Abstract
We report here that Janus kinase 3 (Jak3) is a primary response gene for interleukin-6 (IL-6) in macrophage differentiation, and ectopic overexpression of Jak3 accelerates monocytic differentiation of normal mouse bone marrow cells stimulated with cytokines. Furthermore, we show that incubation of normal mouse bone marrow cells with a JAK3-specific inhibitor results in profound inhibition of myeloid colony formation in response to granulocyte-macrophage colony-stimulating factor or the combination of stem cell factor, IL-3, and IL-6. In addition, mutagenesis of the Jak3 promoter has revealed that Sp1 binding sites within a -67 to -85 element and a signal transducer and activator of transcription (Stat) binding site at position -44 to -53 are critical for activation of Jak3 transcription in murine M1 myeloid leukemia cells stimulated with IL-6. Electrophoretic mobility shift assay (EMSA) analysis has demonstrated that Sp1 can bind to the -67 to -85 element and Stat3 can bind to the -44 to -53 STAT site in IL-6-stimulated M1 cells. Additionally, ectopic overexpression of Stat3 enhanced Jak3 promoter activity in M1 cells. This mechanism of activation of the murine Jak3 promoter in myeloid cells is distinct from a recently reported mechanism of activation of the human JAK3 promoter in activated T cells.
Introduction
The development of hematopoietic precursor cells into terminally differentiated cells such as granulocytes and macrophages requires integration of signals for cell survival, growth, and differentiation. These signals are induced by a network of cytokines, which bind to their receptors and initiate signals that can couple the processes of growth and differentiation.1 Many cytokines transmit their signals through a family of cytoplasmic tyrosine kinases known as Janus kinases (Jak kinases).2 Gene targeting studies in mice have illustrated the importance of the Jak family members in hematopoiesis. Lymphoid development is impaired in mice deficient in Jak13 and Jak3,4-6 and erythropoiesis is lost in Jak2-deficient mice.7 Among the Jak family members, Jak3 is unique because it is expressed mostly in hematopoietic cells and its transcription is regulated by cytokines such as granulocyte colony-stimulating factor8 (G-CSF), which can induce terminal differentiation of myeloid cells. In addition, Jak3 transcription was shown to be up-regulated by interleukin-2 (IL-2) and IL-4, which mediate proliferative signals in T and B cells.9-13 Interestingly, overexpression of Jak3 in either 32Dcl3 cells or primary mouse bone marrow (BM) cells in the presence of G-CSF or granulocyte-macrophage (GM)-CSF results in an early onset of growth arrest of the cells in the G1 phase of the cell cycle, which is accompanied by terminal granulocytic differentiation.8 It thus appears that Jak3 can transmit either proliferative or growth arrest signals depending on the cell type and the nature of the cytokine used. In this study, we present evidence that Jak3 also plays an important role in myeloid differentiation along the monocytic lineage. Furthermore, detailed analysis of the promoter/enhancer region of Jak3 has revealed that regulation of Jak3 transcription during IL-6-induced macrophage differentiation is achieved through a mechanism that is distinct from the mechanism of activation of the JAK3 promoter (JAK3pr) in activated T cells that has recently been described.14
Materials and methods
Cycloheximide assays
M1 cells were treated with recombinant murine IL-6 (100 ng/mL; Stem Cell Technologies, Vancouver, BC, Canada) in the presence or absence of cycloheximide (10 μg/mL). Total cellular RNA was extracted at 0, 2, 4, 6, 8, and 24 hours following cytokine treatment by the guanidinium thiocyanatephenol-chloroform method.15 RNA was subjected to electrophoresis on 1% formaldehyde-agarose gels and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH). Filters were prehybridized for 4 hours in 50% formamide containing 5 × standard sodium citrate solution (SSC), 1 × Denhardt solution, 250 mg/mL denatured salmon sperm DNA, and 50 mM sodium pyrophosphate, and hybridized to nick-translated 32P-labeled Jak3 probe for 16 hours at 42°C. Filters were washed at a final stringency of 0.1 × SSC, 0.1% sodium dodecyl sulfate (SDS) at 55°C before exposure to X-ray film with 2 intensifying screens.
Actinomycin D assays
M1 cells were plated in IL-6-free medium or in medium containing IL-6 (100 ng/mL) for 24 hours. Actinomycin D (10 μg/mL) was then added to the cultures and RNA was extracted 0, 2, 4, and 6 hours after addition of actinomycin D. Northern blot assays were carried out as described in “Cycloheximide assays.”
Nuclear run-on transcription assays
M1 cells (2 × 107) were stimulated with IL-6 for 0, 2, or 4 hours and then lysed with lysis buffer (0.5% nonidet P-40, 10 mM Tris [tris(hydroxymethyl)aminomethane, pH 7.4], 10 mM NaCl, 3 mM MgCl2). The nuclear pellet was obtained by centrifugation at 325g at 4°C. Nuclear run-on transcription was performed at 37°C for 45 minutes in a final volume of 75 μL in reaction buffer containing 10% glycerol, 10 mM Tris (pH 8.0), 5 mM MgCl2,25mM MnCl2, 150 mM KCl, 5 mM dithiothreitol (DTT), 1 mM nucleoside triphosphates (adenosine, guanosine, and cytosine triphosphate), and 150 μCi (5.55 MBq) 32P-ribo uridine triphosphate (rUTP). Reaction was terminated by the addition of 1 mL RNAzol reagent (Biotecx Laboratories, Houston, TX) followed by ethanol precipitation. The precipitated RNA was dissolved in 200 μL 100-mM Tris-HCl (pH 7.5), followed by addition of 200 μL 3.5-mM MgCl2, 1 mM DTT, 100 mM CaCl2, and 1.5 U RNase-free DNase I. After incubation at 30°C for 10 minutes, RNA was isolated by phenol extraction (2 times) and ethanol precipitation. Nytran membranes containing 5 μg linearized plasmid specific for full-length murine Jak3 or glucose-3-phosphate dehydrogenase (G3PDH) were hybridized with 2 × 106 cpm of nuclear run-on products in 1 mL hybridization solution (3 × SSC, 5 × Denhardt solution, 1 mM sodium pyrophosphate, 20 mM sodium phosphate [pH 7.0], 1% SDS, 100 μg/mL yeast tRNA, and 50% formamide) at 42°C for 36 hours. Hybridization strips were then washed twice in 2 × SSC at room temperature for 10 minutes each, washed once in 2 × SSC containing 10 μg/mL RNase A at 37°C for 30 minutes, followed by 2 washes in 0.2 × SSC, 0.1% SDS at 42°C for 10 minutes each, and finally one wash in 0.1 × SSC, 0.1% SDS at 42°C for 10 minutes. Strips were then exposed either to X-ray film at -80°C or Fuji illuminating screens at room temperature. When the Fuji screens were used, quantitation was performed using the Fuji Phosporimager (Edison, NJ).
Plasmid construction
To obtain a genomic clone of Jak3 containing the promoter/enhancer region, a 129J mouse genomic library in λDASHII phage vector was screened using a 32P-labeled 5′ fragment of Jak3 cDNA. A clone that contained 5.7 kb of sequence upstream of the initiator ATG was subcloned into pBR322 and sequenced completely (GenBank accession number, U71201). The promoter/enhancer sequences upstream of the ATG codon were then cloned into the pGL3-Basic promoterless vector upstream of the luciferase reporter gene (Promega, Madison, WI).
Construction of 5′ deletion mutants and linker scanning mutants of the Jak3 promoter
The 5′ deletion constructs were created either through restriction enzyme digestion with Af l II, SbfI, or BstZ17I, or via a polymerase chain reaction (PCR)-based approach. The linker scanning mutants of the Jak3 promoter were created using a PCR-based technique in which consecutive 18-base pair stretches of wild-type Jak3 promoter sequence were replaced with an NdeI-XhoI-SalI polylinker (CATATGCTCGAGGTCGAC). This technique has been described previously.16 All mutations were verified by sequencing.
Construction of site-specific mutants of the Jak3 promoter
The site-specific mutants that disrupted the Sp1 and signal transducer and activator of transcription (Stat) binding sites were created by a PCR-based approach. Using Jak3pr -3805 as a template, a 5′ primer containing mutations in either the Sp1 or STAT binding sites was hybridized to position -100 of the Jak3 promoter. The 3′ primer in the PCR reaction corresponded to the 5′ untranslated region (UTR) of Jak3 just upstream of the initial ATG. The resultant PCR product was blunt-ended into the SmaI site of pGL3-Basic. The primers used to make the mutations in either the Sp1 or STAT binding sites are as follows: -100 Sp1 mutant (mut) 5′-GTCTCCCCCGGGCTTCCCGAACCGAACCGAACCCAGGGCGCCCTGACT-3′ and -100 STAT mut 5′-GTCTCCCCCGGGCTTCCCGCCCCGCCCCGCCCCCAGGGCGCCCTGACTTTCCGTCCATGACGGCGG-3′. Mutations are in boldface and were verified by sequencing.
Tissue culture, transient transfections, and luciferase assays
Murine M1 myeloid leukemia cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated horse serum. Transient transfection of M1 cells was achieved by electroporation, as described previously.17 Following electroporation, M1 cells were left unstimulated or stimulated with 100 ng/mL recombinant murine IL-6 (Stem Cell Technologies). Reporter or effector DNA (20 μg) was used in each transfection. The Renilla luciferase plasmid pRL-CH110 (2 μg) was used as control for transfection efficiency, and this plasmid has been described previously.18 The Dual Luciferase Reporter Assay Kit (Promega) was used to determine firefly and Renilla luciferase activity. Firefly luciferase expression levels were normalized to the levels of Renilla expression for each transfection. Wild-type Stat3 (WT Stat3) or constitutively active Stat3 (Stat3-C)19 were subcloned into the pRC CMV expression vector, and 20 μg of each expression vector was used in transfection studies.
BM infection, selection, and expansion
Myeloblast-enriched BM cells were obtained from femurs of sodium caseinate-injected, 6- to 8-week-old C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME). Cell preparations consisting of myeloblast-enriched cells were infected with the retroviral vectors pMSCV-neo or pMSCV-Jak3-neo as described previously.8
Colony-forming assays
BM cells were extracted from femurs and tibiae of C57BL/6 mice (Jackson Laboratory), cultured in RPMI 1640 medium with 10% fetal calf serum, and treated with either 100 μg/mL WHI-P154 (Calbiochem, La Jolla, CA), a specific pharmacologic inhibitor of JAK3,20-22 or dimethyl sulfoxide (DMSO) for 24 hours and plated on methylcellulose (Stem Cell Technologies) supplemented with either 10 ng/mL recombinant murine GM-CSF alone or a combination of 10 ng/mL recombinant murine IL-6, 50 ng/mL recombinant murine stem cell factor (SCF), and 10 ng/mL recombinant murine IL-3 (all from Stem Cell Technologies). Cultures were seeded in triplicate using 35-mm plastic Petri dishes (Sarstedt, Newton, NC), and colony-forming units were determined after one week.
Electrophoretic mobility shift assays (EMSAs)
Following overnight stimulation in 100 ng/mL recombinant murine IL-6, nuclear extracts were obtained from 107 M1 cells as described previously.23 Unstimulated M1 cells were also pelleted for preparation of nuclear extracts. Electrophoretic mobility shift assays were performed as described previously.23 The probes used in the EMSA analysis corresponded to positions -38 to -58 and -39 to -89 of the Jak3 promoter. The -38 to -58 probe was of the following sequence, and its complement: 5′-CCGTCATTTACGGAAAGTCAG-3′. The -39 to -89 probe was of the following sequence, and its complement: 5′-CGTCATTTACGGAAAGTCAGGGCCCCTGGGGGCGGGGCGGGGCGGGAAGC-3′. The anti-Sp1 (PEP2) and anti-Stat3 (H-190) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Results
Jak3is a primary response gene for IL-6
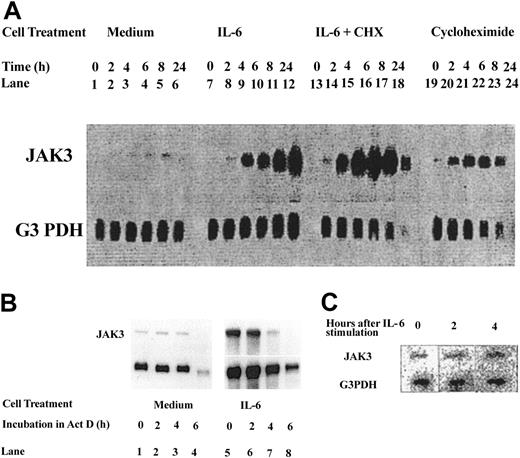
In studies using the murine myeloid cell line 32Dcl3, which undergoes granulocytic differentiation in response to G-CSF,24-26 we previously reported that Jak3 is a primary response gene for G-CSF8 and ectopic overexpression of Jak3 in 32Dcl3 cells or normal mouse BM cells results in acceleration of granulocytic differentiation. To determine whether Jak3 also plays a role in macrophage differentiation, we have used the murine myeloblastic leukemia cell line M1, which undergoes differentiation into macrophages upon stimulation with IL-6.27 Results presented in Figure 1A show that stimulation with IL-6 results in an upregulation of Jak3 RNA within 2 hours that continues through 24 hours of stimulation (lanes 7-12). To determine whether the induction of Jak3 was dependent upon new protein synthesis, M1 cells were incubated in the presence or absence of cycloheximide for times ranging from 0 to 24 hours. These studies show that cycloheximide failed to inhibit induction of Jak3 RNA, and in fact superinduction of Jak3 RNA was seen with IL-6 and cycloheximide (Figure 1, lanes 13-18). These results show that the induction of Jak3 mRNA by IL-6 does not require new protein synthesis.
Jak3is a primary response gene for IL-6. (A) M1 cells were treated with medium alone (lanes 1-6), medium containing IL-6 in the absence or presence of cycloheximide (CHX, 10 μg/mL; lanes 7-18), or medium containing cycloheximide alone (lanes 19-24). Total cellular RNA was extracted at 0, 2, 4, 6, 8, and 24 hours following cytokine treatment and 20 μg RNA was used for Northern analysis. The level of expression of Jak3 was determined by hybridization to a 32P-labeled probe (upper panel). To control for the amount of RNA loaded, the membrane was stripped and reprobed with a 32P-labeled G3PDH probe (lower panel). (B) Cells were plated in IL-6-free medium (lanes 1-4) or in medium containing IL-6 (lanes 5-8) for 24 hours. Actinomycin D (Act D) was then added to the cultures and incubation was further continued for the indicated hours. RNA (20 μg) from each sample was subjected to Northern blot analysis. (C) M1 cells were stimulated with IL-6 for the indicated hours. Cells were pelleted and the labeled RNA was subjected to nuclear run-on assays as described in “Materials and methods.”
Jak3is a primary response gene for IL-6. (A) M1 cells were treated with medium alone (lanes 1-6), medium containing IL-6 in the absence or presence of cycloheximide (CHX, 10 μg/mL; lanes 7-18), or medium containing cycloheximide alone (lanes 19-24). Total cellular RNA was extracted at 0, 2, 4, 6, 8, and 24 hours following cytokine treatment and 20 μg RNA was used for Northern analysis. The level of expression of Jak3 was determined by hybridization to a 32P-labeled probe (upper panel). To control for the amount of RNA loaded, the membrane was stripped and reprobed with a 32P-labeled G3PDH probe (lower panel). (B) Cells were plated in IL-6-free medium (lanes 1-4) or in medium containing IL-6 (lanes 5-8) for 24 hours. Actinomycin D (Act D) was then added to the cultures and incubation was further continued for the indicated hours. RNA (20 μg) from each sample was subjected to Northern blot analysis. (C) M1 cells were stimulated with IL-6 for the indicated hours. Cells were pelleted and the labeled RNA was subjected to nuclear run-on assays as described in “Materials and methods.”
To rule out the possibility that the accumulation of Jak3 mRNA in response to IL-6 stimulation is due to its stabilization, we conducted actinomycin D chase experiments. M1 cells were incubated with or without IL-6 for 12 hours to allow for accumulation of Jak3 message prior to addition of actinomycin D. Cells were removed at 0, 2, 4, and 6 hours after actinomycin D addition, total RNA was extracted, and the levels of RNA were examined by Northern blot analysis. Results shown in Figure 1B demonstrate that Jak3 mRNA has a long half-life in M1 cells (Figure 1B, lanes 1-4), and the addition of actinomycin D did not lead to an alteration in the half-life of Jak3 mRNA in the presence of IL-6 (Figure 1B, lanes 5-8). These results show that the up-regulation of Jak3 RNA in M1 cells stimulated with IL-6 is not due to stabilization of the mRNA.
We also performed nuclear run-on transcription assays to determine whether the increase in Jak3 mRNA in response to IL-6 was due to a direct increase in transcription. Figure 1C shows an increase in Jak3 transcription within 2 hours following IL-6 stimulation and a 5-fold increase in levels of Jak3 RNA by 4 hours following IL-6 stimulation. Levels of G3DPH were unchanged in stimulated and unstimulated cells. These results indicate that the increased levels of Jak3 mRNA seen following stimulation of M1 cells with IL-6 are due to a direct increase in transcription. These results allow us to conclude that Jak3 is a primary response gene for IL-6, as was seen with G-CSF.
Ectopic expression of Jak3 in normal mouse BM cells results in accelerated macrophage differentiation in response to GM-CSF
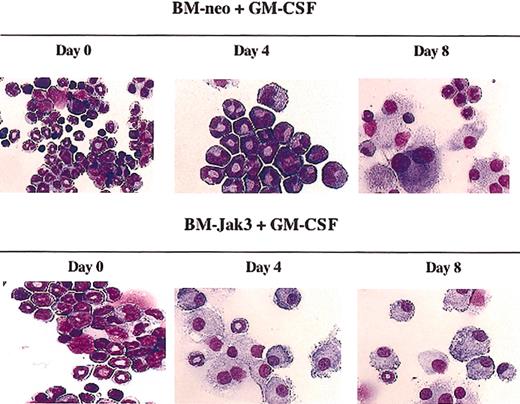
We next examined whether Jak3 plays a role in the monocytic differentiation of normal mouse BM cells. The M1 myeloid leukemia cell system has been used successfully in the past to identify genes that play a role in monocytic differentiation that can be readily extrapolated to normal BM cells.28 However, it is important to note that, unlike with M1 cells, IL-6 alone is not sufficient to induce differentiation of normal mouse BM cells.29 Myeloid differentiation of normal mouse BM cells does occur in the presence of IL-6 but requires the addition of macrophage colony-stimulating factor (M-CSF), or SCF and IL-3.30 Myeloid differentiation can also occur in the presence of GM-CSF alone. Examining the consequences of Jak3 overexpression in one of these contexts would allow an assessment of whether Jak3 plays a functional role in monocytic differentiation, a possibility that was suggested by the observation that Jak3 is strongly up-regulated during IL-6-induced monocytic differentiation of M1 cells. Accordingly, we isolated a myeloblast-enriched population of BM cells from C57BL/6 mice and infected these cells with the retroviral vector pMSCV-Jak3/neo or a control vector, pMSCV-neo, as desc ribed in “Materials and methods.” Following infection, the cells were seeded in methylcellulose in the presence of G418 to allow for the selection of retrovirus-infected cells, which were then transferred to liquid culture and induced for myeloid differentiation with GM-CSF. Figure 2 shows that BM-neo cells could undergo terminal macrophage differentiation by day 8 following the addition of GM-CSF. In contrast, morphologically differentiated macrophages could be readily detected by day 4 in BM cells infected with the Jak3 expression vector. We have previously reported similar accelerated differentiation of myeloid precursors into Gr1+ granulocytes in response to GM-CSF and have reported that overexpression of Jak3 in GM-CSF-stimulated normal BM cells results in acceleration of the onset of G0/G1 phase cell-cycle arrest.8 As described previously, Jak3 overexpression reduced the percentage of cells in the S phase of the cell cycle at day 4 following GM-CSF stimulation from 30% to 13%, and increased the percentage of cells arrested in the G0/G1 phase from 57% to 73%.8 The observation presented here that Jak3 overexpression can also accelerate macrophage differentiation in response to GM-CSF, together with our observation that Jak3 is a primary response gene for IL-6 during macrophage differentiation of M1 cells, suggests that Jak3 may play an important role in macrophage differentiation in addition to its already established role in granulocytic differentation.8
Ectopic expression of Jak3 in normal mouse BM cells results in accelerated macrophage differentiation in response to GM-CSF. Following infection of myeloblast-enriched BM cells from wild-type (C57BL/6) with pMSCV-neo or pMSCV-Jak3 retroviral vectors, cells were induced for differentiation with GM-CSF. Morphology was assessed by May-Grunwald and Giemsa staining at the indicated time points. Original magnification, × 20.
Ectopic expression of Jak3 in normal mouse BM cells results in accelerated macrophage differentiation in response to GM-CSF. Following infection of myeloblast-enriched BM cells from wild-type (C57BL/6) with pMSCV-neo or pMSCV-Jak3 retroviral vectors, cells were induced for differentiation with GM-CSF. Morphology was assessed by May-Grunwald and Giemsa staining at the indicated time points. Original magnification, × 20.
Effect of Jak3 inhibition on myeloid colony formation of normal mouse BM cells
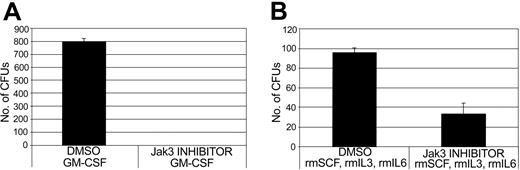
Because overexpression of Jak3 was found to accelerate myeloid differentiation of normal BM cells, we were interested in ascertaining whether inhibition of Jak3 in these cells could inhibit myeloid differentiation. BM cells obtained from C57BL/6 mice were cultured for 24 hours in the presence of DMSO or WHI-P154 ([4-(3′-Bromo-4′-hydroxyphenyl)-amino-6,7-dimethoxyquinazoline]), a compound known to specifically inhibit Jak3.20-22 The cells were then plated in methylcellulose culture medium and were stimulated with either GM-CSF alone or the combination of SCF, IL-3, and IL-6 to permit colony formation. GM-CSF supports the development of granulocyte and macrophage colonies from normal BM cells, whereas SCF, IL-3, and IL-6 support the development of colonies containing all myeloid lineages.30 After one week, colonies on both DMSO-treated and WHI-P154-treated plates were counted. As shown in Figure 3A, incubation in the presence of Jak3 inhibitor resulted in a block to myeloid colony formation induced by GM-CSF. Likewise, incubation of BM cells in the presence of Jak3 inhibitor reduced myeloid colony formation induced by SCF, IL-3, and IL-6 by more than 60% (Figure 3B). These results provide further evidence of an important role for Jak3 in myeloid differentiation of normal BM cells.
Pharmacologic inhibition of Jak3 results in reduced myeloid colony formation in normal mouse BM cells stimulated with either GM-CSF alone or the combination of SCF, IL-3, and IL-6. Normal BM cells were obtained from wild-type (C57BL/6) mice and were cultured for 24 hours in the presence of either DMSO or WHI-P154, a specific inhibitor of Jak3. 20-22 Cells were then plated on methylcellulose to permit colony formation and stimulated with either GM-CSF or a combination of SCF, IL-3, and IL-6. After one week, colonies were counted and the number of colonies counted for DMSO-treated and Jak3 inhibitor-treated plates is shown. (A) Colony counts for GM-CSF-treated cells, plus or minus 1 standard deviation. (B) Colony counts for cells treated with SCF, IL-3, and IL-6, plus or minus 1 standard deviation. rm indicates recombinant murine.
Pharmacologic inhibition of Jak3 results in reduced myeloid colony formation in normal mouse BM cells stimulated with either GM-CSF alone or the combination of SCF, IL-3, and IL-6. Normal BM cells were obtained from wild-type (C57BL/6) mice and were cultured for 24 hours in the presence of either DMSO or WHI-P154, a specific inhibitor of Jak3. 20-22 Cells were then plated on methylcellulose to permit colony formation and stimulated with either GM-CSF or a combination of SCF, IL-3, and IL-6. After one week, colonies were counted and the number of colonies counted for DMSO-treated and Jak3 inhibitor-treated plates is shown. (A) Colony counts for GM-CSF-treated cells, plus or minus 1 standard deviation. (B) Colony counts for cells treated with SCF, IL-3, and IL-6, plus or minus 1 standard deviation. rm indicates recombinant murine.
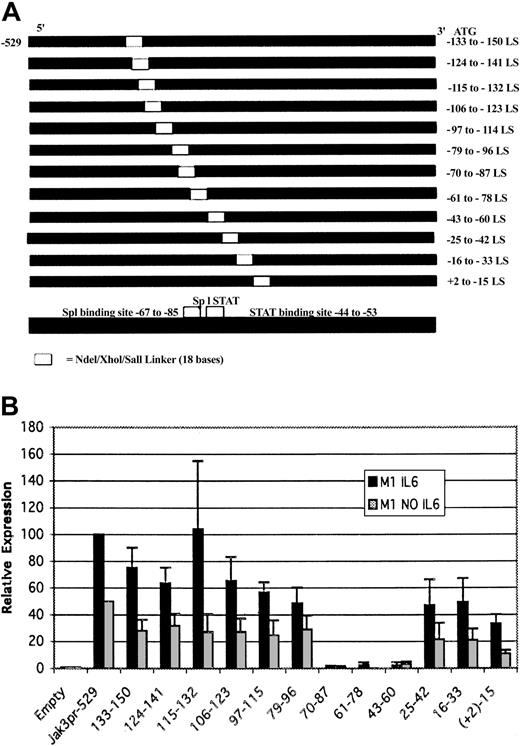
Effect of 5′ deletions and linker scanning mutations on Jak3 promoter activity
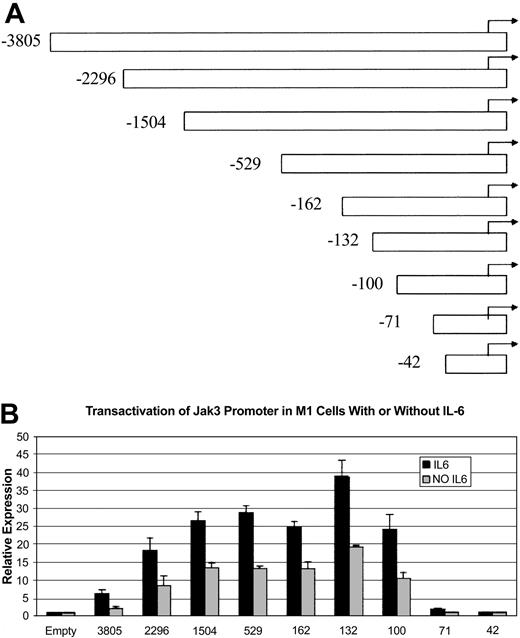
To study the mechanisms by which IL-6 up-regulates Jak3 expression, we isolated a genomic clone of the Jak3 gene that was sequenced completely and found to contain 5.7 kb of sequence upstream of the initiator ATG. We then compared our genomic clone with the Jak3 cDNA described by Gurniak and Berg.31 Consistent with previous reports that the 5′ UTR of the murine Jak3 gene is interrupted by an intron, our sequence analysis revealed that sequences present in the Jak3 cDNA described by Gurniak and Berg31 flanked a 1.7-kb intron in the 5′ UTR of Jak3. Using this genomic DNA clone, we created several 5′ deletion mutants of the Jak3 promoter and subcloned these deletion mutants and the fragment containing 5.7 kb of sequence upstream of the initiator ATG into the pGL3-Basic luciferase reporter vector. These constructs were designated as Jak3pr -3805, Jak3pr -2296, and others to indicate the position of the 5′ end of the deletion relative to the approximate start site of transcription, which is located at the 5′-most extent of the UTR of the Jak3 cDNA described by Gurniak and Berg.31 Each of these constructs is depicted in Figure 4A and includes the complete 5′ UTR of the Jak3 gene. However, since the 5′ UTR is interrupted by a 1.7-kb intron, we created an additional construct that did not include the intron sequence. Because luciferase reporter assays performed with both intronless and intron-containing Jak3pr constructs showed that the presence of the intron sequence in the 5′ UTR had no effect on generation of luciferase activity, we proceeded with our analysis using the constructs that included the intron sequence, in an effort to preserve the original structure of the genomic DNA. The Jak3pr -3805 construct and the 5′ deletion constructs were then transiently transfected into unstimulated M1 cells or M1 cells stimulated with IL-6, and luciferase activity was measured. Figure 4B shows that some of the constructs transfected into M1 cells stimulated with IL-6 generated luciferase activity 20- to 40-fold greater than background, while the same constructs transfected into unstimulated M1 cells generated luciferase activity 10- to 20-fold greater than background. Most noticeable, though, was the observation that there was a sharp decline in promoter activity between Jak3pr -100 and Jak3pr -71, suggesting the presence of an activating element or elements between these 2 positions. The Jak3pr -42 construct failed to generate any luciferase activity at all.
Jak3promoter/reporter constructs and sequence requirements for the activity of theJak3promoter in M1 cells. (A) The coding region of Jak3 was replaced with the coding region of the firefly luciferase gene when a 5.7-kb genomic clone and several 5′ deletion constructs were subcloned into the pGL3-Basic luciferase reporter vector. The 5′ deletion constructs used in the luciferase reporter assays are depicted. Constructs were numbered according to the approximate start of transcription, which is at the 5′-most extent of the Jak3 cDNA described by Gurniak and Berg.31 (B) The pGL3-Basic Jak3pr -3805 construct and several 5′ deletion constructs were transiently transfected into M1 cells that were either unstimulated or stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation.
Jak3promoter/reporter constructs and sequence requirements for the activity of theJak3promoter in M1 cells. (A) The coding region of Jak3 was replaced with the coding region of the firefly luciferase gene when a 5.7-kb genomic clone and several 5′ deletion constructs were subcloned into the pGL3-Basic luciferase reporter vector. The 5′ deletion constructs used in the luciferase reporter assays are depicted. Constructs were numbered according to the approximate start of transcription, which is at the 5′-most extent of the Jak3 cDNA described by Gurniak and Berg.31 (B) The pGL3-Basic Jak3pr -3805 construct and several 5′ deletion constructs were transiently transfected into M1 cells that were either unstimulated or stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation.
Analysis of the sequences between positions -42 and -100 of the Jak3 promoter using the Mat Inspector (Genomatix, Munich, Germany) and TFSEARCH (Real World Computing Partnership, Japan) transcription factor search programs revealed several binding sites of interest. The most notable elements were a stretch of consecutive binding sites for the zinc-finger transcription factor Sp1 located from positions -67 to -85 and gamma-interferon activated sequence (GAS) elements of the consensus sequence TTNCNNNAA between positions -44 to -53. All STATs except STAT2 are known to bind to GAS elements of the form TTNCNNNAA.32,33
To precisely identify the sequences required for Jak3pr activity, a linker scanning mutagenesis strategy was used. The strongly activating Jak3pr -529 construct was used as a template and consecutive 18-base stretches within the region of interest were replaced with an NdeI/XhoI/SalI polylinker. The advantages of such a strategy are that the size and topology of the promoter and the spatial relationships between regulatory elements are maintained, thus minimizing the occurrence of artifacts that can be generated by gross deletions.34 Linker scanning mutants were constructed that spanned from immediately upstream to immediately downstream of the -42 to -100 region of interest. The linker scanning constructs that were used are shown in Figure 5A. Each construct was subcloned into the pGL3-Basic luciferase reporter vector. The Jak3pr -529 construct and 12 linker scanning constructs with mutations spanning positions +2 to -150 in the Jak3 promoter were transiently transfected into M1 cells. The results of the luciferase reporter assays performed with the linker scanning mutants in either unstimulated M1 cells or M1 cells stimulated with IL-6 are presented in Figure 5B. In both unstimulated and stimulated M1 cells, Jak3pr -61 to -78 and -70 to -87 were associated with declines in luciferase activity to near background levels, indicating that Sp1 binding sites play an important role in Jak3 transcription. Disruption of the GAS element also had a pronounced effect on the luciferase activity generated in M1 cells. These studies with the linker scanning mutants suggest that the -44 to -53 GAS motif and the Sp1 binding sites may both be required for Jak3 transcription in M1 cells.
Linker-scanning mutagenesis reveals that sequences between positions -43 to -87 are required for activity of theJak3promoter in M1 cells. (A) Shown is a representation of the linker scanning mutants of the Jak3 promoter that was used for luciferase assays. All linker scanning mutants shown above use the Jak3pr -529 construct as a template. The linker scanning mutants span the region between +2 and -150. (B) Linker scanning mutants of the Jak3 promoter and the strongly activating -529 5′ deletion construct were transiently transfected into either unstimulated M1 cells or M1 cells stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation. For M1 cells stimulated with IL-6, the value for the -529 construct was close to 100-fold over empty vector, so this value was normalized to 100 and all other values were expressed as a percentage of the -529 value. For unstimulated M1 cells, the value for the -529 construct was close to 50-fold over empty vector, so this value was normalized to 50 and again all other values were expressed as a percentage of the -529 value.
Linker-scanning mutagenesis reveals that sequences between positions -43 to -87 are required for activity of theJak3promoter in M1 cells. (A) Shown is a representation of the linker scanning mutants of the Jak3 promoter that was used for luciferase assays. All linker scanning mutants shown above use the Jak3pr -529 construct as a template. The linker scanning mutants span the region between +2 and -150. (B) Linker scanning mutants of the Jak3 promoter and the strongly activating -529 5′ deletion construct were transiently transfected into either unstimulated M1 cells or M1 cells stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation. For M1 cells stimulated with IL-6, the value for the -529 construct was close to 100-fold over empty vector, so this value was normalized to 100 and all other values were expressed as a percentage of the -529 value. For unstimulated M1 cells, the value for the -529 construct was close to 50-fold over empty vector, so this value was normalized to 50 and again all other values were expressed as a percentage of the -529 value.
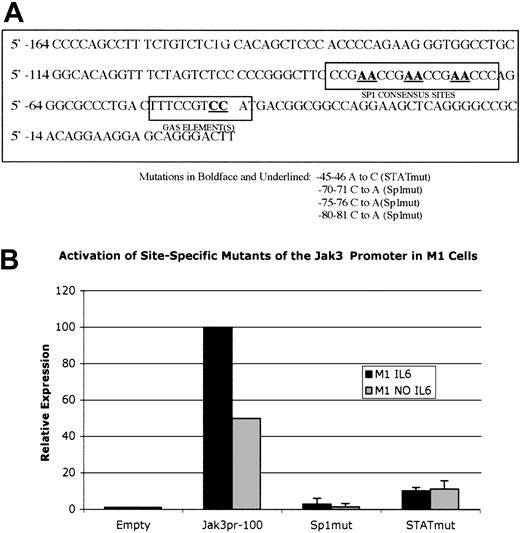
Role of Sp1 binding sites and the GAS element on Jak3pr activity
To verify the importance of Sp1 and GAS elements, we constructed additional Jak3pr mutants in which point mutations were introduced into these elements. Using the strongly activating Jak3pr -100 construct as a template, the -100 Sp1 mut and the -100 Stat mut constructs were created. The mutated bases are shown in Figure 6A. Luciferase assays performed with the -100 Sp1 mut and -100 Stat mut constructs in M1 cells (Figure 6B) show that there was a near total loss of luciferase activity associated with the -100 Sp1 mut, confirming the critical role of the Sp1 binding sites in Jak3 transcription. The -100 STAT mut construct was associated with a 10-fold reduction of luciferase activity in M1 cells stimulated with IL-6, and a 5-fold reduction in luciferase activity in unstimulated M1 cells.
Site-specific mutation of the Sp1 binding sites between positions -67 to -85 of theJak3promoter or a 2-base change in the -44 to -53 GAS motif markedly reducesJak3pr activity in M1 cells. (A) The -100 Jak3pr construct was used as a template to create -100 Sp1 mut, in which 3 consecutive Sp1 consensus sites are destroyed by making a 2-base change in each site. Additionally, -100 Stat mut was created by making a 2-base change to disrupt the -44 to -53 GAS motif. (B) -100 Sp1 mut and -100 Stat mut were transiently transfected into unstimulated M1 cells or M1 cells stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation. Values for the -100 wild-type construct were normalized to 100 for M1 cells stimulated with IL-6 and normalized to 50 for unstimulated M1 cells.
Site-specific mutation of the Sp1 binding sites between positions -67 to -85 of theJak3promoter or a 2-base change in the -44 to -53 GAS motif markedly reducesJak3pr activity in M1 cells. (A) The -100 Jak3pr construct was used as a template to create -100 Sp1 mut, in which 3 consecutive Sp1 consensus sites are destroyed by making a 2-base change in each site. Additionally, -100 Stat mut was created by making a 2-base change to disrupt the -44 to -53 GAS motif. (B) -100 Sp1 mut and -100 Stat mut were transiently transfected into unstimulated M1 cells or M1 cells stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation. Values for the -100 wild-type construct were normalized to 100 for M1 cells stimulated with IL-6 and normalized to 50 for unstimulated M1 cells.
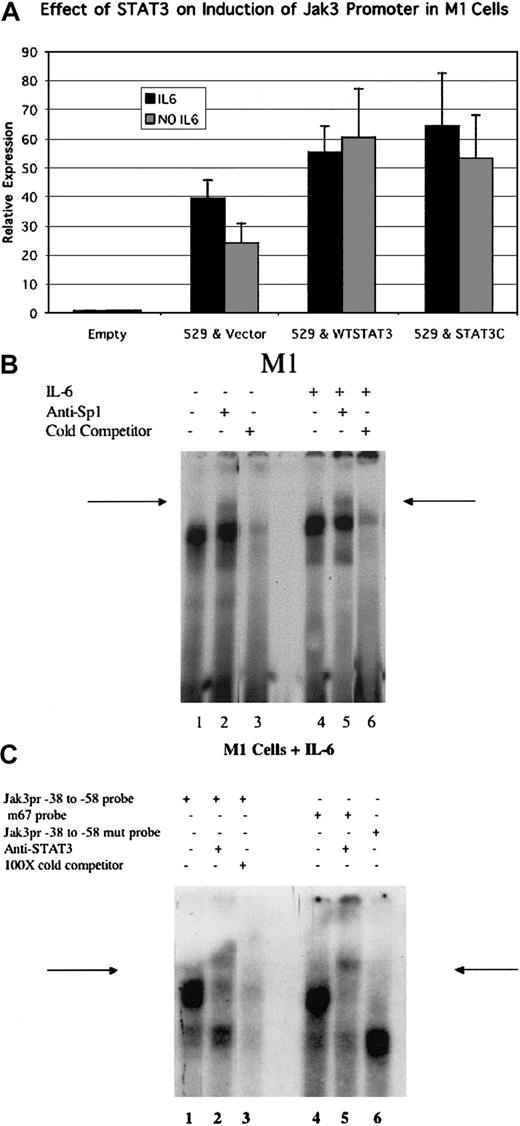
Activation of the Jak3 promoter by Stat3
Given the importance of the -44 to -53 GAS element in the activation of the Jak3 promoter in M1 cells, we sought to test whether cotransfection of a constitutively active form of Stat3 (Stat3-C) along with Jak3pr -529 could enhance Jak3pr activity. Stat3 was an attractive candidate to play a role in Jak3 transcription during monocytic differentiation because overexpression of a dominant-negative Stat3 has been shown to abolish the monocytic differentiation of M1 cells in response to IL-6.35,36 Results presented in Figure 7A show that cotransfection of Stat3-C doubled luciferase activity in unstimulated M1 cells and was associated with a 60% increase in luciferase activity in cells stimulated with IL-6. Because it is known that endogenous Stat3 is activated to a much greater extent in IL-6-stimulated M1 cells than in unstimulated cells,35 it is possible that, upon IL-6 stimulation, endogenous activated Stat3 saturates the promoter to prevent the strong enhancement of luciferase activity that is seen in unstimulated M1 cells by Stat3-C.
Cotransfection of Stat3 enhances Jak3 promoter activity in M1 cells, and Sp1 and Stat3 proteins present in nuclear lysates of M1 cells bind to a radiolabeled probe corresponding to theJak3pr sequence. (A) Either wild-type Stat3 or a constitutively active mutant of Stat3 (Stat3-C) were cotransfected with the -529 Jak3pr construct into either unstimulated M1 cells or M1 cells stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation. (B) EMSA analysis using a radiolabeled probe corresponding to the -39 to -89 sequence of the Jak3 promoter and nuclear lysates from M1 cells is depicted. Lanes 1 to 3 demonstrate gel shifts obtained by using nuclear lysates from unstimulated M1 cells. A band supershift is seen with the addition of anti-Sp1 antibody (arrow, lane 2). The band shift was competed away with the addition of 100-fold excess of cold -39 to -89 probe (lane 3). Lanes 4 to 6 demonstrate gel shifts obtained by using nuclear lysates from M1 cells stimulated with IL-6. A band supershift is seen with the addition of anti-Sp1 antibody (arrow, lane 5). The band shift was competed away with the addition of 100-fold excess of cold -39 to -89 probe (lane 6). (C) Lanes 1 to 3 demonstrate band shifts obtained using a radiolabeled probe corresponding to the -38 to -58 sequence of the Jak3 promoter. A band supershift is seen with the addition of anti-STAT3 antibody (arrow, lane 2). The band shift was competed away with excess of cold -38 to -58 probe (lane 3). The band shift obtained using the m67 STAT3 consensus oligonucleotide19 is seen in lane 4, and a band supershift is seen with the addition of anti-STAT3 antibody (arrow, lane 5). A different complex is obtained by using a labeled probe containing -45 and -46 A to C mutations that disrupt the -44 to -53 GAS element (lane 6).
Cotransfection of Stat3 enhances Jak3 promoter activity in M1 cells, and Sp1 and Stat3 proteins present in nuclear lysates of M1 cells bind to a radiolabeled probe corresponding to theJak3pr sequence. (A) Either wild-type Stat3 or a constitutively active mutant of Stat3 (Stat3-C) were cotransfected with the -529 Jak3pr construct into either unstimulated M1 cells or M1 cells stimulated with IL-6. Following a 20-hour incubation period, cells were lysed and analyzed for luciferase activity. Relative expression refers to the fold increase in luciferase activity over vector alone. Values expressed are the average of 3 independent experiments plus or minus one standard deviation. (B) EMSA analysis using a radiolabeled probe corresponding to the -39 to -89 sequence of the Jak3 promoter and nuclear lysates from M1 cells is depicted. Lanes 1 to 3 demonstrate gel shifts obtained by using nuclear lysates from unstimulated M1 cells. A band supershift is seen with the addition of anti-Sp1 antibody (arrow, lane 2). The band shift was competed away with the addition of 100-fold excess of cold -39 to -89 probe (lane 3). Lanes 4 to 6 demonstrate gel shifts obtained by using nuclear lysates from M1 cells stimulated with IL-6. A band supershift is seen with the addition of anti-Sp1 antibody (arrow, lane 5). The band shift was competed away with the addition of 100-fold excess of cold -39 to -89 probe (lane 6). (C) Lanes 1 to 3 demonstrate band shifts obtained using a radiolabeled probe corresponding to the -38 to -58 sequence of the Jak3 promoter. A band supershift is seen with the addition of anti-STAT3 antibody (arrow, lane 2). The band shift was competed away with excess of cold -38 to -58 probe (lane 3). The band shift obtained using the m67 STAT3 consensus oligonucleotide19 is seen in lane 4, and a band supershift is seen with the addition of anti-STAT3 antibody (arrow, lane 5). A different complex is obtained by using a labeled probe containing -45 and -46 A to C mutations that disrupt the -44 to -53 GAS element (lane 6).
EMSA analysis demonstrates that Sp1 and Stat3 bind to Jak3 promoter
To demonstrate binding of Sp1 to the -67 to -85 binding elements in the Jak3 promoter, we performed EMSA analysis using a 32P-labeled oligonucleotide probe that corresponded to positions -39 to -89 of the Jak3 promoter using nuclear extracts obtained from M1 cells (Figure 7B). In both unstimulated (Figure 7B, lane 2, arrow) and IL-6-stimulated (lane 5, arrow) cells, a DNA-protein complex was observed that, upon the addition of anti-Sp1 antibody, resulted in a supershifted complex. The observation that Sp1 can bind to the Jak3 promoter in both unstimulated and IL-6-stimulated cells demonstrates that Sp1 binds to the Jak3 promoter constitutively rather than inducibly.
We also performed EMSA analysis using a radiolabeled oligonucleotide that corresponded to positions -38 to -58 of the Jak3 promoter that encompassed the GAS element. As a positive control, we used the m67 STAT3 consensus oligonucleotide, which has been shown to bind STAT3 with high affinity.19 As shown in Figure 7C, nuclear lysates from IL-6-stimulated M1 cells formed a prominent complex with the -38 to -58 Jak3 promoter oligonucleotide (lane 1) that comigrated with the prominent complex formed with the m67 STAT3 consensus probe (lane 4). This complex could be supershifted by anti-Stat3 antibody when bound to either of the probes (Figure 7C, lanes 2,5, arrows). EMSA analysis was also performed using a radiolabeled Jak3 promoter -38 to -58 mutant oligonucleotide that lacked a Stat consensus site due to consecutive A to C mutations at positions -45 and -46. This oligonucleotide (Figure 7C, lane 6) yielded a different prominent complex from that obtained using the wild-type Jak3pr -38 to -58 oligonucleotide or the m67 STAT3 consensus oligonucleotide, confirming that -45A and -46A were essential for formation of complexes containing Stat3. Similar EMSA analysis was also performed with unstimulated M1 cells, but binding of proteins present in the nuclear lysates of unstimulated M1 cells to both the probes was very weak, probably because IL-6 stimulation is needed for robust Stat3 activation in M1 cells.35 This suggests that Stat3 may regulate Jak3pr activity in an inducible manner. However, since mutation of the -44 to -53 GAS element in unstimulated M1 cells causes a 5-fold reduction in Jak3 promoter activity and cotransfection of wild-type as well as constitutively activated Stat3 doubles Jak3pr activity in unstimulated M1 cells, it is possible that Stat3 plays a significant role in basal activation of the Jak3 promoter in unstimulated M1 cells. This activity appears to be further enhanced following IL-6 stimulation, which results in the enhanced phosphorylation of Stat3.35
Discussion
Our studies demonstrate that Jak3 mRNA is strongly induced during IL-6-induced macrophage differentiation of M1 cells. Our results also show that deregulated expression of Jak3 in normal mouse BM cells accelerates macrophage differentiation induced by GM-CSF. In addition, treatment of normal mouse BM cells with a JAK3-specific pharmacologic inhibitor resulted in a profound inhibition of myeloid colony formation in response to either GM-CSF alone or the combination of SCF, IL-3, and IL-6. These results, together with our previous observation that deregulated expression of Jak3 in normal BM cells accelerates differentiation of Gr1+ granulocytes in response to G-CSF or GM-CSF,8 suggest that Jak3 plays a direct role in mediating myeloid differentiation of normal mouse BM cells.
Jak3 knock-out mice and JAK3-deficient humans have well-characterized defects in the lymphoid compartment. Jak3 knockout mice exhibit severe defects in B- and T-cell development,4-6,37 and can survive only in a pathogen-free environment, consistent with a severe combined immunodeficiency (SCID)-like phenotype. Humans deficient in JAK3 are afflicted with SCID38-40 and have greatly reduced numbers of T cells with normal or increased numbers of B cells.41 Defects in the myeloid compartment of Jak3-deficient mice are more subtle, but careful analysis has revealed the existence of dysregulated myelopoiesis in these animals. Grossman et al42 report that mice deficient in Jak3 display increased immature neutrophil and monocyte counts in the peripheral blood, splenomegaly, and infiltration of organs with immature myeloid cells. They also find that the presence of Jak3-deficient T cells in the periphery was necessary for this myeloid expansion, although it could not be determined whether Jak3-deficient T cells were sufficient for this phenotype.42 In fact, the possibility was raised that Jak3 expression in the myeloid compartment may be required to halt a myeloproliferative response initiated by autoreactive T cells.42 Such a role for Jak3 would be consistent with our previous observation that Jak3 is associated with cell-cycle arrest8 and our new observation that Jak3 is required for formation of terminally differentiated myeloid colonies. In addition to the defects in myelopoiesis observed in Jak3-deficient mice, new evidence presented in Saemann et al43 has revealed functional defects in human dendritic cells treated with WHI-P154, the same pharmacologic inhibitor of JAK3 used in our studies presented here. It is known that engagement of CD40 on peripheral blood monocytes by CD154 on T cells results in JAK3 phosphorylation and subsequent STAT5 activation.44 When human dendritic cells were treated with WHI-P154, there was a marked reduction in production of costimulatory molecules and antigen-presenting molecules, and dendritic cells exposed to the JAK3 inhibitor induced a state of hyporeactivity in alloreactive T cells.43 These results suggest that intact JAK3 signaling is required for monocytes to engage in a fully functional immune response. Our observation that myeloid colony formation in BM progenitors is profoundly impaired upon treatment with a JAK3 inhibitor indicates that Jak3 may be necessary even earlier to facilitate the terminal differentiation of myeloid cells prior to a requirement for their functionality. Although the work we present here suggests an important role for Jak3 in myeloid differentiation, what is not seen in either mice or humans deficient in Jak3 is a total loss of the myeloid compartment. This is consistent with the possibility that other Jak kinase family members can compensate for the loss of Jak3 during myeloid differentiation. There is a high degree of redundancy in the hematopoietic system, especially in the myeloid compartment. Even elimination of as important a mediator as G-CSF still leaves the mouse with 30% of normal circulating neutrophil levels.45 Overlapping functions of the Jak kinase family members may serve to mask individual contributions, much as major deficiencies in myeloid differentiation became apparent with knock out of the gp130 receptor46 but were not evident with the knock out of IL-6,47 just one of its many ligands.
Our studies presented here also attempt to provide an insight into the mechanisms that bring about activation of Jak3 transcription in response to IL-6 stimulation. Since Jak1, Jak2, and tyrosine kinase 2 (Tyk2) are ubiquitously expressed, inducible expression of Jak3 makes it unique among members of the Janus kinase family.41,48 Our studies show that Jak3 transcription in M1 cells stimulated with IL-6 requires the binding of Sp1 to consensus binding sites located at positions -67 to -85 of the Jak3 promoter. Sp1 binding is apparently constitutive, as it also occurs in autonomously proliferating M1 cells. Sp1 is expressed at especially high levels in developing granulocytes49 and has been shown to induce activation of the promoters for numerous genes specifically expressed in myeloid cells.17,50-58 It has been demonstrated that there is no change in the ability of Sp1 to bind DNA during the course of G-CSF-induced granulocytic differentiation of 32Dcl3 cells,56 suggesting that Sp1 may depend on posttranslational modification or binding to inducible partners to preferentially activate transcription in response to a cytokine such as IL-6. Our results presented here suggest that one such partner for Sp1 is Stat3, which is consistent with previous reports that have demonstrated a requirement for Sp1 and a member of the STAT family for maximal transcriptional activation of cytokine-induced genes.59-61
Our studies show that Stat3 is capable of binding to the -44 to -53 GAS element of the Jak3 promoter, and mutation of this GAS element results in a 10-fold reduction in luciferase activity of IL-6-stimulated cells, but only a 5-fold reduction in luciferase activity of unstimulated cells, suggesting that IL-6-stimulated cells are particularly sensitive to disruption of the GAS element. This is not surprising since Stat3 is strongly phosphorylated in response to IL-6 stimulation.35 Furthermore, cotransfection of a constitutively active form of Stat3 enhanced Jak3pr activity 60% in IL-6-stimulated M1 cells, and doubled Jak3pr activity in unstimulated M1 cells, once again indicating that Stat3 plays a role in mediating Jak3 transcription.
Our report here for the first time details a molecular mechanism underlying transcriptional activation of a member of the Jak kinase family during myeloid differentiation. Interestingly, since binding of members of the IL-6 family of cytokines can activate Jak1, Jak2, and Tyk262-64 as well as Stat3,35 it is quite possible that the phosphorylation of Stat3 and its subsequent translocation to the nucleus to activate transcription of Jak3 represent an example of regulation of a Jak kinase by a Jak/Stat signaling pathway distinct from its own. Such a model suggests a new degree of interrelatedness among multiple Jak/Stat pathways.
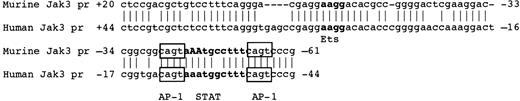
Recently, an analysis of the human JAK3 promoter was conducted to gain insight into the up-regulation of JAK3 transcription following activation of human T cells.14 A sequence homology search between mouse and human JAK3 promoter sequences reveals strong homology between the 2 promoter sequences in only a single stretch of 88 bases. As shown in Figure 8, the 2 sequences exhibited 81% identity in this region, with 72 of 88 bases matching. The homologous region corresponds to positions -61 to +20 in the murine Jak3 promoter and positions -44 to +44 in the human JAK3 promoter. Interestingly, this region encompasses the -44 to -53 GAS element found by us to be the Stat3 binding site essential for promoter activity in M1 cells. Mutation of this element by Aringer et al14 had no effect on the activity of the Jak3 promoter in activated Jurkat cells. Instead, T-cell-specific activation is in part achieved through the use of the 2 activator protein 1 (AP-1) sites that flank the GAS element. Although the -67 to -85 Sp1 binding sites fall just outside of the region of highest homology between the murine and human Jak3 promoters, there is a similar GC-rich region with a consensus SP1 binding site near position -60 of the human JAK3 promoter. A 5′ deletion mutant that eliminated this GC-rich region resulted in a loss of promoter activity,14 suggesting that SP1 is important in regulating basal activity of the JAK3 promoter in both activated T cells and myeloid cells. The proximal ETS site determined by Aringer et al14 through site-specific mutation to be essential for JAK3 promoter activity in human T cells is also present in the murine Jak3 promoter. The (+2) to -15 linker scanning mutant disrupted this site and was associated with a 60% reduction in promoter activity. However, this site is clearly more important in activated T cells, since the site-specific mutation performed by Aringer et al14 resulted in a total loss of JAK3 promoter activity.
The human and murineJAK3promoters share an 88-bp region of homology but use different transcription factor binding sites within that region to activate transcription ofJAK3. The sequence alignment of the human and murine Jak3 promoters is shown. The sequence of the human Jak3 promoter has been described in Aringer et al.14 The Stat binding site that is necessary for transcription of the murine Jak3 gene in myeloid cells and the AP-1 and ETS sites that are necessary for maximal transcription of the human JaK3 gene in T cells are highlighted. Stat and ETS binding sites are in boldface, and the AP-1 binding sites are boxed. Positions -45A and -46A of the murine Jak3 promoter are denoted with capital letters. When these 2 bases are mutated together (-45 and -46 A to C), activity of the Jak3 promoter in M1 cells is markedly reduced.
The human and murineJAK3promoters share an 88-bp region of homology but use different transcription factor binding sites within that region to activate transcription ofJAK3. The sequence alignment of the human and murine Jak3 promoters is shown. The sequence of the human Jak3 promoter has been described in Aringer et al.14 The Stat binding site that is necessary for transcription of the murine Jak3 gene in myeloid cells and the AP-1 and ETS sites that are necessary for maximal transcription of the human JaK3 gene in T cells are highlighted. Stat and ETS binding sites are in boldface, and the AP-1 binding sites are boxed. Positions -45A and -46A of the murine Jak3 promoter are denoted with capital letters. When these 2 bases are mutated together (-45 and -46 A to C), activity of the Jak3 promoter in M1 cells is markedly reduced.
Although both promoters appear to use the Sp1 binding sites for basal transcription, activation of the human JAK3 promoter upon T-cell activation appears to be mediated predominantly by ETS binding sites that are not essential in the activation of the murine Jak3 promoter in IL-6-stimulated myeloid cells. The most striking difference between mechanisms of activation of the 2 promoters is the essential requirement for an intact Stat binding site in the murine promoter in myeloid cells, a site which is dispensable for up-regulation of JAK3 transcription upon T-cell activation. Whether this is a cell-type-specific mechanism or a stimulus-specific mechanism is not completely clear, since it is possible that use of the STAT binding site could occur in T cells upon cytokine stimulation, in contrast to the lack of use of the STAT site upon activation by CD3 cross-linking. Alternatively, since activation of Jak3 is so closely associated with the process of growth arrest and differentiation in myeloid cells,8 it is possible that STAT3-mediated activation of Jak3 is a differentiation-specific mechanism, while ETS-mediated activation of JAK3 is a proliferation- or survival-specific mechanism. Consistent with the potential for ETS-mediated activation of a JAK3 proliferative signal in T cells, ETS-/- T cells demonstrate decreased proliferative capacity and undergo apoptosis in response to the activation signals that should initiate progression through the cell cycle.65 The JAK3 promoter appears to have evolved in a remarkably versatile manner, which likely reflects the diversity of roles that JAK3 plays in different cell types. It will be important to carefully examine the mechanisms for regulation of JAK3 in the context of its various important roles in lymphopoiesis, myelopoiesis, and immune regulation.
Prepublished online as Blood First Edition Paper, February 19, 2004; DOI 10.1182/blood-2003-06-2165.
Supported by grants CA68239, CA79086, ES09225, and R24 CA88261 from the National Institutes of Health (E.P.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Atul Kumar for obtaining the genomic clone of Jak3 containing the upstream promoter/enhancer region. We also thank Dr Anita L. Korapati for constructing the constitutively active mutant of Stat3.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal