Abstract
The BCR/ABL fusion protein is found in more than 90% of patients with chronic myeloid leukemia (CML) as well as in a subset of patients with acute B-cell leukemia. We have previously described a transgenic model for an inducible and reversible acute B-cell leukemia caused by p210 BCR/ABL. Here, we describe a new model of an inducible BCR/ABL disease by directing the expression of the oncogene to megakaryocytic progenitor cells within the murine bone marrow using the tetracycline-responsive expression system under the control of human CD34 regulatory elements. The predominant feature was the development of a chronic thrombocytosis. The condition progressed with the development of splenomegaly accompanied by lymphadenopathy in some mice. Affected animals demonstrated a dramatic increase in the number of megakaryocytes in the bone marrow and the spleen. Immunohistochemistry demonstrated that the reporter gene was expressed in hematopoietic stem cells (HSCs), common myeloid progenitor (CMP) cells, as well as in megakaryocytic/erythroid progenitor cells (MEPs). Although these mice did not display the increase in granulopoiesis commonly found in chronic myeloid leukemia (CML), the phenotype closely resembles a myeloproliferative disorder affecting the megakaryocytic lineage observed in some patients with the BCR/ABL P210 translocation.
Introduction
The Philadelphia chromosome results from a reciprocal translocation that fuses chromosomes 9 and 22, thereby generating a fusion between parts of the breakpoint cluster region gene BCR with the gene encoding the nonreceptor tyrosine kinase ABL. The molecular consequence of this fusion, the BCR/ABL protein, is a constitutively active tyrosine kinase that is associated with different forms of leukemia.1-5 The most common form, P210 BCR/ABL, is the hallmark of chronic myeloid leukemia (CML) in which it is found in more than 90% of patients. CML is a disorder of hematopoietic stem cells that is characterized not only by an increase of myeloid cells and platelets in the peripheral blood and the bone marrow but also by extramedullary hematopoiesis. In addition and consistent with a defect in a pluripotent stem cell, thrombocytosis is seen in most patients.
We have described a model of an inducible p210 BCR/ABL-dependent acute pre-B cell leukemia (ALL) using an inducible system in mice.6 In this model, the long terminal repeat (LTR) of the mouse mammary tumor virus (MMTV) was used to direct the expression of the tetracycline transactivator protein tTA to early B-cell progenitors within the murine bone marrow, thus accounting for the B-cell phenotype. We reasoned that expression of BCR/ABL in stem cells and early progenitor cells might induce a disease with closer resemblance to human CML. In previous studies we have shown that regulatory elements contained within 141 kilobase (kb) of the locus of the human CD34 gene were sufficient to drive expression of a reporter gene in hematopoietic stem cells.7-9 Subsequently, we developed a transgenic line of mice (hCD34tTA) in which the same human CD34 regulatory elements were used to express tTA in bone marrow progenitors.10 Here, we show that by using recently developed methods for isolation and characterization of hematopoietic progenitors by multicolor fluorescence-activated cell sorting (FACS),11 we were able to define the expression of reporter genes responding to this hCD34tTA activator line precisely to hematopoietic stem cells (HSCs), common myeloid progenitors (CMPs), and most strongly to megakaryocytic/erythroid progenitors (MEPs). We then bred these hCD34tTA transactivator mice with a line of transgenic mice that carries the sequence for P210 BCR/ABL under the control of a tetracycline responsive element (TRE).6 In contrast to the B-cell leukemia, which developed within 2 weeks in MMTVtTA-BCR/ABL mice, a different phenotype was observed when the P210 BCR/ABL oncogene was expressed in the bone marrow by regulatory elements of the human CD34 gene in this hCD34tTA line. The expression of BCR/ABL to MEP led to thrombocytosis and an increase in the number of megakaryocytes in the bone marrow and the spleen. Consistent with this expression, the number of megakaryocytic progenitors in the bone marrow was significantly increased as shown by colony-forming assays. CML, as the name indicates, is characterized by a marked increase in the number of myeloid cells. However, there is also an increase in the platelet count in many patients with CML, consistent with the fact that CML is a clonal stem cell disorder.12 The transgenic mice in this study consistently developed a phenotype with preferential expansion of the megakaryocytic lineage, demonstrating that the phenotype of P210 BCR/ABL disease in mice largely results from the type of hematopoietic progenitor in which the oncogene is expressed.
Materials and methods
Transgenic constructs, establishment of mouse lines, and genotyping of transgenic mice
The TRE-BCR/ABL transgenic construct and mouse line, the hCD34tTA transactivator line and the Tet-O-cre mice were described previously.6,10 In the hCD34tTA line, we used homologous recombination in bacteria to insert the tTA coding region into a 141-kb human CD34 p1-derived artificial chromosome (PAC) so that tTA translation was initiated at the position normally used by human CD34 to minimize alteration of regulation of the gene.7,13 Transgenic mice were identified by Southern blot analysis of tail DNA after EcoRI digestion.6 We used a 450-base pair (bp) fragment from the joining region of BCR/ABL generated by polymerase chain reaction (PCR)14 and the 1020-bp tTA fragment isolated from the transactivator plasmid pUHD15-315 after digestion with BamHI and EcoRI for detection of the transgenes. Tet-O-cre transgenic mice were identified by PCR with primers specific for the cre gene. The upstream primer sequence was 5′-ATGTTCAATTTACTGACCG and the sequence of the downstream primer was 5′-CGCCGCATAACCAGTGAAAC. Amplification conditions consisted of 35 cycles of 30 seconds of denaturation at 94°C, 30 seconds of annealing at 51°C, and 30 seconds of extension at 72°C, yielding an amplification product representing the first 355 bp within the cre recombinase gene.
Monitoring of peripheral blood for signs of disease
Total differential and white blood cell counts were performed every other week from double transgenic mice and single transgenic or nontransgenic littermates kept in the same cages. Peripheral blood films were stained with Wright-Giemsa for differential analysis. Determination of the number of platelets was performed on 1:5 phosphate-buffered saline (PBS)-diluted whole blood with a Coulter Counter ACT Diff (Beckman Coulter, Fullterton, CA).
Immunohistochemistry
Mice were killed, and organs were fixed in neutral-buffered 10% formalin at room temperature for 16 hours prior to embedding in paraffin and sectioning. Sections were deparaffinized and then subjected to antigen retrieval in a microwave oven. Endogenous peroxidase was blocked by incubation of slides in 1% hydrogen peroxide. Staining for Gr-1 was performed with a monoclonal antibody (Pharmingen, San Diego, CA) and the Vectastain Universal ABC kit (Vector Laboratories, Burlingame, CA). Stable diaminobenzidine (ResGen, Huntsville, AL) was used as a chromogen substrate, and the sections were counterstained with hematoxylin solution (Sigma, St Louis, MO). Bone marrow cells were isolated by flushing femurs and tibia with phosphate-buffered saline. Hematopoietic stem cells (HSCs) and progenitor cells were stained and sorted as previously described.9 For sorting myeloid progenitors, bone marrow cells were stained with biotinylated rat anti-interleukin 7 receptor α (IL-7Rα) chain (A7R34), and rat antibodies specific for the following lineage markers: CD3 (KT31.1), CD4 (GK1.5), CD8 (53-6.7), B220 (6B2), Gr-1 (8C5), Ter119, and CD19 (1D3) (Pharmingen). IL-7Rα+Lin+ cells were removed with sheep anti-rat IgG-conjugated magnetic beads (Dynabeads M-450; Dynal A.S., Oslo, Norway), and the remaining cells were stained with avidin-phycoerythrin (PE)-Cy5 (Tricolor) (Caltag, Burlingame, CA). Cells were then stained with PE-conjugated anti-FcγRII/III (2.4G2), fluorescein isothiocyanate (FITC)-conjugated anti-CD34 (RAM34) (Pharmingen), or allophycocyanin (APC)-Cy7-conjugated anti-Sca-1 (E13-161-7) and APC-conjugated anti-c-Kit (2B8) monoclonal antibodies. Myeloid progenitors were sorted as Lin-Sca-1-c-Kit+CD34+FcγRII/IIIlo (CMPs), Lin-Sca-1-c-Kit+CD34+FcγRII/IIIhi (granulocyte macrophage progenitors [GMPs]), and Lin-Sca-1-c-Kit+CD34-FcγRII/IIIlo (MEPs) as described previously.11 HSCs were sorted as Lin-Sca-1hic-KithiIL-7R- populations.16 Cells were sorted by using a highly modified double laser (488 nm/350 nm Enterprise II + 647 nm Spectrum) high-speed FACS (Moflo-MLS; Cytomation, Fort Collins, CO). Cytospins were prepared, and the cells were fixed in 4% paraformaldehyde for 2 minutes at 4°C and stored in 70% ethyl alcohol (EtOH) at -20°C. Permeabilization of cells was performed by a 5-minute immersion in 0.1% Triton/PBS at room temperature. The slides were incubated for 30 minutes in 1% hydrogen peroxidase to quench endogenous peroxidase. The Vectastain ABC Elite kit for rabbit IgG (Vector Laboratories) was used for immunohistochemistry following the manufacturer's instruction, and a polyclonal antiserum17 (Babco, Princeton, NJ) was used at a dilution of 1:3000 to detect cre recombinase protein. The values of the expression of the Tet-O-Cre in HSCs and MEPs were determined using a scale in which “no staining” represented a level of staining that was not above background (nontransgenic mice), and “high” was the highest level (observed in MEPs). Values were scored by blinded readers. The antibody is specific in that no staining was observed above background (normal rabbit serum) in nontransgenic mice.
Isolation of RNA and Northern blot analysis
RNA from murine tissue was isolated according to the CsCl method18 and blotted to a positively charged nylon membrane (ICN, East Hills, NY). Expression analysis was performed as described.6 To ensure uniform loading levels and integrity of RNA samples, we stripped and rehybridized blots to a probe specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).19
Expression analysis by quantitative real-time PCR
Multiplex PCR with amplification of 18S RNA in the same tube for quantitation of BCR/ABL expression, TaqMan analysis, and subsequent calculations were performed with an ABI Prism 7700 sequence detection system (Perkin Elmer, Foster City, CA), which detects the signal from a fluorogenic internal probe. cDNA (100 ng) for each sample was subjected to PCR. The sequence of the BCR/ABL forward primer was 5′-CGTCCACTCAGCCACTGGAT; the sequence of the reverse primer was with primers 5′-TAACCAGTGGCTTCACTCAGACCCTGAGG. The sequence of the double-labeled oligonucleotide used as probe for BCR/ABL was FAMAGCGGCCAGTATCATCTGACTTTGAGC-TAMRA. Amplification of 18S RNA was performed in the same reaction tubes as internal standard with an alternatively labeled probe to discern from the cre probe (multiplex PCR). All experiments were performed in duplicates for each standard and tissue.
Flow cytometry
Bone marrow cells were flushed from tibias and femurs with PBS and filtered through a cell strainer. Enucleated red blood cells were lysed with lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM Na2-EDTA (ethylenediaminetetraacetic acid) pH 7.3), and the remaining white blood cells were washed once with PBS. We isolated cells from lymph nodes and spleen by squeezing tissue with the plunger of a syringe through a cell strainer (Becton Dickinson Labware, Franklin Lakes, NJ). Cells were incubated with the appropriate antibodies in PBS containing 5% calf serum for 30 minutes on ice in a total volume of 50 μL. One mL of PBS was added and the suspension was underlayed with 0.5 mL calf serum and centrifuged for 5 minutes at 1000 rpm, and the supernatant was discarded. The cell pellet was washed once with PBS, and the staining for the secondary antibody was performed. All analyses were performed on a dual-laser FACS (Becton Dickinson). The data were analyzed with the CellQuest program. The following antibodies were used: antimurine B220 (Caltag, San Francisco, CA), all other antibodies (murine CD34, CD41, Ter119, Gr-1, Mac-1) were purchased from Pharmingen.
Colony-forming unit (CFU) assay and megakaryocyte assay (CFU-MK)
Bone marrow was flushed with Iscove modified Dulbecco medium (IMDM) containing 5% of fetal bovine serum (FBS) from femurs and tibias of double-transgenic and single-transgenic control animals. Cells were inoculated in Methocult GF M3434 (Stem Cell Technologies, Vancouver, BC) supplemented with 30% fetal calf serum, 1% bovine serum albumin, 20 ng/mL of stem cell factor (SCF), 20 ng/mL IL-3, 10 ng/mL IL-11, 10 ng/mL granulocyte macrophage colony-stimulating factor (GM-CSF), 1 U/mL erythropoietin, 10 ng/mL thrombopoietin (R&D Systems, Minneapolis, MN), 2 mM l-glutamine (Stem Cell Technologies), 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO BRL, Carlsbad, CA) for colony assay for granulocyte macrophage colony-forming unit (CFU-GM), erythroid burst-forming unit (BFU-E), and granulocyte, erythroid, megakaryocyte, and macrophage colony-forming unit (CFU-GEMM). MegaCult-C (Stem Cell Technologies) was used for the growth of megakaryocyte progenitor colonies strictly following the manufacturer's protocol. CFU-MKs were stained for acetyl cholinesterase20 after 6 to 8 days, and positive colonies consisting of 3 to 50 cells were counted.
Results
Human CD34 regulatory elements contained in the CD34tTa strain direct expression to HSCs, CMPs, and MEPs
To express P210 BCR/ABL in hematopoietic stem and selected progenitor cells within murine bone marrow, we developed a new line of transgenic mice, hCD34tTA (Figure 1). These mice contain a transgene in which a 141-kb human CD34 PAC clone has been modified by insertion of the tTA sequence at the location of the human CD34 translational start site.10 We previously demonstrated that the hCD34tTA transgene directs inducible expression of a cre reporter gene to cells within the bone marrow of transgenic mice.10 The experiments described here were performed to identify the cell type specificity and to assess whether expression is directed to hematopoietic stem cells and progenitor cells. Functionality of the transactivator line was assessed in vivo by crossbreeding to an indicator line of mice, which carry a reporter gene under the control of a tetracycline-responsive element (TRE). In this study we used mice in which expression of the gene for cre recombinase is induced by binding of functional tTA protein to the TRE.10 To identify the cells that express Cre, we performed 5-color fluorescence-activated cell sorting11,16 on bone marrow cells. HSCs, CMPs, GMPs, and MEPs were isolated from double-transgenic mice (hCD34tTA × Tet-O-cre) and from nontransgenic littermates. Expression of cre protein was detected by immunohistochemistry using a polyclonal antiserum raised against the cre protein. The intensity of staining as judged by the intensity of the colored substrate of individual cells was used to grade the level of expression (no expression, low and medium expression, and high expression). We found staining in 93% of all HSCs, with the majority of these cells (79%) staining low positive. Staining was also seen in 94% of all CMPs, with 48% of the cells showing low staining and 46% expressing high levels of cre. Staining was also observed in 85% of all MEPs, among which 60% of the cells showed intensive staining, identifying this population as the one with high expression in the majority of cells (Figure 2). No staining was seen in GMPs and in cells from control animals.
Transgenic constructs used to express heterologous genes in early hematopoietic progenitors. (A) The diagram at the top demonstrates the structure of the human CD34 PAC clone with 110 kb 5′ and 24 kb 3′ sequence, as well as the 8 exons encompassing 25 kb. (B) The structure of the tetracycline transactivator (tTA) cassette, which was inserted by homologous recombination in bacteria into human CD34 exon 1. Shown are the tTA cDNA followed by an internal ribosome entry site (IRES). The 2 different responder transgenic constructs, expressed from a tetracycline regulatory element (TRE) are shown as follows: (C) one that expresses Cre, and (D) a second expressing the human P210 BCR/ABL fusion protein. Details of these constructs and their function in mice have been published.6-10
Transgenic constructs used to express heterologous genes in early hematopoietic progenitors. (A) The diagram at the top demonstrates the structure of the human CD34 PAC clone with 110 kb 5′ and 24 kb 3′ sequence, as well as the 8 exons encompassing 25 kb. (B) The structure of the tetracycline transactivator (tTA) cassette, which was inserted by homologous recombination in bacteria into human CD34 exon 1. Shown are the tTA cDNA followed by an internal ribosome entry site (IRES). The 2 different responder transgenic constructs, expressed from a tetracycline regulatory element (TRE) are shown as follows: (C) one that expresses Cre, and (D) a second expressing the human P210 BCR/ABL fusion protein. Details of these constructs and their function in mice have been published.6-10
Expression of thecretransresponder gene in bone marrow progenitors response to the human CD34tTA transgene. Multicolor FACS11 was used to separate bone marrow of human CD34tTA × tet-O-cre double transgenic mice (tg) into (A) hematopoietic stem cells (HSC), (B) common myeloid progenitors (CMP), (C) granulocyte/macrophage progenitors (GMP), and (D) megakaryocyte/erythrocyte progenitors (MEP). (E) HSCs and (F) mixed progenitor cells (including CMP, GMP, and MEP) from a wild-type animal (wt) were isolated and used as control. The expression of cre protein was demonstrated by immunohistochemistry with a polyclonal antiserum directed against the protein. Expression was found in HSCs, CMPs, and MEPs, whereas GMPs and cells from wild-type animals were negative. Original magnification, × 100.
Expression of thecretransresponder gene in bone marrow progenitors response to the human CD34tTA transgene. Multicolor FACS11 was used to separate bone marrow of human CD34tTA × tet-O-cre double transgenic mice (tg) into (A) hematopoietic stem cells (HSC), (B) common myeloid progenitors (CMP), (C) granulocyte/macrophage progenitors (GMP), and (D) megakaryocyte/erythrocyte progenitors (MEP). (E) HSCs and (F) mixed progenitor cells (including CMP, GMP, and MEP) from a wild-type animal (wt) were isolated and used as control. The expression of cre protein was demonstrated by immunohistochemistry with a polyclonal antiserum directed against the protein. Expression was found in HSCs, CMPs, and MEPs, whereas GMPs and cells from wild-type animals were negative. Original magnification, × 100.
Induction of BCR/ABL driven by CD34tTA induces expansion of the megakaryocytic lineage
Double transgenic mice were generated by crossbreeding hCD34tTA mice with TRE-P210 BCR/ABL mice under continuous administration of tetracycline through the drinking water6,10 to prevent expression of the oncogene during embryogenesis. Expression of BCR/ABL was induced by withdrawal of the antibiotic at 4 weeks of age. In contrast to the disease that developed in BCR/ABL mice in which expression was directed by the MMTV-tTa transactivator line6 leading to acute B-cell leukemia within 2 to 3 weeks after induction, significant changes in the number of peripheral white blood cells were not noticed, and most of the animals remained disease free for 5 months, when an increase in the number of platelets was found. Three mice out of a total of 9 in this cohort were found in moribund condition; altogether, 5 of 12 double-transgenic mice progressed to a moribund condition (Figure 3; Table 1). In addition, 2 of 3 animals that died within 2 to 4.5 months after induction had also developed thrombocytosis (Table 1). The number of platelets in mice varies depending on the genetic background. In this study with mice from a FVB/N background, it was found to range from 5 × 105 to 1.2 × 106 platelets/mm3 (Figure 3). On necropsy, histologic examination revealed elevated numbers of megakaryocytes in the bone marrow and the spleen in 11 of 12 mice (Table 1; Figure 4). In addition, variable splenomegaly ranging from mild (1.5-fold increase in weight) to severe (5-fold increase in size) was found in 50% of the animals, and 33% had developed moderate enlargement of cervical lymph nodes (up to 0.7 cm in diameter). Furthermore, invasion of hematopoietic cells that stained positive for the myeloid marker Gr-1 into the liver was observed (Figure 4H). The histologic-determined expansion of megakaryocytes in bone marrow and spleen was confirmed by FACS analysis with staining for glycoprotein IIb integrin (CD41). This integrin is the platelet receptor for fibrinogen and other extracellular matrix molecules and is expressed on megakaryocytes and platelets during adult hematopoiesis22 (Figure 5A-B). The mean value of the number of cells that stained positive for CD41 in the bone marrow and spleen of double transgenic mice was 25.5% ± 6.4% versus 12.2% ± 4.3% in single transgenic and wild-type littermates, a difference that was statistically highly significant (P < .0001) using a one-sided Student t test (Figure 5). We confirmed expression of the BCR/ABL transgene by Northern blot analysis and quantitative real time PCR in the bone marrow, spleen, and lymph node of diseased mice (Figure 6).
Elevation in peripheral blood platelet counts in hCD34tTA × BCR/ABL mice. Controls (□) were a nontransgenic littermate and 2 single BCR/ABL transgenic mice. The numbers of platelets in the control animals did not change over time as shown and were comparable to the values obtained at a single time point in 8 additional control animals (1 nontransgenic littermate, 1 hCD34tTA single transgenic, and 6 BCR/ABL single transgenic mice). Expression of BCR/ABL in the double transgenic hCD34tTA ×BCR/ABL animals (▪; F280, C814, C815, and C819 in Table 1) caused a significant increase in the number of platelets in the peripheral blood of double transgenic animals, up to more than 5 × 106/mm.3
Elevation in peripheral blood platelet counts in hCD34tTA × BCR/ABL mice. Controls (□) were a nontransgenic littermate and 2 single BCR/ABL transgenic mice. The numbers of platelets in the control animals did not change over time as shown and were comparable to the values obtained at a single time point in 8 additional control animals (1 nontransgenic littermate, 1 hCD34tTA single transgenic, and 6 BCR/ABL single transgenic mice). Expression of BCR/ABL in the double transgenic hCD34tTA ×BCR/ABL animals (▪; F280, C814, C815, and C819 in Table 1) caused a significant increase in the number of platelets in the peripheral blood of double transgenic animals, up to more than 5 × 106/mm.3
Clinical characteristics and peripheral blood counts in induced human CD34tTA × P210 BCR/ABL double transgenic animals
Animal . | Clinical condition . | Platelets . | WBC count, cells/μL . | Hgb level, g/dL . | Splenomegaly . | Anemia . | Time of analysis, mo . |
|---|---|---|---|---|---|---|---|
| F274 | Moribund | > 2.5 × 106 | 12 000 | 13.0 | 3.5 × | ND | 2.5 (death) |
| F277 | Hepatomegaly | Normal level | 8 300 | 13.0 | 2 × | ND | 12 |
| F280 | Moribund, LN | > 5 × 106 | 3 800 | 12.0 | 1.5 × | + | 5 (death) |
| C814 | Moribund, LN | > 5 × 106 | 4 900 | 12.0 | 1.5 × | + | 5 (death) |
| Hepatomegaly, | |||||||
| C815 | LN enlarged | 1.8 × 106 | 1 600 | 8.7 | 5 × | + | 14 (death) |
| C818 | Killed | Normal level | 5 800 | 15.0 | - | - | 15 |
| C819 | Killed | > 3 × 106 | 3 700 | 13.5 | - | - | 10 |
| C1161 | Moribund | > 2 × 106 | 3 700 | 13.0 | 3 × | ND | 2 (death) |
| C1152 | Moribund, LN | Normal level | 5 800 | 12.0 | - | + | 4.5 (death) |
| C1153 | Killed | Normal level | 5 200 | 13.0 | - | - | 6 |
| C1113 | Killed | Normal level | 4 800 | 15.0 | - | - | 7 |
| C1143 | Killed, neutrophilia | Normal level | 17 100 | 13.6 | - | - | 6 |
Animal . | Clinical condition . | Platelets . | WBC count, cells/μL . | Hgb level, g/dL . | Splenomegaly . | Anemia . | Time of analysis, mo . |
|---|---|---|---|---|---|---|---|
| F274 | Moribund | > 2.5 × 106 | 12 000 | 13.0 | 3.5 × | ND | 2.5 (death) |
| F277 | Hepatomegaly | Normal level | 8 300 | 13.0 | 2 × | ND | 12 |
| F280 | Moribund, LN | > 5 × 106 | 3 800 | 12.0 | 1.5 × | + | 5 (death) |
| C814 | Moribund, LN | > 5 × 106 | 4 900 | 12.0 | 1.5 × | + | 5 (death) |
| Hepatomegaly, | |||||||
| C815 | LN enlarged | 1.8 × 106 | 1 600 | 8.7 | 5 × | + | 14 (death) |
| C818 | Killed | Normal level | 5 800 | 15.0 | - | - | 15 |
| C819 | Killed | > 3 × 106 | 3 700 | 13.5 | - | - | 10 |
| C1161 | Moribund | > 2 × 106 | 3 700 | 13.0 | 3 × | ND | 2 (death) |
| C1152 | Moribund, LN | Normal level | 5 800 | 12.0 | - | + | 4.5 (death) |
| C1153 | Killed | Normal level | 5 200 | 13.0 | - | - | 6 |
| C1113 | Killed | Normal level | 4 800 | 15.0 | - | - | 7 |
| C1143 | Killed, neutrophilia | Normal level | 17 100 | 13.6 | - | - | 6 |
All mice except for C818 had elevated numbers of megakaryocytes in the bone marrow and spleen. Clinical condition refers to the condition of the animals at the time of analysis. Killed indicates mice that were sacrificed for analysis prior to the development of an obvious phenotype; LN, enlargement of lymph nodes; ND, not determined at time of death; -, feature did not develop; +, feature developed.
Increase in platelets in the peripheral blood and in megakaryocytes in bone marrow and spleen of induced double transgenic hCD34tTA × BCR/ABL mice. Cells in panels were stained with Wright Giemsa, except for panel H, which demonstrates Gr-1 staining (brown color) with a hematoxylin counterstain. (A) Peripheral blood and (B) bone marrow from a nontransgenic littermate are shown. Thrombocytosis developed within several months after induction of BCR/ABL expression (C-D, peripheral blood). Consistent with this finding in peripheral blood was an increase in the number of megakaryocytes in the bone marrow. (E) Stains of a cytospin of bone marrow cells and (F) a bone marrow section from another animal are shown. High numbers of megakaryocytes were also found in the spleens of induced animals (G) as well as invasion of myeloid cells into the liver (H). Original magnification, × 20.
Increase in platelets in the peripheral blood and in megakaryocytes in bone marrow and spleen of induced double transgenic hCD34tTA × BCR/ABL mice. Cells in panels were stained with Wright Giemsa, except for panel H, which demonstrates Gr-1 staining (brown color) with a hematoxylin counterstain. (A) Peripheral blood and (B) bone marrow from a nontransgenic littermate are shown. Thrombocytosis developed within several months after induction of BCR/ABL expression (C-D, peripheral blood). Consistent with this finding in peripheral blood was an increase in the number of megakaryocytes in the bone marrow. (E) Stains of a cytospin of bone marrow cells and (F) a bone marrow section from another animal are shown. High numbers of megakaryocytes were also found in the spleens of induced animals (G) as well as invasion of myeloid cells into the liver (H). Original magnification, × 20.
Flow cytometry of bone marrow cells from an animal that died of BCR/ABL disease demonstrates an increased number of megakaryocytic cells. The number of megakaryocytes staining positive for CD41 in hCD34tTA × BCR/ABL (+/+) was significantly increased compared with bone marrow cells from control animals. The top panel demonstrates surface expression of CD41 in the horizontal axis. The graph below demonstrates the difference between 10 double-transgenic induced mice versus bone marrow cells from 11 control animals, including 2 nontransgenic littermates with the same genetic background (FVB), 1 hCD34tTA single transgenic, and 8 BCR/ABL single transgenic mice. No difference was found between single transgenic and the nontransgenic wild-type controls. Percentages in top panels indicate percent positive for CD41.
Flow cytometry of bone marrow cells from an animal that died of BCR/ABL disease demonstrates an increased number of megakaryocytic cells. The number of megakaryocytes staining positive for CD41 in hCD34tTA × BCR/ABL (+/+) was significantly increased compared with bone marrow cells from control animals. The top panel demonstrates surface expression of CD41 in the horizontal axis. The graph below demonstrates the difference between 10 double-transgenic induced mice versus bone marrow cells from 11 control animals, including 2 nontransgenic littermates with the same genetic background (FVB), 1 hCD34tTA single transgenic, and 8 BCR/ABL single transgenic mice. No difference was found between single transgenic and the nontransgenic wild-type controls. Percentages in top panels indicate percent positive for CD41.
BCR/ABL RNA is expressed in hematopoietic tissues of hCD34tTA-BCR/ABL mice. (A) RNA expression of the oncogene is shown in hematopoietic tissues, including bone marrow, spleen, and thymus, but not from liver or lung detected by Northern blot analysis. RNAfrom the lymph node of a MMTVtTA-BCR/ABL animal that died of pre-B-cell leukemia6 was used as a positive control, whereas bone marrow from a single-transgenic littermate served as a negative control. Total RNA (20 μg) was loaded in each lane and probed with a BCR/ABL-specific probe, followed by hybridization to GAPDH. (B) Result is shown of quantitative real-time PCR to detect and quantify the expression of BCR/ABL in the same animal whose tissue was used for Northern blot analysis in panel A (C814) and from a second double transgenic animal that died of BCR/ABL related disease (C1152).
BCR/ABL RNA is expressed in hematopoietic tissues of hCD34tTA-BCR/ABL mice. (A) RNA expression of the oncogene is shown in hematopoietic tissues, including bone marrow, spleen, and thymus, but not from liver or lung detected by Northern blot analysis. RNAfrom the lymph node of a MMTVtTA-BCR/ABL animal that died of pre-B-cell leukemia6 was used as a positive control, whereas bone marrow from a single-transgenic littermate served as a negative control. Total RNA (20 μg) was loaded in each lane and probed with a BCR/ABL-specific probe, followed by hybridization to GAPDH. (B) Result is shown of quantitative real-time PCR to detect and quantify the expression of BCR/ABL in the same animal whose tissue was used for Northern blot analysis in panel A (C814) and from a second double transgenic animal that died of BCR/ABL related disease (C1152).
Invasion of tissue by megakaryocytes at advanced stage of induction
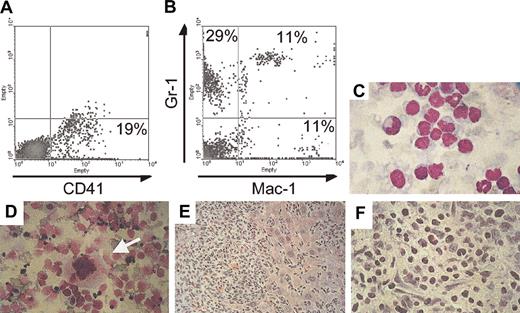
One double-transgenic animal (C815) developed thrombocytosis within 5 months after induction of BCR/ABL expression and was found an additional 9 months later in moribund condition with massive abdominal swelling, anemia (hemoglobulin [Hgb] 8.7 versus 15 for healthy mice) and decreased number of peripheral white blood cells (1600 cells/μL, among which 27% were neutrophils). Necropsy revealed massive hepatomegaly and ascites with hematopoietic cells destroying the architecture of the liver and the presence of megakaryocytes in this organ and in ascites (Figure 7). FACS analysis of ascitic fluid demonstrated that 19% of the cells stained positive for CD41, but most of the cells stained positive for Mac-1 and/or Gr-1 (Figure 7). In addition and consistent with previous findings in other affected mice (Figure 5), FACS analysis showed high numbers of CD41+ cells in the bone marrow (33%), and microscopic examination demonstrated the presence of many megakaryocytes in the spleen (Figure 7). On the basis of the long latency of the disease, it is likely that secondary mutations in BCR/ABL-expressing cells occurred, causing progression to this accelerated phenotype.
Invasion of nonhematopoietic tissues with megakaryocytes in an animal that died of advanced BCR/ABL disease. (A,B) FACS analysis of cells that were isolated from the ascitic fluid stained for the megakaryocyte- and platelet-specific marker CD41 and for the myeloid markers Gr-1 and Mac-1. (C) Histology of the ascitic cells stained with Wright Giemsa demonstrates cells at various stages of maturation. (D) Megakaryocytes (arrow) were identified in cytospin preparations of liver cells. The liver was massively enlarged and invaded by malignant hematopoietic cells (E) that destroyed the architecture of the organ. (F) The liver at higher magnification is shown. Original magnification of panels C-F, × 60. Percentages in A and B indicate percent positive for the respective markers.
Invasion of nonhematopoietic tissues with megakaryocytes in an animal that died of advanced BCR/ABL disease. (A,B) FACS analysis of cells that were isolated from the ascitic fluid stained for the megakaryocyte- and platelet-specific marker CD41 and for the myeloid markers Gr-1 and Mac-1. (C) Histology of the ascitic cells stained with Wright Giemsa demonstrates cells at various stages of maturation. (D) Megakaryocytes (arrow) were identified in cytospin preparations of liver cells. The liver was massively enlarged and invaded by malignant hematopoietic cells (E) that destroyed the architecture of the organ. (F) The liver at higher magnification is shown. Original magnification of panels C-F, × 60. Percentages in A and B indicate percent positive for the respective markers.
Number of progenitor cells from the megakaryocytic lineage is increased
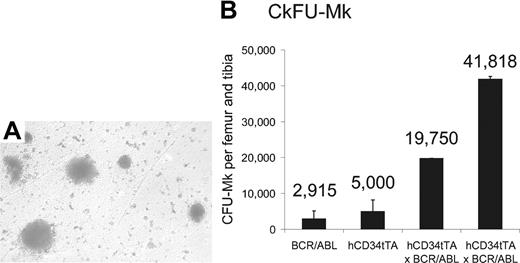
On the basis of the results described in “Invasion of tissue by megakaryocytes at advanced stage of induction” in the paragraph above, we hypothesized that the high levels of expression of BCR/ABL in MEPs induced proliferation of progenitor cells of the megakaryocytic lineage in the bone marrow. To test this hypothesis, we performed colony-forming unit assays for megakaryocytic progenitor cells (CFU-MK) on bone marrow cells from double transgenic animals and control mice. We observed a significant increase of megakaryocytic progenitor colonies in the bone marrow of CD34tTA-BCR/ABL mice compared with nontransgenic littermates (Figure 8). In addition, as immunohistochemistry had demonstrated expression of the cre reporter protein in CMP, we performed CFU-GEMM and CFU-GM assays. However, consistent with a phenotype only affecting the megakaryocytic lineage in vivo, no difference in numbers of these colonies was observed. Specifically, CFU-GEMMs were 71 053 ± 4823 and 58 751 ± 8247 and CFU-GMs were 40 763 ± 7643 and 33 548 ± 16 002 per femur and tibia derived from 2 hCD34tTA × BCR/ABL induced double transgenic and control animals (1 hCD34tTA single transgenic and 2 BCR/ABL single transgenics), respectively.
Increase of megakaryocytes and their progenitors in the bone marrow of double transgenic mice. CFU-MK assay with subsequent staining for megakaryocyte-specific acetylcholinesterase was performed on bone marrow cells from 1 BCR/ABL single transgenic, 1 hCD34tTA single transgenic, and 2 hCD34tTA-BCR/ABL double transgenic mice. Panel A shows the typical megakaryocytic morphology of the CFU in agar from a double transgenic animal (original magnification, × 100), and (B) shows the number of CFU-MKs per femur and tibia. The error bars indicate the standard deviation on the basis of counting duplicate wells. The number of colonies found in CD34tTA-BCR/ABL bone marrow cells was increased 7-fold.
Increase of megakaryocytes and their progenitors in the bone marrow of double transgenic mice. CFU-MK assay with subsequent staining for megakaryocyte-specific acetylcholinesterase was performed on bone marrow cells from 1 BCR/ABL single transgenic, 1 hCD34tTA single transgenic, and 2 hCD34tTA-BCR/ABL double transgenic mice. Panel A shows the typical megakaryocytic morphology of the CFU in agar from a double transgenic animal (original magnification, × 100), and (B) shows the number of CFU-MKs per femur and tibia. The error bars indicate the standard deviation on the basis of counting duplicate wells. The number of colonies found in CD34tTA-BCR/ABL bone marrow cells was increased 7-fold.
Discussion
Inducible mouse models of human leukemia provide a useful tool to study disease mechanisms and to identify possible drug targets. We developed a model of an inducible P210 BCR-ABL-dependent acute pre-B-cell lymphoblastic leukemia (ALL) using the tet-off system.6 The development of a B-cell phenotype as opposed to chronic myeloid disease was attributed to the expression pattern of the MMTV-LTR, mainly in the B-cell lineage. Here, we describe the phenotype of transgenic mice, in which the cell type-specific expression of BCR/ABL is determined by regulatory elements of the human CD34 gene. Using a transgenic construct that consists of 141-kb genomic sequence of the human CD34 locus and the inserted sequence of the transactivator protein tTA to induce expression of a reporter gene, we were able to show that the reporter is expressed in hematopoietic stem cells, common myeloid progenitor cells, and progenitors of megakaryocytes and erythroid cells.
Double transgenic mice consistently developed a myeloproliferative disorder, mainly affecting the megakaryocytic lineage with extramedullary hematopoiesis, expansion of CD41+ cells in the bone marrow and spleen as well as proliferation of bone marrow-derived CFU-MKs with a mean latency of 5 months. In keeping with the morphology, the number of CD41+ cells as determined by flow cytometry was significantly elevated in these mice. The finding that myeloproliferation in these mice was preferentially detected in the megakaryocytic lineage as opposed to the granulocytic lineage was somewhat unexpected. However, consistent with this result, BCR-ABL expression was higher in the MEP population than in CMPs from which it is derived11 as demonstrated by immunohistochemical detection of the expression pattern of the cre reporter gene. A recent study has characterized monopotent megakaryocyte-committed progenitors (MKPs) downstream of CMPs and MEPs. MKPs are thought to arise from myeloid progenitors with the sole potential for the megakaryocytic lineage,23 and it is possible that our transgene is expressed in this progenitor. In another study using retroviral transduction to deliver P210 BCR/ABL into CD34+ cord blood cells resulted in the outgrowth of erythroid colonies with suppression of granulopoiesis.24 We do not think that the increased platelet count is a result of the mice being moribund, because (1) only 5 of 12 mice were moribund; (2) the mice were not anemic, which would be likely if they were severely dehydrated, resulting in falsely increased blood counts, while they in fact had a selective increase in platelets and not any other cell type; and (3) the mice demonstrated a marked increase in megakaryocytes (Figure 4) and CFU-MKs (Figure 8), indicating that they had increased platelet precursors.
The expression pattern of the human CD34tTA transgene is slightly different from that of the unmodified hCD34 construct.9 This change might be due to the effects of the tTA protein on cells. Toxicity of the tTA protein, which consists of the tet repressor and the transcriptional activation domain of VP16, was initially suspected as the cause for low expression levels25 and for “squelching”26 by the VP16 transactivator domain, leading to death of some of the cells that express tTA protein. Nevertheless, the phenotype does fit in the spectrum of features that are seen in patients with Philadelphia chromosome-positive (Ph+) disease. Although CML is a clonal disorder of pluripotent hematopoietic cells27 which is morphologically characterized by myeloproliferation with preferential expansion of the granulocytic compartment and extramedullary hematopoiesis, up to 50% of patients with CML may present with thrombocytosis.28 Histologic studies have shown that patients with CML have significantly elevated numbers of megakaryocytes in the bone marrow.28 It could be argued that the phenotype of the mice resembled essential thrombocythemia closer than CML. However, Ph+ patients with thrombocytosis are not clearly distinguishable from patients with CML, and according to recent a suggestion should be classified as CML.29 Although isolated thrombocytosis without significant leukocytosis is infrequent in Ph+ patients, several cases have been described in the literature.30,31
In addition to the cell type that expresses the BCR/ABL oncogene, the location of the breakpoint of the bcr gene within the BCR/ABL fusion gene is considered as an important factor in the development of thrombocytosis in patients with Ph+ disease. The results of several studies have shown that patients with the B3A2 translocation have higher platelet counts than patients with the B2A2 translocation.32,33 These reports are of particular interest as the animal model described in this study uses the B3A2 form. Thrombocytosis was also observed in a transgenic model that used the tec promoter to express the BCR/ABL oncogene with the B3A2 translocation.34
BCR/ABL is a very potent oncogene; therefore, the relatively mild nature of the phenotype in double transgenic animals compared with the disease that develops in animals of the same transgenic TRE-BCR/ABL line in which expression is driven by the MMTV-tTA transactivator line is surprising. The leukemogenic potential of BCR/ABL is determined by the tyrosine kinase activity of the oncogene, which differs among the 3 different isoforms, with P190 having the highest, followed by P210, and P230 with the lowest.35 However, the expression level of the oncogene is also an important factor. Studies for minimal residual disease (MRD) in patients with CML after allogeneic bone marrow transplantation have shown that the number of transcripts can predict the outcome of treatment.36 The same conclusion was drawn from studies on patients treated with interferon, in which low levels or absence of MRD measured with a highly sensitive quantitative real-time PCR for BCR/ABL transcripts were associated with continuing remission.37,38 Those studies had quantified the transcript level for BCR/ABL to the level for abl, which served as an internal control, thus giving an accurate ratio of both transcripts on an individual cell basis. We conclude from the results of Northern blot analysis comparing the expression levels in tissues from CD34tTA-BCR/ABL mice versus MMTVtTA-BCR/ABL animals (Figure 6) that the expression level in CD34tTA animals is much lower, most likely causing the considerable latency period and mild phenotype.
Although the relative levels of BCR/ABL expression may be a determinant in the severity of the phenotype, another hypothesis is that the type of disease observed in BCR/ABL models is determined by the type of progenitor cell in which the regulatory cassette is expressed. A number of other transgenic models of BCR/ABL disease have been generated, but to date a detailed description of the pattern of expression of the transgene in different hematopoietic subsets to correlate with phenotype has not yet been defined.34,39-44 However, so far the available data are consistent with this model. For example, expressing high levels of BCR/ABL in early B cells resulted in a model of B-cell ALL,6 and in this study we have expressed relatively low levels of BCR/ABL in MEPs and observe a milder myeloproliferative syndrome characterized by megakarycytosis. This hypothesis is further supported by studies in which we used regulatory elements of the murine stem cell leukemia (SCL) gene45,46 to express tTA in HSCs and myeloid progenitors. Preliminary results indicate that all animals develop granulocytosis more typical of chronic-phase CML.47 Further analysis using multiple tetracycline transactivator strains expressing in different hematopoietic precursors will enable us to address this hypothesis in a more definitive manner in the future.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2003-03-0768.
Supported by the Jose Carreras Leukemia Foundation (international fellowship JC 2000) (C.S.H.), a fellowship from the Deutsche Forschungsgemeinschaft (DFG) (S.K.), a fellowship from The Golden Family Foundation Award (H.S.R.), and the National Institutes of Health (grant DK48 660) (D.G.T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ”advertisement” in accordance with 18 U.S.C. section 1734.
We thank Kristin Geary and Maris Fenyus for animal husbandry and genotyping, Joel Lawitts and the Beth Israel Deaconess Medical Center Transgenic Facility for production of transgenic mice, Nikla Emamboklu for assistance with immunohistochemical analysis of Cre, Maris Handley from the Dana-Farber Flow Cytometry Core Facility for assistance with cell sorting, Ramesh Shivdasani for assistance with quantitation of megakaryocytes and platelets and other suggestions, other members of the Tenen Laboratory and Rick van Etten for valuable advice, and Alison Lugay and Mary Singleton for assistance with the manuscript.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal