Abstract
We have reported marked mitochondrial DNA (mtDNA) sequence heterogeneity among individual CD34 clones from adult bone marrow (BM) and the age-dependent accumulation of mtDNA mutations in this mitotically active tissue. Here, we show direct evidence of clonal expansion of cells containing mtDNA mutations and that the mtDNA sequence may be easily determined by using peripheral blood (PB) as a CD34 cell source. Analysis of 594 circulating CD34 clones showed that 150 (25%) had mtDNA sequences different from the same donor's corresponding aggregate sequence. Examination of single granulocytes indicated that 103 (29%) from the same 6 individuals showed mtDNA heterogeneity, with sequences distinct from the corresponding aggregate tissue sequence and from the sequences of other single granulocytes. Circulating and BM CD34 cells showed virtually identical patterns of mtDNA heterogeneity, and the same changes were seen in progeny granulocytes as in their progenitors, indicating that blood sampling could be used in studies to determine whether mtDNA reflects an individual's cumulative or recent exposure to mutagens; as a marker of individual hematopoietic progenitors, stem cells, and their expansion; and for the detection of minimal residual disease in hematologic malignancies of CD34 cell origin. (Blood. 2004;103:4466-4477)
Introduction
Mitochondria are the only organelles of animal cells other than the nucleus that contain DNA as well as their own machinery for RNA and protein synthesis.1,2 Mitochondrial DNA (mtDNA) is present in multiple copies (usually 103 to 104 copies per cell).3 Human mtDNA is a circular molecule of 16 569 base pairs containing 37 genes, which include 2 ribosomal RNAs and 22 tRNAs, required for translation of the mtDNA, and 13 polypeptides that encode subunits of the respiratory chain.2,3 Mitochondria function in intracellular signaling and apoptosis, in intermediary metabolism, and in the metabolism of amino acids, lipids, cholesterol, steroids, heme, and nucleotides.1 Mitochondrial genetics differs from Mendelian genetics of the nuclear genome in 3 major characteristics: maternal inheritance, heteroplasmy, and mitotic segregation.2 In comparison to the nuclear genome, mtDNA has a modified genetic code,4 a paucity of introns, and lack of histone protection.5 The repair capacity of mtDNA is limited, and the proximity of mtDNA to sites of reactive oxygen species generation suggests that mitochondrial DNA may be more susceptible to mutation than in nuclear DNA. Although the limited repair capacity hypothesis has been validated experimentally in some experimental systems, recent data have shown that base excision repair mechanisms do occur in mammalian mtDNA.6,7
Because of its abundance, inherent variability, and ability to survive extreme environmental conditions, mtDNA has been widely used for forensic identification and in anthropologic studies. Furthermore, hundreds of human diseases have been associated with maternally inherited, specific mtDNA deletions and mutations.8 Somatically acquired mtDNA mutations also have been linked to aging and degenerative diseases, cancer, and autoimmunity.9 A large deletion of mtDNA is a hallmark of Pearson syndrome, a constitutional disorder that includes sideroblastic anemia.10 Mutations of mtDNA were reported in apparently acquired sideroblastic anemia and in myelodysplastic syndromes in general.11 Although we were unable to confirm these results by amplification and direct sequencing of the entire mtDNA genome in patients and healthy control subjects, we coincidentally observed numerous sequence changes in bulk samples of bone marrow (BM) cells from our healthy control subjects as well as in patients.12 When we undertook to investigate the possibility that mtDNA mutations might accumulate in human CD34 cells by examination of a portion of the mtDNA control region in individual clonogenic cells, we unexpectedly found marked heterogeneity among marrow CD34 cells, not present in umbilical cord blood.13 Age-dependent accumulation of mtDNA mutations and the replacement of one DNA base pair by another, leading to heteroplasmy and then homoplasmy of mtDNA sequences in a single cell, appeared to be relatively common in marrow progenitor and presumably stem cells.13 Although aging has been linked to mitochondrial senescence, mutant mtDNA genomes have been assumed to be lost by dilution from rapidly dividing tissues such as bone marrow.8 Single cell analyses have indicated similar high levels of mtDNA heterogeneity and homoplasmy in other tissues.14,15
We hypothesized that mtDNA mutations might arise and become homoplasmic in hematopoietic progenitor and stem cells during life, and mutations might then become fixed after clonal expansion of individual CD34 cells. In the current study, we show that the mutational spectra of marrow hematopoietic progenitor cells is preserved in circulating CD34 cells and present direct evidence that the clonal expansion of age-related mtDNA mutations occurs in hematopoietic progeny.
Materials and methods
Tissue samples
BM and peripheral blood (PB) from 6 healthy adult donors were collected after informed consent was obtained following protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute (Tables 1 and 2). Sequence analyses of the mtDNA control region from BM CD34 cells of these healthy donors have been published13 and are included in the current manuscript for comparative purposes only.
Plating efficiency and grade of CD34 clones after culture and immediate recovery of single granulocytes
. | PB CD34 clone . | . | . | . | . | . | . | BM CD34 clone . | . | . | . | . | . | . | Single granulocyte mtDNA PCR . | . | . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | Grade, no. . | . | . | . | . | . | . | Grade, no. . | . | . | . | . | . | . | . | PCR efficiency . | . | ||||||||||||||
| . | 1 . | 2 . | 3 . | 4 . | Subtotal, no. . | Microplates*/total wells . | PEf, % . | 1 . | 2 . | 3 . | 4 . | Subtotal, no. . | Microplates*/total wells . | PEf, % . | Granulocytes, no. . | No. positive . | % positive . | ||||||||||||||
| 1 (47/F) | 13 | 54 | 128 | 85 | 280 | 5/480 | 58.3 | 33 | 21 | 30 | 38 | 122 | 4/384 | 31.8 | 480 | 51 | 10.6 | ||||||||||||||
| 2 (38/F) | 33 | 61 | 64 | 34 | 192 | 4/384 | 50.0 | — | — | — | — | — | — | — | 1440 | 37 | 2.6 | ||||||||||||||
| 3 (43/M) | 24 | 20 | 23 | 26 | 93 | 3/288 | 32.3 | 279 | 149 | 91 | 127 | 646 | 20/1920 | 33.6 | 480 | 50 | 10.4 | ||||||||||||||
| 4 (34/M) | 38 | 38 | 58 | 66 | 200 | 5/480 | 41.7 | — | — | — | — | — | — | — | 768 | 111 | 14.5 | ||||||||||||||
| 5 (54/M) | 35 | 50 | 61 | 36 | 182 | 5/480 | 37.9 | 306 | 177 | 119 | 140 | 742 | 20/1920 | 38.6 | 768 | 42 | 5.5 | ||||||||||||||
| 6 (34/F) | 37 | 46 | 52 | 92 | 227 | 5/480 | 47.3 | — | — | — | — | — | — | — | 768 | 64 | 8.3 | ||||||||||||||
| Total | 180 | 269 | 386 | 339 | 1174 | 27/2592 | 45.3 | 618 | 347 | 240 | 305 | 1510 | 44/4224 | 35.7 | 4704 | 355 | 7.5 | ||||||||||||||
. | PB CD34 clone . | . | . | . | . | . | . | BM CD34 clone . | . | . | . | . | . | . | Single granulocyte mtDNA PCR . | . | . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | Grade, no. . | . | . | . | . | . | . | Grade, no. . | . | . | . | . | . | . | . | PCR efficiency . | . | ||||||||||||||
| . | 1 . | 2 . | 3 . | 4 . | Subtotal, no. . | Microplates*/total wells . | PEf, % . | 1 . | 2 . | 3 . | 4 . | Subtotal, no. . | Microplates*/total wells . | PEf, % . | Granulocytes, no. . | No. positive . | % positive . | ||||||||||||||
| 1 (47/F) | 13 | 54 | 128 | 85 | 280 | 5/480 | 58.3 | 33 | 21 | 30 | 38 | 122 | 4/384 | 31.8 | 480 | 51 | 10.6 | ||||||||||||||
| 2 (38/F) | 33 | 61 | 64 | 34 | 192 | 4/384 | 50.0 | — | — | — | — | — | — | — | 1440 | 37 | 2.6 | ||||||||||||||
| 3 (43/M) | 24 | 20 | 23 | 26 | 93 | 3/288 | 32.3 | 279 | 149 | 91 | 127 | 646 | 20/1920 | 33.6 | 480 | 50 | 10.4 | ||||||||||||||
| 4 (34/M) | 38 | 38 | 58 | 66 | 200 | 5/480 | 41.7 | — | — | — | — | — | — | — | 768 | 111 | 14.5 | ||||||||||||||
| 5 (54/M) | 35 | 50 | 61 | 36 | 182 | 5/480 | 37.9 | 306 | 177 | 119 | 140 | 742 | 20/1920 | 38.6 | 768 | 42 | 5.5 | ||||||||||||||
| 6 (34/F) | 37 | 46 | 52 | 92 | 227 | 5/480 | 47.3 | — | — | — | — | — | — | — | 768 | 64 | 8.3 | ||||||||||||||
| Total | 180 | 269 | 386 | 339 | 1174 | 27/2592 | 45.3 | 618 | 347 | 240 | 305 | 1510 | 44/4224 | 35.7 | 4704 | 355 | 7.5 | ||||||||||||||
Numbers in columns indicate cells or clones observed microscopically after 5-day suspension culture of CD34 cells from PB and BM or immediate recovery of sorted granulocytes. PB indicates peripheral blood; BM, bone marrow; grade 1, less than 5 cells/well; grade 2, 6 to 10 cells/well; grade 3, 11 to 20 cells/well; grade 4, more than 21 cells/well; PEf, plating efficiency: number of wells with clones/total wells.
Number of 96-well microplates.
CD34 clones from peripheral blood and bone marrow and single granulocytes for mtDNA analysis, successfully assayed by nested PCR for mtDNA sequence
. | Control region . | . | . | . | . | . | . | . | . | . | . | Coding region (CO1 and Cytb) . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | BM CD34 clone*, no. . | . | . | . | . | PB CD34 clone, no. . | . | . | . | . | Single granulocyte, no. . | BM CD34 clone, no. . | . | . | . | . | PB CD34 clone, no. . | . | . | . | . | ||||||||||||||||||
| . | G1 . | G2 . | G3 . | G4 . | Subtotal . | G1 . | G2 . | G3 . | G4 . | Subtotal . | . | G1 . | G2 . | G3 . | G4 . | Subtotal . | G1 . | G2 . | G3 . | G4 . | Subtotal . | ||||||||||||||||||
| 1 (47/F) | 28 | 20 | 30 | 37 | 115 | 13 | 30 | 47 | 30 | 120 | 51 | 28 | 20 | 29 | 17 | 94 | 13 | 30 | 35 | 17 | 95 | ||||||||||||||||||
| 2 (38/F) | 23 | 3 | 4 | 6 | 36 | 23 | 24 | 23 | 24 | 94 | 37 | — | — | — | — | — | — | — | — | — | — | ||||||||||||||||||
| 3 (43/M) | 30 | 30 | 30 | 30 | 120 | 24 | 20 | 23 | 26 | 93 | 50 | 25 | 26 | 24 | 20 | 95 | 24 | 20 | 23 | 26 | 93 | ||||||||||||||||||
| 4 (34/M) | 26 | 28 | 28 | 29 | 111 | 23 | 24 | 24 | 24 | 95 | 111 | — | — | — | — | — | — | — | — | — | — | ||||||||||||||||||
| 5 (54/M) | 26 | 28 | 30 | 30 | 114 | 24 | 24 | 24 | 24 | 96 | 42 | 25 | 29 | 30 | 12 | 96 | 24 | 24 | 24 | 24 | 96 | ||||||||||||||||||
| 6 (34/F) | 25 | 30 | 30 | 30 | 115 | 24 | 24 | 24 | 24 | 96 | 64 | — | — | — | — | — | — | — | — | — | — | ||||||||||||||||||
| Total | 158 | 139 | 152 | 162 | 611 | 131 | 146 | 165 | 152 | 594 | 355 | 78 | 75 | 83 | 49 | 285 | 61 | 74 | 82 | 67 | 284 | ||||||||||||||||||
. | Control region . | . | . | . | . | . | . | . | . | . | . | Coding region (CO1 and Cytb) . | . | . | . | . | . | . | . | . | . | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | BM CD34 clone*, no. . | . | . | . | . | PB CD34 clone, no. . | . | . | . | . | Single granulocyte, no. . | BM CD34 clone, no. . | . | . | . | . | PB CD34 clone, no. . | . | . | . | . | ||||||||||||||||||
| . | G1 . | G2 . | G3 . | G4 . | Subtotal . | G1 . | G2 . | G3 . | G4 . | Subtotal . | . | G1 . | G2 . | G3 . | G4 . | Subtotal . | G1 . | G2 . | G3 . | G4 . | Subtotal . | ||||||||||||||||||
| 1 (47/F) | 28 | 20 | 30 | 37 | 115 | 13 | 30 | 47 | 30 | 120 | 51 | 28 | 20 | 29 | 17 | 94 | 13 | 30 | 35 | 17 | 95 | ||||||||||||||||||
| 2 (38/F) | 23 | 3 | 4 | 6 | 36 | 23 | 24 | 23 | 24 | 94 | 37 | — | — | — | — | — | — | — | — | — | — | ||||||||||||||||||
| 3 (43/M) | 30 | 30 | 30 | 30 | 120 | 24 | 20 | 23 | 26 | 93 | 50 | 25 | 26 | 24 | 20 | 95 | 24 | 20 | 23 | 26 | 93 | ||||||||||||||||||
| 4 (34/M) | 26 | 28 | 28 | 29 | 111 | 23 | 24 | 24 | 24 | 95 | 111 | — | — | — | — | — | — | — | — | — | — | ||||||||||||||||||
| 5 (54/M) | 26 | 28 | 30 | 30 | 114 | 24 | 24 | 24 | 24 | 96 | 42 | 25 | 29 | 30 | 12 | 96 | 24 | 24 | 24 | 24 | 96 | ||||||||||||||||||
| 6 (34/F) | 25 | 30 | 30 | 30 | 115 | 24 | 24 | 24 | 24 | 96 | 64 | — | — | — | — | — | — | — | — | — | — | ||||||||||||||||||
| Total | 158 | 139 | 152 | 162 | 611 | 131 | 146 | 165 | 152 | 594 | 355 | 78 | 75 | 83 | 49 | 285 | 61 | 74 | 82 | 67 | 284 | ||||||||||||||||||
Numbers in columns indicate CD34 clones and individual granulocytes successfully subjected to nested PCR for mtDNA sequences from the control and the CO1 and Cytb coding regions. G indicates grade; CO1, cytochrome c oxidase 1; Cytb, cytochrome b; and—, not applicable.
The same CD34 clones are used as in our recent publication.13
Single cell sorting for CD34 cells and granulocytes
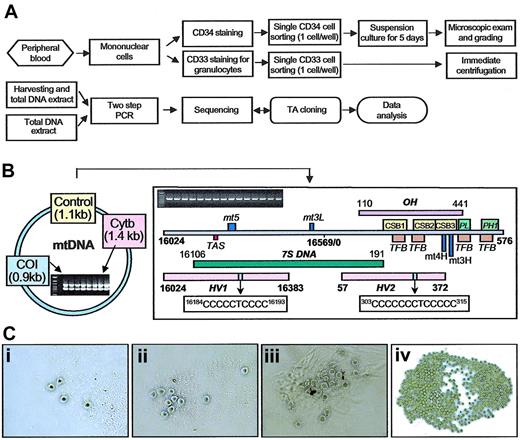
Mononuclear cells from BM and PB were separated by density gradient centrifugation and washed twice in phosphate-buffered saline (PBS). The number of cells suspended in PBS was adjusted to 2 × 107 cells/mL. Anti-CD34 phycoerythrin (PE)-conjugated monoclonal antibody (10 μL) and anti-CD33 fluorescein isothiocyanate (FITC)-conjugated antibody (10 μL) (BD Bioscience, San Jose, CA) were added to each 12 × 75-mm tube containing 100 μL cell suspension. After incubation for 30 minutes at 4°C, cells were washed with cold PBS and resuspended in 0.5 mL PBS. Cell sorting was performed on a MoFlo Cytometer (Dako-Cytomation, Ft Collins, CO), using 100 mW of the 488 nm line of an argon laser (I-90; Coherent, Palo Alto, CA) for excitation. Forward scatter was the triggering parameter. Fluorescence of FITC was detected by using a 530/20 bandpass filter, and the fluorescence of PE was detected by using a 580/30 bandpass filter. Single cell deposition was accomplished by using the CyClone automated cloner (Dako-Cytomation) in the 0.5 single drop mode with gating based on forward scatter and fluorescence. Individual CD34 cells were placed into each well of a 96-well microplate (Nalge Nunc International, Rochester, NY) containing 100 μL culture media, and single granulocytes were deposited in each well of a MicroAmp optical 96-well reaction plate (Applied Biosystems, Foster City, CA) containing 30 μL1 × Tris (tris(hydroxymethyl)aminomethane) EDTA (ethylenediaminetetraacetic acid) (TE) buffer (Figure 1A).
Work flow chart, linearized map of mtDNA control region, 2-step PCR amplification for target genes and circulating CD34 clones. (A) Flow chart for mtDNA analysis of single circulating CD34 cells and granulocytes (described in “Materials and methods”). Evidence of mtDNA heterogeneity was further confirmed by reamplification of the original lysate. (B) Linearized map and function location of mtDNA control region between nucleotides 16024 to 16569 and 1 to 576 (D-loop) and 2-step PCR amplification of control region (1.12kb) and CO1 (0.91 kb) and Cytb (1.39 kb) genes; HV1 (hypervariable segment 1, nucleotides 16024-16383), HV2 (hypervariable segment 2, nucleotides 57-372), OH (H-stand origin, nucleotides), CSB (conserved sequence block), mt5 (control element), mt3L (L-strand control element), TAS (termination-associated sequence), PL (L-strand promoter), PH1 (major H-strand promoter), TFB (mitochondrial transcription factor binding site), mt4H (H-strand control element), and mt3H (H-strand control element). The homopolymeric C tracts located on HV1 (nucleotides 16184-16193; 5CT4C) and HV2 (nucleotides 303-315; 7CT5C). (C) The number and morphology of circulating CD34 clones after 5-day suspension culture. Each picture shows a different grade: (i) less than 5 cells per well (grade 1), (ii) between 6 and 10 cells per well (grade 2), (iii) 11 to 20 cells per well (grade 3), (iv) more than 21 cells per well (grade 4). Original magnification for panels Ci-Ciii, × 200; for Civ, × 100.
Work flow chart, linearized map of mtDNA control region, 2-step PCR amplification for target genes and circulating CD34 clones. (A) Flow chart for mtDNA analysis of single circulating CD34 cells and granulocytes (described in “Materials and methods”). Evidence of mtDNA heterogeneity was further confirmed by reamplification of the original lysate. (B) Linearized map and function location of mtDNA control region between nucleotides 16024 to 16569 and 1 to 576 (D-loop) and 2-step PCR amplification of control region (1.12kb) and CO1 (0.91 kb) and Cytb (1.39 kb) genes; HV1 (hypervariable segment 1, nucleotides 16024-16383), HV2 (hypervariable segment 2, nucleotides 57-372), OH (H-stand origin, nucleotides), CSB (conserved sequence block), mt5 (control element), mt3L (L-strand control element), TAS (termination-associated sequence), PL (L-strand promoter), PH1 (major H-strand promoter), TFB (mitochondrial transcription factor binding site), mt4H (H-strand control element), and mt3H (H-strand control element). The homopolymeric C tracts located on HV1 (nucleotides 16184-16193; 5CT4C) and HV2 (nucleotides 303-315; 7CT5C). (C) The number and morphology of circulating CD34 clones after 5-day suspension culture. Each picture shows a different grade: (i) less than 5 cells per well (grade 1), (ii) between 6 and 10 cells per well (grade 2), (iii) 11 to 20 cells per well (grade 3), (iv) more than 21 cells per well (grade 4). Original magnification for panels Ci-Ciii, × 200; for Civ, × 100.
Suspension culture
Individual CD34 cells placed into separate wells of 96-well plates were cultured in serum-free medium containing 100 ng/mL stem cell factor, 100 ng/mL Flt-3, 100 ng/mL thrombopoietin, and 50 ng/mL granulocyte colony-stimulating factor (G-CSF) (all from Stem Cell Technologies, Vancouver, British Columbia, Canada). After culture for 5 days, each well of the microtiter plate was examined with an inverted microscope (Olympus IX50, Melville, NY) to determine growth and plating efficiency of the single CD34 cells. Growth was quantified and graded with the following scoring system according to cell number in each CD34 clone: grade 1, 5 or less cells/well; grade 2, 6 to 10 cells/well; grade 3, 11 to 20 cells/well; grade 4, 21 or more cells/well (Figure 1C). Plating efficiency was defined as the number of positive (cells were present) wells/total wells × 100. Each CD34 clone was harvested from the well by vigorous pipetting and dispensed into a 1.5-mL microcentrifuge tube and rinsed with 200 μL PBS. Cells were collected after centrifugation at 300g for 5 minutes, and then washed with PBS. Cell pellets were stored at -80°C.
DNA extraction from individual CD34 clones and single granulocytes
The amount of 30 μL 1 × TE buffer was placed in each 1.5-mL tube containing one cell pellet. The cells were lysed by incubation at 95°C for 10 minutes with occasional shaking to liberate the total DNA.16 Following single granulocyte deposition into each well of a 96-well microplate containing 30 μL 1 × TE buffer, total DNA was extracted from single granulocytes by incubation at 95°C for 10 minutes in the 96-well reaction plate (Applied Biosystems) with use of the GeneAmp PCR system 9700 (Applied Biosystems). The lysate was briefly centrifuged and stored at -20°C.
PCR amplification of the mtDNA control region and the CO1 and Cytb gene coding regions
Cell lysates of individual CD34 clones and single granulocytes were subjected to amplification of mtDNA with use of the LA PCR (polymerase chain reaction) kit (TaKaRa LA Taq, Madison, WI). Two-step nested PCR amplification was performed with outer and inner pairs of primers to generate sufficient template from CD34 clones and single granulocyte for sequencing of the mtDNA control region and CO1 and Cytb gene coding regions. Primer pairs for targeted mtDNA amplification are shown in Table 3. The primary PCR mixture contained 400 μM of each deoxynucleotide triphosphate (dNTP), 2 U LA Taq (TaKaRa LA Taq), 0.8 μM outer primers, 3 μL and 10 μL cell lysates from individual CD34 clones and single granulocyte, respectively. PCR amplification was carried out in a MicroAmp optical 96-well reaction plate (Applied Biosystems) with use of the GeneAmp PCR system 9700 (Applied Biosystems) as follows: 1 cycle of 95°C for 1 minute; then 35 cycles of 95°C for 30 seconds, 52°C for 50 seconds, and 72°C for 1 minute with 10-second increase per cycle; ending with 1 cycle of 72°C for 5 minutes. The secondary PCR was performed in 50 μL reaction mixture containing 400 μM of each dNTP, 2 U LA Taq, 0.8 μM inner nested primers, and 2 μL primary PCR product under the same amplification conditions as described earlier. Secondary PCR samples were electrophoresed on 1% agarose gels and stained with ethidium bromide to assess the purity and size of the DNA fragments and were subsequently purified by using the QIA quick PCR purification kit (Qiagen, Valencia, CA). Controls, reaction mixtures without DNA templates, were subjected to the same PCR amplification conditions and in all cases were confirmed to be negative. To prevent DNA cross-contamination, special precautions were taken during the procedures of cell harvesting, DNA extraction, PCR amplification, and DNA sequencing.
Primer sets for 2-step nested PCR and direct sequencing of mtDNA control region and CO1 and Cytb genes
Amplicon mtDNA region (np) (size, kb); mtDNA region (np) . | . | Sequence (5′ to 3′) for 2-step nested PCR . | . | Sequencing primers (5′ to 3′) . |
|---|---|---|---|---|
| 1 (1.12); control region (16024-16569; 1-576) | ||||
| F15574 (O) | CGCCTACACAATTCTCCGATC (O) | F15971 | TTAACTCCACCATTAGCACC | |
| R921 (O) | ACTTGGGTTAATCGTGTGACC (O) | R1 | CAGTGTATTGCTTTGAGGAGG | |
| F15971 (I) | TTAACTCCACCATTAGCACC (I) | SR | GCATGGAGAGCTCCCGTGAGTGG | |
| R1 (l) | CAGTGTATTGCTTTGAGGAGG (I) | SF | CATCTGGTTCCTACTTCAGGGTC | |
| 2 (1.39); Cytb (14688-15996) | ||||
| F 14622 (O) | CCACAAACCCCATTACTAAACCCAC (O) | F14688 | CTACAACCACGACCAATGATATG | |
| R16084 (O) | CGGTTGTTGATGGGTGAGTC (O) | R15996 | GCTTTGGGTGCTAATGGTGGAG | |
| F14663 (l) | GCATACATCATTATTCTCGCACGG (I) | |||
| R16057 (l) | GGGTGGTACCCAAATCTGCTTCC (I) | |||
| 3 (0.91); CO1 (6611-7470) | ||||
| F6541 (O) | GCAACCTCAACACCACCTTCTTCG (O) | F6645 | CTACCAGGCTTCGGAATAATCTCCC | |
| R7613 (O) | GTAGACCTACTTGCGCTGCATGTGC (O) | R7425 | GTTCTTCGAATGTGTGGTAGGGTG | |
| F6586 (l) | CCATTCTATACCAACACCTATTCTG (I) | |||
| R7498 (l) | CCATGGGGTTGGCTTGAAACCAGC (I) |
Amplicon mtDNA region (np) (size, kb); mtDNA region (np) . | . | Sequence (5′ to 3′) for 2-step nested PCR . | . | Sequencing primers (5′ to 3′) . |
|---|---|---|---|---|
| 1 (1.12); control region (16024-16569; 1-576) | ||||
| F15574 (O) | CGCCTACACAATTCTCCGATC (O) | F15971 | TTAACTCCACCATTAGCACC | |
| R921 (O) | ACTTGGGTTAATCGTGTGACC (O) | R1 | CAGTGTATTGCTTTGAGGAGG | |
| F15971 (I) | TTAACTCCACCATTAGCACC (I) | SR | GCATGGAGAGCTCCCGTGAGTGG | |
| R1 (l) | CAGTGTATTGCTTTGAGGAGG (I) | SF | CATCTGGTTCCTACTTCAGGGTC | |
| 2 (1.39); Cytb (14688-15996) | ||||
| F 14622 (O) | CCACAAACCCCATTACTAAACCCAC (O) | F14688 | CTACAACCACGACCAATGATATG | |
| R16084 (O) | CGGTTGTTGATGGGTGAGTC (O) | R15996 | GCTTTGGGTGCTAATGGTGGAG | |
| F14663 (l) | GCATACATCATTATTCTCGCACGG (I) | |||
| R16057 (l) | GGGTGGTACCCAAATCTGCTTCC (I) | |||
| 3 (0.91); CO1 (6611-7470) | ||||
| F6541 (O) | GCAACCTCAACACCACCTTCTTCG (O) | F6645 | CTACCAGGCTTCGGAATAATCTCCC | |
| R7613 (O) | GTAGACCTACTTGCGCTGCATGTGC (O) | R7425 | GTTCTTCGAATGTGTGGTAGGGTG | |
| F6586 (l) | CCATTCTATACCAACACCTATTCTG (I) | |||
| R7498 (l) | CCATGGGGTTGGCTTGAAACCAGC (I) |
CO1 indicates cytochrome c oxidase 1; Cytb, cytochrome b; np, nucleotide position; F, forward primer; O, outer primer; R, reverse primer; l, inner primer; SR, reverse sequence primer; SF, forward sequence primer.
Two-step nested PCR amplification of mtDNA CO1 and Cytb genes was performed in the same manner as described earlier; corresponding primers for each gene amplification are listed in Table 3.
Sequence analysis
The amplified mtDNA PCR products were directly sequenced by using the BigDye Terminator v3.1 ready reaction kit (Applied Biosystems) and the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequencing primers used for each mtDNA product are shown in Table 3. Experimentally obtained mtDNA sequences were compared with the Revised Cambridge Reference Sequence (RCRS) (http://www.mitomap.org/mitomap/mitoseq.html)17 by using the Blast2 program (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) and the database search tool, MitoAnalyzer (http://www.cstl.nist.gov/biotech/strbase/mitoanalyzer.html, 2001)18 to determine the polymorphisms and mutations that differed from the Revised Cambridge Reference Sequence and whether the differences caused amino acid changes in the resultant polypeptides. All differences detected through automated procedures were manually confirmed. To exclude potential artifacts, PCR amplifications from original cell lysates were replicated 1 or 2 additional times and resequenced. When the same nucleotide changes were reproduced in all independent PCR amplifications, they were considered to be confirmed. Several sequence differences, which were not reproducible on replicated PCR of mtDNA extracted from CD34 cells and granulocytes, were discarded, although they may have represented mutation events at very low copy number.
TA cloning
In other experiments to develop a heteroplasmic Standard Reference Material, 285-bp amplicons were generated by gene amplification of wild-type mtDNA and mtDNA altered in a single base and then mixed in varying proportions.19 By sequencing the mixtures, one could detect the minor species of mtDNA when it was present above about the 20% level. Therefore, mixed nucleotide signals on sequencing electropherograms, when observed in the current study, were assumed to represent at least 20% heteroplasmy but could have also resulted from gene amplification artifacts. To confirm heteroplasmy and mixed nucleotide signals in the mtDNA control region, PCR products were directly inserted into the pCR 2.1-TOPO vector and transformed into competent Escherichia coli (TOP10 cells) using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Recombinant plasmids isolated from 8 to 12 white colonies were sequenced.
Statistical analysis
A chi-square test was used to determine statistical differences in the frequency of heteroplasmy in adult bone marrow and circulating blood mtDNA. The one-way ANOVA (analysis of variance) test was performed to examine whether the total rate and unique differences observed in cord blood CD34 clones, adult BM CD34 clones, circulating CD34 clones, and single granulocytes produced significant statistical differences in mtDNA heterogeneity; a P value less than .05 was considered significant.
Results
PB CD34 cell clones
After sorting, single CD34 cells from PB were cultured in individual wells of 96-well plates in serum-free medium containing selected hematopoietic growth factors. CD34 cell-derived colonies were classified according to the cell number per well (Figure 1C). Although there was some variation of plating efficiency of CD34 cells obtained from 6 healthy donors, overall average plating efficiency in PB was 45% ± 9.3% (mean ± SD), comparable and not statistically different from our previous results with BM from these same donors (36% ± 3.6%; Table 1).
Single granulocyte mtDNA PCR
Individual granulocytes from the 6 donors were isolated by sorting, and the mtDNA control region was amplified by using a nested PCR procedure (Table 1). From 4704 granulocytes, 355 (8%) yielded a PCR product of the correct size; however, there was marked variation of PCR efficiency from the various donors. Any observed polymorphisms and mutations were confirmed by reamplification and sequencing (described in “Materials and methods”).
Aggregate genotype of the mtDNA control region
To identify mtDNA heterogeneity in individual CD34 clones and single granulocytes, we first determined the aggregate cell mitochondrial genotype of each donor, as compared to the Revised Cambridge Reference Sequence. As we previously observed, there was marked variation in the number of nucleotide changes among individual donors, with a range from 6 (donor 1) to 24 (donor 4) (11.8 ± 5.8; Table 4). A total of 71 mtDNA sequence variants were found in aggregate cells from the 6 volunteers; 69 variants were already listed in the polymorphism database (http://www.mitomap.org/cgi-bin/mitomap/tbl4gen.pl), and the 2 new nucleotide variants (478A>G and 517A>G) were found in donors 2 and 6, respectively. Donors 2 and 3 had length variations of the homopolymeric C tracts at nucleotide positions 16183 to 16193 (16189T>C, 12C) and 303 to 315 (Table 4). Most of the substitutions were transitions, but a few were transversions, in agreement with findings in the literature.20
Nucleotide sequence changes of mtDNA control region from aggregate cells
Donor (age, y/sex) and polymorphism . | Affected mtDNA gene . |
|---|---|
| 1 (47/F) | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 16192C>T | HV1, 7S |
| 16270C>T | HV1, 7S |
| 2 (38/F) | |
| 73A>G | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 478A>G† | |
| 16093T>G | HV1 |
| 16158A>G | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16183A>C | HV1, 7S |
| 16189T>C (12C) | HV1, 7S |
| 16219A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| 3 (43/M) | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 152T>C | HV2, 7S, OH |
| 195T>C | HV2, OH |
| 263A>G | HV2, OH |
| 9CT6C*, 8CT6C* | HV2, OH, CSB2 |
| 514delC | — |
| 515delA | — |
| 16223C>T | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16294C>T | HV1, 7S |
| 16390G>A | 7S |
| 4 (34/M) | |
| 93A>G | HV2, 7S |
| 95A>C | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 189A>G | HV2, 7S, OH |
| 236T>C | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 247G>A | HV2, OH, TFB1 |
| 263A>G | HV2, OH |
| 514delC | — |
| 515delA | — |
| 16093T>C | HV1 |
| 16129G>A | HV1, 7S |
| 16148C>T | HV1, 7S |
| 16168C>T | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16187C>T | HV1, 7S |
| 16188C>G | HV1, 7S |
| 16189T>C | HV1, 7S |
| 16223C>T | HV1, 7S |
| 16230A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16293A>G | HV1, 7S |
| 16311T>C | HV1, 7S |
| 16320C>T | HV1, 7S |
| 5 (54/M) | |
| 73A>G | HV2, 7S |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 514delC | — |
| 515delA | — |
| 16126T>C | HV1, 7S |
| 16294C>T | HV1, OH |
| 16296C>T | HV1, OH |
| 16519T>C | 7S |
| 6 (34/F) | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 517A>G† | — |
| 16270C>T | HV1, 7S |
| 16292C>T | HV1, 7S |
| 16362T>C | HV1, 7S |
Donor (age, y/sex) and polymorphism . | Affected mtDNA gene . |
|---|---|
| 1 (47/F) | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 16192C>T | HV1, 7S |
| 16270C>T | HV1, 7S |
| 2 (38/F) | |
| 73A>G | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 478A>G† | |
| 16093T>G | HV1 |
| 16158A>G | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16183A>C | HV1, 7S |
| 16189T>C (12C) | HV1, 7S |
| 16219A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| 3 (43/M) | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 152T>C | HV2, 7S, OH |
| 195T>C | HV2, OH |
| 263A>G | HV2, OH |
| 9CT6C*, 8CT6C* | HV2, OH, CSB2 |
| 514delC | — |
| 515delA | — |
| 16223C>T | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16294C>T | HV1, 7S |
| 16390G>A | 7S |
| 4 (34/M) | |
| 93A>G | HV2, 7S |
| 95A>C | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 189A>G | HV2, 7S, OH |
| 236T>C | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 247G>A | HV2, OH, TFB1 |
| 263A>G | HV2, OH |
| 514delC | — |
| 515delA | — |
| 16093T>C | HV1 |
| 16129G>A | HV1, 7S |
| 16148C>T | HV1, 7S |
| 16168C>T | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16187C>T | HV1, 7S |
| 16188C>G | HV1, 7S |
| 16189T>C | HV1, 7S |
| 16223C>T | HV1, 7S |
| 16230A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16293A>G | HV1, 7S |
| 16311T>C | HV1, 7S |
| 16320C>T | HV1, 7S |
| 5 (54/M) | |
| 73A>G | HV2, 7S |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 514delC | — |
| 515delA | — |
| 16126T>C | HV1, 7S |
| 16294C>T | HV1, OH |
| 16296C>T | HV1, OH |
| 16519T>C | 7S |
| 6 (34/F) | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 517A>G† | — |
| 16270C>T | HV1, 7S |
| 16292C>T | HV1, 7S |
| 16362T>C | HV1, 7S |
HV2 indicates hypervariable segment 2; 7S, 7S DNA; OH, H-strand origin; CSB2, conserved sequence block II; HV1, hypervariable segment 1; TAS, termination-association sequence; TFB1, mitochondrial transcription factor 1 binding site; and —, not applicable.
Homopolymeric C tract localized between nucleotide 303 and 315 (eg, 8CT6C defined CCCCCCCCTCCCCCC).
New mtDNA polymorphisms (not listed in MitoMap database).
Aggregate genotype of the CO1 and Cytb genes in the mtDNA coding region
A total of 17 mtDNA nucleotide changes (5.7 ± 3.2) were noted in the CO1 and Cytb genes among 3 donors (Table 5), 15 of these nucleotide variants were already listed in a published polymorphism database (http://www.mitomap.org/cgi-bin/mitomap/tbl4gen.pl), and 2 new sequence variations that were not previously recorded, including unpublished mtDNA polymorphisms, were found. These 2 mutations were silent changes and not predicted to produce any amino acid change (Table 5).
Nucleotide sequence changes of mtDNA coding region (CO1 and Cytb genes) from aggregate cells
Donor (age, y/sex) and mtDNA gene . | Nucleotide change . | Amino acid change . |
|---|---|---|
| 1 (47/F) | ||
| CO1 | 7028C>T | Silent |
| Cytb | 15326A>G | Thr-Ala |
| 3 (43/M) | ||
| CO1 | 6663A>G | Ile-Val |
| CO1 | 7028C>T | Silent |
| CO1 | 7175T>C | Silent |
| CO1 | 7256C>T | Silent |
| CO1 | 7274C>T | Silent |
| Cytb | 15301G>A | Silent |
| Cytb | 15326A>G | Thr-Ala |
| Cytb | 15784T>C* | Silent |
| 5 (54/M) | ||
| CO1 | 7022T>C* | Silent |
| CO1 | 7028C>T | Silent |
| Cytb | 14766C>T | Silent |
| Cytb | 14905G>A | Silent |
| Cytb | 15326A>G | Thr-Ala |
| Cytb | 15452C>A | Leu-Ile |
| Cytb | 15607A>G | Silent |
Donor (age, y/sex) and mtDNA gene . | Nucleotide change . | Amino acid change . |
|---|---|---|
| 1 (47/F) | ||
| CO1 | 7028C>T | Silent |
| Cytb | 15326A>G | Thr-Ala |
| 3 (43/M) | ||
| CO1 | 6663A>G | Ile-Val |
| CO1 | 7028C>T | Silent |
| CO1 | 7175T>C | Silent |
| CO1 | 7256C>T | Silent |
| CO1 | 7274C>T | Silent |
| Cytb | 15301G>A | Silent |
| Cytb | 15326A>G | Thr-Ala |
| Cytb | 15784T>C* | Silent |
| 5 (54/M) | ||
| CO1 | 7022T>C* | Silent |
| CO1 | 7028C>T | Silent |
| Cytb | 14766C>T | Silent |
| Cytb | 14905G>A | Silent |
| Cytb | 15326A>G | Thr-Ala |
| Cytb | 15452C>A | Leu-Ile |
| Cytb | 15607A>G | Silent |
Ala represents alanine; Ile, isoleucine; Leu, leucine; Thr, threonine; Val, valine.
New mtDNA polymorphisms (not listed in MitoMap database).
mtDNA heterogeneity in the mtDNA control region
To assess the polymorphic spectra of the individual cell mtDNA sequences and their clonal expansion among CD34 clones and in single granulocytes, we first examined the 1121-bp mtDNA control region, known to contain multiple mutational hot spots (Figure 1B).20,21 An average of 100 CD34 clones per donor was subjected to sequencing, resulting in a total number of 594 PB CD34 clones from the 6 donors (Table 2). A total of 355 single granulocytes from the same 6 donors were used directly for sequencing analysis to determine evidence for clonal expansion of progenitor cells bearing polymorphisms (Table 2). These sequences were compared with the 611 BM CD34 clones examined in our previous work.13
Analysis of 594 circulating CD34clones revealed that a total of 150 clones (25.3% ± 8.2%) had mtDNA heterogeneity distinct from the donor's corresponding aggregate mtDNA sequences (Tables 6-7). The frequent patterns of mtDNA heterogeneity in circulating CD34 clones consisted of 1 or 2 nucleotide changes (substitutions, insertions, or deletions) in addition to the polymorphisms detected in the respective aggregate mtDNA. Among them, most differences were due to single nucleotide transitions at various positions and length alterations in the homopolymeric C tract localized between nucleotides 303 to 315 (Figure 2A). The heterogeneous mtDNA of circulating CD34 clones from the 6 donors was classified into several polymorphic patterns according to the following nucleotide changes; 5, 3, 2, 5, 9, and 6 patterns in donors 1 to 6, respectively (Table 6). The mean proportion of unique polymorphisms in the mtDNA among PB CD34 clones was 5.1% ± 2.4% (Table 6).
Mutational spectra of mtDNA control region in individual CD34 clones and single granulocytes
. | BM CD34 clones‡ . | . | . | . | PB CD34 clones . | . | . | . | Single granulocytes . | . | . | . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | mtDNA sequence differences from aggregate sequence* . | . | Heterogeneity . | . | mtDNA sequence differences from aggregate sequence* . | . | Heterogeneity . | . | mtDNA sequence differences from aggregate sequence* . | . | Heterogeneity . | . | |||||||||
| . | . | Clone no. . | Frequency, no. (%) . | Unique†no. (%) . | . | Clone no. . | Frequency, no. (%) . | Unique†no. (%) . | . | Clone no. . | Frequency, no. (%) . | Unique†no. (%) . | |||||||||
| 1 (47/F) | |||||||||||||||||||||
| Aggregate sequence | 85 | 30 (26.1) | 8 (7.0) | Aggregate sequence | 90 | 30 (25.0) | 5 (4.2) | Aggregate sequence | 39 | 12 (23.1) | 6 (11.5) | ||||||||||
| + 8CT6C§, 9CT6C§ | 22 | — | — | + 8CT6C§, 9CT6C§ | 23 | — | — | + 9CT6C§, 10CT6C§ | 7 | — | — | ||||||||||
| + 9CT6C§ | 2 | — | — | + 9CT6C§, 10CT6C§ | 4 | — | — | + 7CT6C§ | 1 | — | — | ||||||||||
| + 7CT6C§ | 1 | — | — | + 16129G>A/G | 1 | — | — | + 10CT6C§, 11CT6C | 1 | — | — | ||||||||||
| + 189A>G/A | 1 | — | — | + 16129G>A | 1 | — | — | + 146T>C | 1 | — | — | ||||||||||
| + 204T>C | 1 | — | — | + 16256C>T/C | 1 | — | — | + 349C>T/C, 368A>G | 1 | — | — | ||||||||||
| + 277C>T | 1 | — | — | Subtotal | 120 | — | — | + 369C>T | 1 | — | — | ||||||||||
| + ins 514CA | 1 | — | — | Subtotal | 51 | — | — | ||||||||||||||
| + 16114C>T | 1 | — | — | ||||||||||||||||||
| Subtotal | 115 | — | — | ||||||||||||||||||
| 2 (38/F) | |||||||||||||||||||||
| Aggregate sequence | 27 | 9 (25.0) | 5 (13.9) | Aggregate sequence | 85 | 9 (9.6) | 3 (3.2) | Aggregate sequence | 24 | 13 (35.1) | 3 (8.1) | ||||||||||
| + 16184C>CC (11C) | 5 | — | — | + 16184C>CC (11C) | 5 | — | — | + 16184C>CC (11C) | 11 | — | — | ||||||||||
| + 16131T>C/T | 1 | — | — | + 16184C>CC | 3 | — | — | + 16184C>CC C | 1 | — | — | ||||||||||
| CC(13)C | (13C) | ||||||||||||||||||||
| + 16145G>A | 1 | — | — | + 478A>G/A | 1 | — | — | + 16459C>T | 1 | — | — | ||||||||||
| + | 1 | — | — | Subtotal | 94 | — | — | Subtotal | 37 | — | — | ||||||||||
| 16184C>CCCC (13C) | |||||||||||||||||||||
| 73A>G, 263A>G, 191A>AA, 194C>T, 199T>C, 207G>A, 8CT6C§, 489T>C, 16147C>T, 16173C>T, 16245C>T, 16362T>C | 1 | — | — | ||||||||||||||||||
| Subtotal | 36 | — | — | ||||||||||||||||||
| 3 (43/M) | |||||||||||||||||||||
| Aggregate sequence | 96 | 24 (20.0) | 7 (5.8) | Aggregate sequence | 66 | 27 (29.0) | 2 (2.2) | Aggregate sequence | 34 | 16 (32.0) | 10 (20.0) | ||||||||||
| + 9CT6C§, 10CT6C§* | 11 | — | — | + 9CT6C§, 10CT6C§ | 26 | — | — | + 9CT6C§, 10CT6C§ | 4 | — | — | ||||||||||
| + 9CT6C§ | 6 | — | — | + 8CT6C§, 9CT6C§, 16124T>C/T | 1 | — | — | + 8CT6C§ | 5 | — | — | ||||||||||
| + 8CT6C§ | 2 | — | — | Subtotal | 93 | — | — | + 8CT6C§, 9CT6C§, 104C>T | 1 | — | — | ||||||||||
| + 182C>T/C, 8CT6C§, 9CT6C§ | 2 | — | — | + 8CT6C§, 9CT6C§, 181A>A/G | 1 | — | — | ||||||||||||||
| + del 71G, 9CT6C§, 10CT6C§ | 1 | — | — | + 8CT6C§, 9CT6C§, 296C>C/T | 1 | — | — | ||||||||||||||
| + 279T>C/T | 1 | — | — | + 8CT6C§, 9CT6C§, 16339C>T | 1 | — | — | ||||||||||||||
| + 16153G>A | 1 | — | — | + 8CT6C§, 9CT6C§, 16352T>T/C | 1 | — | — | ||||||||||||||
| Subtotal | 120 | — | — | + 9CT6C§, 10CT6C§, 225G>A, 16263T>C | 1 | — | — | ||||||||||||||
| + 8CT6C§, 9CT6C§, 16035G>A | 1 | — | — | ||||||||||||||||||
| Subtotal | 50 | — | — | ||||||||||||||||||
| 4 (34/M) | |||||||||||||||||||||
| Aggregate sequence | 92 | 19 (17.1) | 6 (5.4) | Aggregate sequence | 70 | 25 (26.3) | 5 (5.3) | Aggregate sequence | 77 | 34 (30.6) | 16 (14.4) | ||||||||||
| + 8CT6C§, 9CT6C§ | 11 | — | — | + 8CT6C§, 9CT6C§ | 16 | — | — | + 8CT6C§, 9CT6C§ | 6 | — | — | ||||||||||
| + del514-515CA, ins 514CA∥ | 3 | — | — | + 7CT6C§, 8CT6C§ | 3 | — | — | + 7CT6C§, 8CT6C§ | 3 | — | — | ||||||||||
| + 7CT6C§, 8CT6C§ | 2 | — | — | + 16093T>T/C | 4 | — | — | + 9CT6C§, 10CT6C§ | 6 | — | — | ||||||||||
| + 9CT6C§, 10CT6C§ | 1 | — | — | +8CT6C§, 9CT6C§, 16044T>C/T | 1 | — | — | + 9CT6C§, 10CT6C§, 16093T>T/C | 2 | — | — | ||||||||||
| + 89T>C | 1 | — | — | + 8CT6C§, 8CT7C | 1 | — | — | + 16093T>T/C | 3 | — | — | ||||||||||
| + 8CT6C§, 9CT6C§, 16093T∥ | 1 | — | — | Subtotal | 95 | — | — | + 16093T∥ | 4 | — | — | ||||||||||
| Subtotal | 111 | — | — | + 16093T∥, 16208G>A | 1 | — | — | ||||||||||||||
| + 28A>G/A | 1 | — | — | ||||||||||||||||||
| + 8CT6C§, 9CT6C, 185G>G/A | 1 | — | — | ||||||||||||||||||
| + 195T>T/C | 1 | — | — | ||||||||||||||||||
| + 195T>C, 16175A>C | 1 | — | — | ||||||||||||||||||
| + 277C>T | 1 | — | — | ||||||||||||||||||
| + 315C>CT, 315C∥ | 1 | — | — | ||||||||||||||||||
| + 411C>T | 1 | — | — | ||||||||||||||||||
| + del 452T | 1 | — | — | ||||||||||||||||||
| + 469C>T, 16093T>T/C | 1 | — | — | ||||||||||||||||||
| Subtotal | 111 | — | — | ||||||||||||||||||
| 5 (54/M) | |||||||||||||||||||||
| Aggregate sequence | 57 | 57 (50.0) | 14 (12.3) | Aggregate sequence | 64 | 32 (33.3) | 9 (9.4) | Aggregate sequence | 22 | 20 (47.6) | 14 (33.3) | ||||||||||
| + ins 514CA∥ | 19 | — | — | + ins 514CA∥ | 11 | — | — | + 264C>T | 7 | — | — | ||||||||||
| + 264C>T | 18 | — | — | + 264C>T | 10 | — | — | + 146T>C | 1 | — | — | ||||||||||
| + del514-515CA, ins 514CA∥ | 5 | — | — | + 264C>T/C | 3 | — | — | + 7CT6C§, 8CT6C§ | 1 | — | — | ||||||||||
| + 264C>T/C | 3 | — | — | + 146T>C/T, ins 514CA∥ | 4 | — | — | + 8CT6C | 1 | — | — | ||||||||||
| + 7CT6C§, 8CT6C§ | 2 | — | — | + 94G>A/G | 1 | — | — | + 321T>T/C | 1 | — | — | ||||||||||
| + 146T>C, ins 514CA∥ | 2 | — | — | + 16189T>C | 1 | — | — | + 376A>G | 1 | — | — | ||||||||||
| + 146T>C, 264C>T/C | 1 | — | — | + 16265A>G/A | 1 | — | — | + 582T>T/C, 16075T>C/T | 1 | — | — | ||||||||||
| + 146T>C/T, ins | 1 | — | — | + 357A>AA, ins | 1 | — | — | + 1605T>C/T | 1 | — | — | ||||||||||
| 514CA∥ | 514CA∥, | ||||||||||||||||||||
| 16136T>C/T | |||||||||||||||||||||
| + 146T>C | 1 | — | — | Subtotal | 96 | — | — | + 16098A>A/G | 1 | — | — | ||||||||||
| + 189A>G | 1 | — | — | + 16259C>G | 1 | — | — | ||||||||||||||
| + 264C>T/C, ins | 1 | — | — | + 16274G>A, | 1 | — | — | ||||||||||||||
| 514CA∥ | 16296C>T/C | ||||||||||||||||||||
| + 161T>C/T, | 1 | — | — | + 16292C>T | 1 | — | — | ||||||||||||||
| 264C>T, ins | |||||||||||||||||||||
| 514CA∥ | |||||||||||||||||||||
| + 16189T>C | 1 | — | — | + 16322C>T, | 1 | — | — | ||||||||||||||
| 16366C>T | |||||||||||||||||||||
| + 16296C>C/T | 1 | — | — | + 16422T>C | 1 | — | — | ||||||||||||||
| Subtotal | 114 | — | — | Subtotal | 42 | — | — | ||||||||||||||
| 6 (34/F) | |||||||||||||||||||||
| Aggregate sequence | 102 | 13 (11.3) | 6 (5.2) | Aggregate sequence | 69 | 27 (28.1) | 6 (6.3) | Aggregate sequence | 56 | 8 (12.5) | 5 (7.8) | ||||||||||
| + 8CT6C§, | 5 | — | — | + 8CT6C§, | 18 | — | — | + 8CT6C§, | 4 | — | — | ||||||||||
| 9CT6C§ | 9CT6C§ | 9CT6C§ | |||||||||||||||||||
| + 200A>G/A | 3 | — | — | + 200A>G/A | 4 | — | — | + 28A>G | 1 | — | — | ||||||||||
| + 200A>G | 2 | — | — | + 200A>G | 1 | — | — | + 571C>A | 1 | — | — | ||||||||||
| + 200A>G/A, | 1 | — | — | + 200A>G/A, | 2 | — | — | + 16080A>G/A | 1 | — | — | ||||||||||
| 7CT6C§, | 8CT6C§, | ||||||||||||||||||||
| 8CT6C§ | 9CT6C§ | ||||||||||||||||||||
| + 200A>G/A, | 1 | — | — | + 234A>G/A | 1 | — | — | + 16412G>G/A | 1 | — | — | ||||||||||
| 8CT6C§, | |||||||||||||||||||||
| 9CT6C§ | |||||||||||||||||||||
| + 7CT6C§, | 1 | — | — | + 16111C>T/C | 1 | — | — | Subtotal | 64 | — | — | ||||||||||
| 8CT6C§ | |||||||||||||||||||||
| Subtotal | 115 | — | — | Subtotal | 96 | — | — | ||||||||||||||
| Total | 611 | 152 (24.9) | 46 (7.5) | 594 | 150 (25.3) | 30 (5.1) | 355 | 103 (29.0) | 54 (15.2) | ||||||||||||
. | BM CD34 clones‡ . | . | . | . | PB CD34 clones . | . | . | . | Single granulocytes . | . | . | . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | mtDNA sequence differences from aggregate sequence* . | . | Heterogeneity . | . | mtDNA sequence differences from aggregate sequence* . | . | Heterogeneity . | . | mtDNA sequence differences from aggregate sequence* . | . | Heterogeneity . | . | |||||||||
| . | . | Clone no. . | Frequency, no. (%) . | Unique†no. (%) . | . | Clone no. . | Frequency, no. (%) . | Unique†no. (%) . | . | Clone no. . | Frequency, no. (%) . | Unique†no. (%) . | |||||||||
| 1 (47/F) | |||||||||||||||||||||
| Aggregate sequence | 85 | 30 (26.1) | 8 (7.0) | Aggregate sequence | 90 | 30 (25.0) | 5 (4.2) | Aggregate sequence | 39 | 12 (23.1) | 6 (11.5) | ||||||||||
| + 8CT6C§, 9CT6C§ | 22 | — | — | + 8CT6C§, 9CT6C§ | 23 | — | — | + 9CT6C§, 10CT6C§ | 7 | — | — | ||||||||||
| + 9CT6C§ | 2 | — | — | + 9CT6C§, 10CT6C§ | 4 | — | — | + 7CT6C§ | 1 | — | — | ||||||||||
| + 7CT6C§ | 1 | — | — | + 16129G>A/G | 1 | — | — | + 10CT6C§, 11CT6C | 1 | — | — | ||||||||||
| + 189A>G/A | 1 | — | — | + 16129G>A | 1 | — | — | + 146T>C | 1 | — | — | ||||||||||
| + 204T>C | 1 | — | — | + 16256C>T/C | 1 | — | — | + 349C>T/C, 368A>G | 1 | — | — | ||||||||||
| + 277C>T | 1 | — | — | Subtotal | 120 | — | — | + 369C>T | 1 | — | — | ||||||||||
| + ins 514CA | 1 | — | — | Subtotal | 51 | — | — | ||||||||||||||
| + 16114C>T | 1 | — | — | ||||||||||||||||||
| Subtotal | 115 | — | — | ||||||||||||||||||
| 2 (38/F) | |||||||||||||||||||||
| Aggregate sequence | 27 | 9 (25.0) | 5 (13.9) | Aggregate sequence | 85 | 9 (9.6) | 3 (3.2) | Aggregate sequence | 24 | 13 (35.1) | 3 (8.1) | ||||||||||
| + 16184C>CC (11C) | 5 | — | — | + 16184C>CC (11C) | 5 | — | — | + 16184C>CC (11C) | 11 | — | — | ||||||||||
| + 16131T>C/T | 1 | — | — | + 16184C>CC | 3 | — | — | + 16184C>CC C | 1 | — | — | ||||||||||
| CC(13)C | (13C) | ||||||||||||||||||||
| + 16145G>A | 1 | — | — | + 478A>G/A | 1 | — | — | + 16459C>T | 1 | — | — | ||||||||||
| + | 1 | — | — | Subtotal | 94 | — | — | Subtotal | 37 | — | — | ||||||||||
| 16184C>CCCC (13C) | |||||||||||||||||||||
| 73A>G, 263A>G, 191A>AA, 194C>T, 199T>C, 207G>A, 8CT6C§, 489T>C, 16147C>T, 16173C>T, 16245C>T, 16362T>C | 1 | — | — | ||||||||||||||||||
| Subtotal | 36 | — | — | ||||||||||||||||||
| 3 (43/M) | |||||||||||||||||||||
| Aggregate sequence | 96 | 24 (20.0) | 7 (5.8) | Aggregate sequence | 66 | 27 (29.0) | 2 (2.2) | Aggregate sequence | 34 | 16 (32.0) | 10 (20.0) | ||||||||||
| + 9CT6C§, 10CT6C§* | 11 | — | — | + 9CT6C§, 10CT6C§ | 26 | — | — | + 9CT6C§, 10CT6C§ | 4 | — | — | ||||||||||
| + 9CT6C§ | 6 | — | — | + 8CT6C§, 9CT6C§, 16124T>C/T | 1 | — | — | + 8CT6C§ | 5 | — | — | ||||||||||
| + 8CT6C§ | 2 | — | — | Subtotal | 93 | — | — | + 8CT6C§, 9CT6C§, 104C>T | 1 | — | — | ||||||||||
| + 182C>T/C, 8CT6C§, 9CT6C§ | 2 | — | — | + 8CT6C§, 9CT6C§, 181A>A/G | 1 | — | — | ||||||||||||||
| + del 71G, 9CT6C§, 10CT6C§ | 1 | — | — | + 8CT6C§, 9CT6C§, 296C>C/T | 1 | — | — | ||||||||||||||
| + 279T>C/T | 1 | — | — | + 8CT6C§, 9CT6C§, 16339C>T | 1 | — | — | ||||||||||||||
| + 16153G>A | 1 | — | — | + 8CT6C§, 9CT6C§, 16352T>T/C | 1 | — | — | ||||||||||||||
| Subtotal | 120 | — | — | + 9CT6C§, 10CT6C§, 225G>A, 16263T>C | 1 | — | — | ||||||||||||||
| + 8CT6C§, 9CT6C§, 16035G>A | 1 | — | — | ||||||||||||||||||
| Subtotal | 50 | — | — | ||||||||||||||||||
| 4 (34/M) | |||||||||||||||||||||
| Aggregate sequence | 92 | 19 (17.1) | 6 (5.4) | Aggregate sequence | 70 | 25 (26.3) | 5 (5.3) | Aggregate sequence | 77 | 34 (30.6) | 16 (14.4) | ||||||||||
| + 8CT6C§, 9CT6C§ | 11 | — | — | + 8CT6C§, 9CT6C§ | 16 | — | — | + 8CT6C§, 9CT6C§ | 6 | — | — | ||||||||||
| + del514-515CA, ins 514CA∥ | 3 | — | — | + 7CT6C§, 8CT6C§ | 3 | — | — | + 7CT6C§, 8CT6C§ | 3 | — | — | ||||||||||
| + 7CT6C§, 8CT6C§ | 2 | — | — | + 16093T>T/C | 4 | — | — | + 9CT6C§, 10CT6C§ | 6 | — | — | ||||||||||
| + 9CT6C§, 10CT6C§ | 1 | — | — | +8CT6C§, 9CT6C§, 16044T>C/T | 1 | — | — | + 9CT6C§, 10CT6C§, 16093T>T/C | 2 | — | — | ||||||||||
| + 89T>C | 1 | — | — | + 8CT6C§, 8CT7C | 1 | — | — | + 16093T>T/C | 3 | — | — | ||||||||||
| + 8CT6C§, 9CT6C§, 16093T∥ | 1 | — | — | Subtotal | 95 | — | — | + 16093T∥ | 4 | — | — | ||||||||||
| Subtotal | 111 | — | — | + 16093T∥, 16208G>A | 1 | — | — | ||||||||||||||
| + 28A>G/A | 1 | — | — | ||||||||||||||||||
| + 8CT6C§, 9CT6C, 185G>G/A | 1 | — | — | ||||||||||||||||||
| + 195T>T/C | 1 | — | — | ||||||||||||||||||
| + 195T>C, 16175A>C | 1 | — | — | ||||||||||||||||||
| + 277C>T | 1 | — | — | ||||||||||||||||||
| + 315C>CT, 315C∥ | 1 | — | — | ||||||||||||||||||
| + 411C>T | 1 | — | — | ||||||||||||||||||
| + del 452T | 1 | — | — | ||||||||||||||||||
| + 469C>T, 16093T>T/C | 1 | — | — | ||||||||||||||||||
| Subtotal | 111 | — | — | ||||||||||||||||||
| 5 (54/M) | |||||||||||||||||||||
| Aggregate sequence | 57 | 57 (50.0) | 14 (12.3) | Aggregate sequence | 64 | 32 (33.3) | 9 (9.4) | Aggregate sequence | 22 | 20 (47.6) | 14 (33.3) | ||||||||||
| + ins 514CA∥ | 19 | — | — | + ins 514CA∥ | 11 | — | — | + 264C>T | 7 | — | — | ||||||||||
| + 264C>T | 18 | — | — | + 264C>T | 10 | — | — | + 146T>C | 1 | — | — | ||||||||||
| + del514-515CA, ins 514CA∥ | 5 | — | — | + 264C>T/C | 3 | — | — | + 7CT6C§, 8CT6C§ | 1 | — | — | ||||||||||
| + 264C>T/C | 3 | — | — | + 146T>C/T, ins 514CA∥ | 4 | — | — | + 8CT6C | 1 | — | — | ||||||||||
| + 7CT6C§, 8CT6C§ | 2 | — | — | + 94G>A/G | 1 | — | — | + 321T>T/C | 1 | — | — | ||||||||||
| + 146T>C, ins 514CA∥ | 2 | — | — | + 16189T>C | 1 | — | — | + 376A>G | 1 | — | — | ||||||||||
| + 146T>C, 264C>T/C | 1 | — | — | + 16265A>G/A | 1 | — | — | + 582T>T/C, 16075T>C/T | 1 | — | — | ||||||||||
| + 146T>C/T, ins | 1 | — | — | + 357A>AA, ins | 1 | — | — | + 1605T>C/T | 1 | — | — | ||||||||||
| 514CA∥ | 514CA∥, | ||||||||||||||||||||
| 16136T>C/T | |||||||||||||||||||||
| + 146T>C | 1 | — | — | Subtotal | 96 | — | — | + 16098A>A/G | 1 | — | — | ||||||||||
| + 189A>G | 1 | — | — | + 16259C>G | 1 | — | — | ||||||||||||||
| + 264C>T/C, ins | 1 | — | — | + 16274G>A, | 1 | — | — | ||||||||||||||
| 514CA∥ | 16296C>T/C | ||||||||||||||||||||
| + 161T>C/T, | 1 | — | — | + 16292C>T | 1 | — | — | ||||||||||||||
| 264C>T, ins | |||||||||||||||||||||
| 514CA∥ | |||||||||||||||||||||
| + 16189T>C | 1 | — | — | + 16322C>T, | 1 | — | — | ||||||||||||||
| 16366C>T | |||||||||||||||||||||
| + 16296C>C/T | 1 | — | — | + 16422T>C | 1 | — | — | ||||||||||||||
| Subtotal | 114 | — | — | Subtotal | 42 | — | — | ||||||||||||||
| 6 (34/F) | |||||||||||||||||||||
| Aggregate sequence | 102 | 13 (11.3) | 6 (5.2) | Aggregate sequence | 69 | 27 (28.1) | 6 (6.3) | Aggregate sequence | 56 | 8 (12.5) | 5 (7.8) | ||||||||||
| + 8CT6C§, | 5 | — | — | + 8CT6C§, | 18 | — | — | + 8CT6C§, | 4 | — | — | ||||||||||
| 9CT6C§ | 9CT6C§ | 9CT6C§ | |||||||||||||||||||
| + 200A>G/A | 3 | — | — | + 200A>G/A | 4 | — | — | + 28A>G | 1 | — | — | ||||||||||
| + 200A>G | 2 | — | — | + 200A>G | 1 | — | — | + 571C>A | 1 | — | — | ||||||||||
| + 200A>G/A, | 1 | — | — | + 200A>G/A, | 2 | — | — | + 16080A>G/A | 1 | — | — | ||||||||||
| 7CT6C§, | 8CT6C§, | ||||||||||||||||||||
| 8CT6C§ | 9CT6C§ | ||||||||||||||||||||
| + 200A>G/A, | 1 | — | — | + 234A>G/A | 1 | — | — | + 16412G>G/A | 1 | — | — | ||||||||||
| 8CT6C§, | |||||||||||||||||||||
| 9CT6C§ | |||||||||||||||||||||
| + 7CT6C§, | 1 | — | — | + 16111C>T/C | 1 | — | — | Subtotal | 64 | — | — | ||||||||||
| 8CT6C§ | |||||||||||||||||||||
| Subtotal | 115 | — | — | Subtotal | 96 | — | — | ||||||||||||||
| Total | 611 | 152 (24.9) | 46 (7.5) | 594 | 150 (25.3) | 30 (5.1) | 355 | 103 (29.0) | 54 (15.2) | ||||||||||||
Different indicates different from aggregate cell sequence; BM, bone marrow; PB, peripheral blood; and—, not applicable.
The same results are taken from our recent publication.13
The bold sequences indicate identity of sequence alterations in progeny granulocytes and either BM or PB CD34+ progenitors; +, mtDNA nucleotide changes compared with aggregate cell mtDNA sequence.
Unique indicates uniquely different heterogeneity.
Homopolymeric C tract localized between nucleotides 303 and 315 (eg, 8CT6C defined CCCCCCCCTCCCCCC).
Sequence is the same as the Revised Cambridge Reference Sequence but different from the aggregate sequence.
Summary of mtDNA mutations of CD34 clones and single granulocytes
. | . | Heterogeneity (mutation) . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | Poly C tract . | . | . | . | ||||||
| Specimens . | Assay no. (donor no.) . | Total rate, % (no.) . | Unique, % (no.) . | Substitution, % (no.) . | np 303-315, % (no.) . | np 16184-16189, % (no.) . | NC, % (no.) . | AA change, % (no.) . | ||||||
| CB CD34* | 580 (5) | 1.6 (9) | 1.2 (7) | 0.0 (0) | 0.0 (0) | 0.5 (3) | 0.7 (4) | — | ||||||
| Adult BM CD34* | 611 (6) | 24.9 (152) | 7.5 (46) | 10.5 (64) | 10.8 (66) | 1.1 (7) | 0.0 (0) | — | ||||||
| Adult PB CD34 | 594 (6) | 25.3 (150) | 5.1 (30) | 6.1 (36) | 15.3 (91) | 1.5 (9) | 0.0 (0) | — | ||||||
| Single granulocyte | 355 (6) | 29.0 (103) | 15.2 (54) | 11.8 (42) | 11.0 (39) | 3.4 (12) | 0.0 (0) | — | ||||||
| Adult BM CD34 | 285 (3) | 3.5 (10) | 3.5 (10) | 3.5 (10) | — | — | — | 63.6 (7/11†) | ||||||
| Adult PB CD34 | 284 (3) | 5.3 (15) | 4.2 (12) | 5.3 (15) | — | — | — | 50.0 (8/16†) | ||||||
| Subtotal | 569 (6) | 4.4 (25) | 3.9 (22) | 4.4 (25) | — | — | — | 55.6 (15/27†) | ||||||
. | . | Heterogeneity (mutation) . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | Poly C tract . | . | . | . | ||||||
| Specimens . | Assay no. (donor no.) . | Total rate, % (no.) . | Unique, % (no.) . | Substitution, % (no.) . | np 303-315, % (no.) . | np 16184-16189, % (no.) . | NC, % (no.) . | AA change, % (no.) . | ||||||
| CB CD34* | 580 (5) | 1.6 (9) | 1.2 (7) | 0.0 (0) | 0.0 (0) | 0.5 (3) | 0.7 (4) | — | ||||||
| Adult BM CD34* | 611 (6) | 24.9 (152) | 7.5 (46) | 10.5 (64) | 10.8 (66) | 1.1 (7) | 0.0 (0) | — | ||||||
| Adult PB CD34 | 594 (6) | 25.3 (150) | 5.1 (30) | 6.1 (36) | 15.3 (91) | 1.5 (9) | 0.0 (0) | — | ||||||
| Single granulocyte | 355 (6) | 29.0 (103) | 15.2 (54) | 11.8 (42) | 11.0 (39) | 3.4 (12) | 0.0 (0) | — | ||||||
| Adult BM CD34 | 285 (3) | 3.5 (10) | 3.5 (10) | 3.5 (10) | — | — | — | 63.6 (7/11†) | ||||||
| Adult PB CD34 | 284 (3) | 5.3 (15) | 4.2 (12) | 5.3 (15) | — | — | — | 50.0 (8/16†) | ||||||
| Subtotal | 569 (6) | 4.4 (25) | 3.9 (22) | 4.4 (25) | — | — | — | 55.6 (15/27†) | ||||||
Np indicates nucleotide position; NC, nucleotide change; AA, amino acid; CB, cord blood; and —, not applicable.
Our recent publication data.13
Total number of mutational events.
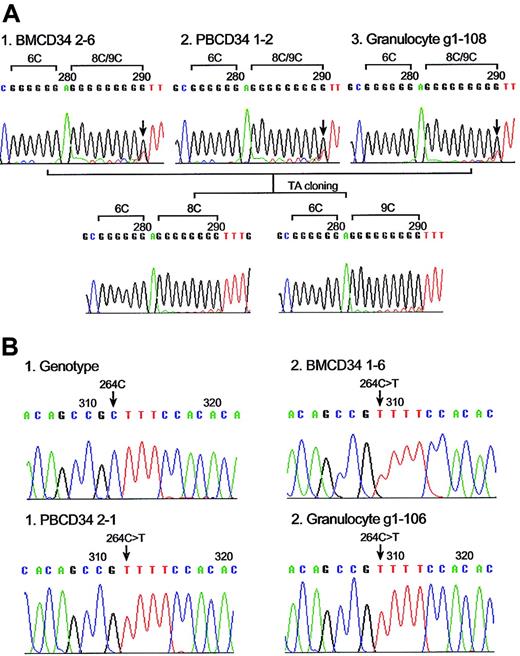
mtDNA mutations and clonal expansions of individual CD34 clones. (A) Reverse-sequence analysis of BM CD34 (clone number 2-6), PB CD34 (clone number 1-2), and single granulocyte (number g1-108) mtDNA from donor 4 showed the same poly C length heteroplasmy (mixture of 8CT6C and 9CT6C) between nucleotide position 303 to 315. Reverse-sequence analysis after TA cloning revealed poly C length heteroplasmy of 8CT6C (bottom left) and 9CT6C (bottom right). (B) Clonal expansion of substitution mutation at mtDNA nucleotide position 264. Sequence chromatogram with aggregate genotype (i) and 264C>T mutation was found in BM CD34 clone (number 1-6) (ii), PB CD34 clone (number 2-1) (iii), and single granulocyte (number g1-106) (iv) from donor 5.
mtDNA mutations and clonal expansions of individual CD34 clones. (A) Reverse-sequence analysis of BM CD34 (clone number 2-6), PB CD34 (clone number 1-2), and single granulocyte (number g1-108) mtDNA from donor 4 showed the same poly C length heteroplasmy (mixture of 8CT6C and 9CT6C) between nucleotide position 303 to 315. Reverse-sequence analysis after TA cloning revealed poly C length heteroplasmy of 8CT6C (bottom left) and 9CT6C (bottom right). (B) Clonal expansion of substitution mutation at mtDNA nucleotide position 264. Sequence chromatogram with aggregate genotype (i) and 264C>T mutation was found in BM CD34 clone (number 1-6) (ii), PB CD34 clone (number 2-1) (iii), and single granulocyte (number g1-106) (iv) from donor 5.
One hundred three of 355 single granulocytes (29.0% ± 9.2%) from the same 6 donors showed mtDNA heterogeneity distinct from the mtDNA sequences of the donor's corresponding aggregate and other single granulocytes (Table 6). The mean proportion of mtDNA polymorphic pattern among single granulocytes was 15.2% (15.2% ± 5.2%).
Mutations in the CO1 and Cytb gene coding regions
We chose CO1 and Cytb as representative coding regions to determine whether the detected mtDNA heterogeneity was restricted to the hypervariable regions or present throughout the mtDNA genome, as well as to assess the possible functional significance of mtDNA mutations in hematopoietic cells. Analysis of 569 CD34 clones from BM (285 clones) and PB (284 clones) of 3 of the 6 donors showed a prevalence of 4.4% (25 clones) with sequences different from the donors' corresponding aggregate sequences (Table 8). All these mutations were base substitutions and mostly transitions. The mean proportion of unique mtDNA mutations was 3.9% (22 clones); half of these substitutions led to amino acid changes (Tables 7-8).
Mutational spectra of mtDNA CO1 and Cytb genes in individual CD34 clones from BM and PB
. | BM CD34 clones . | . | . | . | . | . | . | PB CD34 clones . | . | . | . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | . | . | . | . | . | Heterogeneity . | . | . | . | . | . | . | Heterogeneity . | . | ||||||||||||
| . | mtDNA sequence* . | Gene . | Amino acid change . | Interpretation . | Clone no. . | Frequency, no. (%) . | Unique, no. (%) . | mtDNA sequence* . | Gene . | Amino acid change . | Interpretation . | Clone no. . | Frequency, no. (%) . | Unique, no. (%) . | ||||||||||||
| 1 (47/F) | ||||||||||||||||||||||||||
| Aggregate sequence | 92 | 2 (2.1) | 2 (2.1) | Aggregate sequence | 93 | 2 (2.1) | 2 (2.1) | |||||||||||||||||||
| + 7071T>C/T | CO1 | Met-Thr | Mutation/TS | 1 | — | — | + 6952T>C | CO1 | Val-Ala | Mutation/TS | 1 | — | — | |||||||||||||
| + 14924T>C | Cytb | Ser-Pro | Mutation/TS | 1 | — | — | + 7312T>C | CO1 | Phe-Ser | Mutation/TS | 1 | — | — | |||||||||||||
| Subtotal | 94 | — | — | Subtotal | 95 | — | — | |||||||||||||||||||
| 3 (43/M) | ||||||||||||||||||||||||||
| Aggregate sequence | 92 | 3 (3.2) | 3 (3.2) | Aggregate sequence | 90 | 3 (3.2) | 3 (3.2) | |||||||||||||||||||
| + 6955G>G/A | CO1 | Gly-Asp | Mutation/TS | 1 | — | — | + 6955G>G/A | CO1 | Gly-Asp | Mutation/TS | 1 | — | — | |||||||||||||
| + 6987T>C/T | CO1 | Ser-Pro | Mutation/TS | 1 | — | — | + 14935T>C | Cytb | Silent | Mutation/TS | 1 | — | — | |||||||||||||
| + 7110T>T/C | CO1 | Tyr-His | Mutation/TS | 1 | — | — | + 15724A>G/A | Cytb | Silent | Polymorphism/TS | 1 | — | — | |||||||||||||
| Subtotal | 95 | — | — | Subtotal | 93 | — | — | |||||||||||||||||||
| 5 (54/M) | ||||||||||||||||||||||||||
| Aggregate sequence | 91 | 5 (5.2) | 5 (5.2) | Aggregate sequence | 86 | 10 (10.4) | 7 (7.3) | |||||||||||||||||||
| + 6711T>G/T | CO1 | Tyr-Asp | Mutation/TV | 1 | — | — | + 7022T>T/C | CO1 | Silent | Polymorphism/TS | 4 | — | — | |||||||||||||
| + 6899G>A/G | CO1 | Silent | Mutation/TS | 1 | — | — | + 7075G>A/G | CO1 | Silent | Mutation/TS | 1 | — | — | |||||||||||||
| + 7022T>C/T | CO1 | Silent | Polymorphism/TS | 1 | — | — | + 14864T>C | Cytb | Cys-Arg | Mutation/TS | 1 | — | — | |||||||||||||
| + 7297T>C/T | CO1 | Val-Ala | Mutation/TS | 1 | — | — | + 15132T>C | Cytb | Met-Thr | Mutation/TS | 1 | — | — | |||||||||||||
| + 15607A>A/G, 14905G>G/A | Cytb | Silent | Polymorphism/TS | 1 | — | — | + 15818T>C | Cytb | Tyr-His | Mutation/TS | 1 | — | — | |||||||||||||
| Subtotal | 96 | — | — | + 15452C>C/A | Cytb | Leu-lle | Polymorphism/TV | 1 | — | — | ||||||||||||||||
| 15607A>A/G | Cytb | Silent | Polymorphism/TS | — | — | |||||||||||||||||||||
| + 14905G>G/A | Cytb | Silent | Polymorphism/TS | 1 | — | — | ||||||||||||||||||||
| 15067T>T/C | Cytb | Silent | Polymorphism/TS | — | — | |||||||||||||||||||||
| 15452C>C/A | Cytb | Leu-lle | Polymorphism/TS | — | — | |||||||||||||||||||||
| 15607A>A/G | Cytb | Silent | Polymorphism/TS | — | — | |||||||||||||||||||||
| Subtotal | 96 | — | — | |||||||||||||||||||||||
| Total | 285 | 10 (3.5) | 10 (3.5) | 284 | 15 (5.3) | 12 (4.2) | ||||||||||||||||||||
| Heterogeneity of mtDNA CO1 and Cytb gene in total CD34+ clones from BM and PB | 569 | 25 (4.4) | 22 (3.9) | |||||||||||||||||||||||
. | BM CD34 clones . | . | . | . | . | . | . | PB CD34 clones . | . | . | . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor (age, y/sex) . | . | . | . | . | . | Heterogeneity . | . | . | . | . | . | . | Heterogeneity . | . | ||||||||||||
| . | mtDNA sequence* . | Gene . | Amino acid change . | Interpretation . | Clone no. . | Frequency, no. (%) . | Unique, no. (%) . | mtDNA sequence* . | Gene . | Amino acid change . | Interpretation . | Clone no. . | Frequency, no. (%) . | Unique, no. (%) . | ||||||||||||
| 1 (47/F) | ||||||||||||||||||||||||||
| Aggregate sequence | 92 | 2 (2.1) | 2 (2.1) | Aggregate sequence | 93 | 2 (2.1) | 2 (2.1) | |||||||||||||||||||
| + 7071T>C/T | CO1 | Met-Thr | Mutation/TS | 1 | — | — | + 6952T>C | CO1 | Val-Ala | Mutation/TS | 1 | — | — | |||||||||||||
| + 14924T>C | Cytb | Ser-Pro | Mutation/TS | 1 | — | — | + 7312T>C | CO1 | Phe-Ser | Mutation/TS | 1 | — | — | |||||||||||||
| Subtotal | 94 | — | — | Subtotal | 95 | — | — | |||||||||||||||||||
| 3 (43/M) | ||||||||||||||||||||||||||
| Aggregate sequence | 92 | 3 (3.2) | 3 (3.2) | Aggregate sequence | 90 | 3 (3.2) | 3 (3.2) | |||||||||||||||||||
| + 6955G>G/A | CO1 | Gly-Asp | Mutation/TS | 1 | — | — | + 6955G>G/A | CO1 | Gly-Asp | Mutation/TS | 1 | — | — | |||||||||||||
| + 6987T>C/T | CO1 | Ser-Pro | Mutation/TS | 1 | — | — | + 14935T>C | Cytb | Silent | Mutation/TS | 1 | — | — | |||||||||||||
| + 7110T>T/C | CO1 | Tyr-His | Mutation/TS | 1 | — | — | + 15724A>G/A | Cytb | Silent | Polymorphism/TS | 1 | — | — | |||||||||||||
| Subtotal | 95 | — | — | Subtotal | 93 | — | — | |||||||||||||||||||
| 5 (54/M) | ||||||||||||||||||||||||||
| Aggregate sequence | 91 | 5 (5.2) | 5 (5.2) | Aggregate sequence | 86 | 10 (10.4) | 7 (7.3) | |||||||||||||||||||
| + 6711T>G/T | CO1 | Tyr-Asp | Mutation/TV | 1 | — | — | + 7022T>T/C | CO1 | Silent | Polymorphism/TS | 4 | — | — | |||||||||||||
| + 6899G>A/G | CO1 | Silent | Mutation/TS | 1 | — | — | + 7075G>A/G | CO1 | Silent | Mutation/TS | 1 | — | — | |||||||||||||
| + 7022T>C/T | CO1 | Silent | Polymorphism/TS | 1 | — | — | + 14864T>C | Cytb | Cys-Arg | Mutation/TS | 1 | — | — | |||||||||||||
| + 7297T>C/T | CO1 | Val-Ala | Mutation/TS | 1 | — | — | + 15132T>C | Cytb | Met-Thr | Mutation/TS | 1 | — | — | |||||||||||||
| + 15607A>A/G, 14905G>G/A | Cytb | Silent | Polymorphism/TS | 1 | — | — | + 15818T>C | Cytb | Tyr-His | Mutation/TS | 1 | — | — | |||||||||||||
| Subtotal | 96 | — | — | + 15452C>C/A | Cytb | Leu-lle | Polymorphism/TV | 1 | — | — | ||||||||||||||||
| 15607A>A/G | Cytb | Silent | Polymorphism/TS | — | — | |||||||||||||||||||||
| + 14905G>G/A | Cytb | Silent | Polymorphism/TS | 1 | — | — | ||||||||||||||||||||
| 15067T>T/C | Cytb | Silent | Polymorphism/TS | — | — | |||||||||||||||||||||
| 15452C>C/A | Cytb | Leu-lle | Polymorphism/TS | — | — | |||||||||||||||||||||
| 15607A>A/G | Cytb | Silent | Polymorphism/TS | — | — | |||||||||||||||||||||
| Subtotal | 96 | — | — | |||||||||||||||||||||||
| Total | 285 | 10 (3.5) | 10 (3.5) | 284 | 15 (5.3) | 12 (4.2) | ||||||||||||||||||||
| Heterogeneity of mtDNA CO1 and Cytb gene in total CD34+ clones from BM and PB | 569 | 25 (4.4) | 22 (3.9) | |||||||||||||||||||||||
CO1 indicates cytochrome c oxidase 1; Cytb, cytochrome b; BM, bone marrow; TS, transition; TV, transversion; PB, peripheral blood; and —, not applicable. Sequences in bold indicate the same changes seen in BM CD34 and PB CD34 clones.
Clonal expansion of hematopoietic cells bearing mtDNA mutations
Analysis of single granulocytes revealed that 63 cells of 355 (18%) had the same base alterations present in the donors' respective progenitor cell (BM CD34 and/or PB CD34 cells). In most cases, there were 1 or 2 mutations per cell. These data suggest that each of these mutations had expanded from a single initial mutational event. Most of the clonally expanded mutations present in granulocyte progeny represented length changes in the homopolymeric C tract between nucleotide 303 and 315 of hypervariable region 2 (Figure 2A). Clonal expansion of CD34 cells bearing nucleotide substitution mutations, such as 16093T>T/C and 264C>T, were observed in donors 4 and 5 (Table 6 and Figure 2B).
Characteristics of mtDNA mutations in CD34 cell clones and single granulocytes
On average, approximately 25% of the single granulocytes and CD34 cell clones from adult BM and PB differed from the aggregate mtDNA sequence of each specific donor (Table 7). In contrast, we previously reported that less than 2% of cord blood (CB) CD34 cell clones differed from their respective aggregate cell mtDNA sequence.13 The proportion of cells different from both the aggregate sequence and from other CD34 clones or granulocytes was 7.5%, 5.1%, and 15.2% in BM CD34, PB CD34, and single granulocytes, respectively. The proportion of solitary nucleotide substitutions in BM CD34 cells, circulating CD34 cells, and single granulocytes was similar (these substitutions were not seen in CB CD34 clones). As we had observed for BM CD34,13 most mutations in PB CD34 clones and single granulocytes were localized in the HV2 homopolymeric C tract between nucleotides 303 and 315 (60% or 91 of 151 in PB CD34 clones; 41% or 42 of 103 in single granulocytes; Table 7).
Discussion
Cells can contain thousands of mtDNA molecules, and, when nucleotide changes occur, they will appear first as a single copy DNA among an overwhelming majority of wild-type species. As a result, cells and tissues harbor both wild and mutant mtDNA, a phenomenon known as heteroplasmy.2 A single mutant molecule is unlikely to influence the physiology of the cell, play a role in aging, or cause disease. To affect mitochondrial function and cellular physiology, the nascent somatic mutant must accumulate in the cell to a significant level. The molecular mechanisms responsible for expansion of mutant mtDNA molecules to homoplasmy within a cell are obscure. mtDNA properties of polyploidy and relaxed replication and the need to regulate intracellular mtDNA copy number are believed to contribute to random genetic drift22 and ultimately also to the clonal expansion of mtDNA forms over extended periods of time.23 This physiology has been more amenable to computer simulation than to empirical experimentation.24 Also unclear is the regulation of replicative fidelity in the homopolymeric C tracts, in which we have localized much cell-to-cell heterogeneity and which has been described by others as a mutational “hotspot.”14,15,20 The relative proportion of variable-length poly C tracts appears to be actively maintained during cell division despite evidence of random mtDNA segregation, suggesting de novo regeneration of a specific pattern following cell division by an as yet unknown molecular mechanism.25 Homopolymeric C tract and other hypervariable region variants have been associated with several common human diseases such as diabetes mellitus,26 low birth weight,27 and dilated cardiomyopathy.28 The pathophysiology of mtDNA disease is unclear, and alterations in mtDNA may have tissue-specific consequences. Homopolymeric C tract alterations also cause problems with amplification and sequencing in vitro of the area beyond the C tract and may interfere with the replication of these areas in vivo outside the C tract.
Remarkably, there is evidence that clonal expansion of mtDNA mutations is a widespread process in mammalian and human tissues.14 Our recently published experiments using BM CD34 cells support such a process normally occurring in hematopoietic tissue.13 Because of the marked sequence differences among our healthy donors, we speculated that mtDNA might be highly variable in marrow and vary with donor age as has been described for other tissues.14,15 Even with the analysis restricted to the 1200 base pairs of the mtDNA control region, an average of 25% of individual CD34 clones from adult healthy BM differed in mtDNA sequence from the donor's respective aggregate cell sequence, and almost 8% were uniquely different, in contrast to virtually homogenous mtDNA sequence among healthy umbilical CB CD34 clones.13 These results were unexpected, as rapidly dividing tissues such as bone marrow have not been thought to permit the homoplasmic resolution of mtDNA mutations over time. For BM, accumulation of mtDNA mutations appear to relate to normal aging of the hematopoietic compartment; “normal aging” in this instance may result from secondary physiologic exposure to free oxygen radicals in the mitochondrion, and mtDNA changes, therefore, reflect exposure differences among individual organisms (healthy donors, in our experiments), tissues (marrow), and cells (isolated CD34 cells). “Fixed” specific mtDNA lesions have been speculated to alter the phenotype and function of an expanded population of abnormal cells in clonal hematologic diseases such as myelodysplasia.11
An important practical question motivating the current experiments was whether mutations detected in the BM clones were also present in more accessible circulating CD34 cells. By using the same methodology of single cell sorting, a brief period of cell culture to allow for clonal proliferation, DNA extraction, and mtDNA-specific amplification, followed by direct nucleotide sequencing of approximately 20% of the total mitochondrial genome, we found an equivalent marked degree of mtDNA sequence heterogeneity in blood as in marrow for all donors. Approximately half the observed mtDNA sequence changes in individual circulating CD34 clones and single granulocytes occurred in the homopolymeric C tract (nucleotides 303-315), and most of the remaining were point transitions outside the poly C region. We observed evidence of heteroplasmy in sequence variants among individual circulating CD34 cells and single granulocytes, although low levels of heteroplasmy (in which the minor species is < 20% that of the major heteroplasmic species) would probably be missed. Remarkably, although our healthy donors' blood was sampled 6 months to a year after the marrow was assayed, many of the same mutations and a similar pattern of apparent CD34 cell expansion, marked by mtDNA genetic changes, was apparent in all individuals. mtDNA in blood cells may, therefore, be usable as a natural genetic “marker” of hematopoietic progenitors and stem cells in long-term serial studies. Deserving additional comment is the reproducible finding of a markedly diverse sequence in the hypervariable region of a single BM CD34 cell clone of donor 2, which contrasted with the usual pattern of mtDNA heterogeneity defined by 1 or 2 nucleotide differences in individual cells compared with the bulk mtDNA sequence. Similar extraordinary observations in mouse experiments and in human mitochondrial disease have suggested the possibility of paternal inheritance through recombination between maternal and paternal mtDNA in the fertilized ovum.29,30
Almost 30% of single granulocytes showed mtDNA heterogeneity, in comparison with the aggregate granulocyte mtDNA sequence. Most of the genetic alterations were length changes in the homopolymeric C tract between nucleotide 303 and 315 at hypervariable region 2, although substitutions were also observed (Table 6; Figure 2B). A similar pattern of findings was previously observed for buccal epithelial cells.14,15 Of the nucleic acid changes in granulocytes, most were not seen in CD34 cells of either the donors' BM or PB. These mutations, polymorphisms, and homopolymeric C tract length changes may have arisen during differentiation from primitive progenitor to mature cell, or they simply were not detected because of their rarity in the stem cell pool. Nonetheless, mtDNA sequence analysis showed a consistent pattern of expansion of a minor population of CD34 cells in BM and PB as well as in granulocytes from all donors. This correspondence suggests that mtDNA might be used to estimate the number of active stem cells in the steady state, as has been performed for the cat,31 and also to predict both the appearance and extinction of individual hematopoietic stem cell clones over time. In our laboratory, we are currently studying mtDNA heterogeneity after allogenic stem cell transplantation and will statistically compare the results with donors' unperturbed hematopoietic system. If our results apply to other tissues, in general caution may be needed in forensic identifications and anthropologic studies that depend primarily on the considerable sequence variation found in the 2 hypervariable segments. Other translational experiments are also indicated. For example, if mtDNA heterogeneity is a reflection of environmental mutagen exposure, patients who have undergone chemotherapy and radiation treatment should show an increased prevalence of mtDNA changes when examined by the procedures described here for single cell experiments. Also, fixed mutations in leukemic blast cells might provide a general method of minimal residual disease detection.
Prepublished online as Blood First Edition Paper, March 11, 2004; DOI 10.1182/blood-2003-11-3949.
M.G.S. and S.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This paper is a contribution of the U.S. National Institutes of Health (NIH) and the National Institute of Standards and Technology (NIST) and is not subject to copyright. Certain commercial equipment, instruments, materials, or companies are identified in this paper to specify the experimental procedure. Such identification does not imply recommendation or endorsement by NIH and NIST, nor does it imply that the materials or equipment identified are the best available for this purpose.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal