.
Supported in part by grants from the Association pour la Recherche Contre le Cancer (ARC; grant no. 7394), Télévie 2001 (grant no. 7.4561.01), and the National Cancer Institute (grant nos. 5U10-CA11488-23 through 5U10-CA11488-33).
A complete list of the members of the EORTC-CLG appears in the “Appendix.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank data managers Mrs Christine Waterkeyn, who benefited from a grant of Télévie, and Mr Gabriel Solbu.
The contents of this study are solely the responsibility of the authors and do not represent the official views of the National Cancer Institute (Bethesda, MD). This text presents research results of the Belgian Programme of Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming. The scientific responsibility is assumed by the authors.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal