Abstract
Retinoic acid induces clinical remission in acute promyelocytic leukemia (APL) by triggering differentiation of leukemia promyelocytes. Here, we have characterized a gene encoding a member of the immunoglobulin superfamily, among novel retinoic acid–induced genes identified in APL cells. This protein, which was named JAML (junctional adhesion molecule–like), contains 2 extracellular immunoglobulin-like domains, a transmembrane segment, and a cytoplasmic tail. JAML mRNA is expressed in hematopoietic tissues and is prominently expressed in granulocytes. The fact that JAML protein is localized at the cell plasma membrane in the areas of cell-cell contacts, whereas it is not detected at free cell borders, suggests that JAML is engaged in homophilic interactions. Furthermore, a conserved dimerization motif among JAM members was shown to be important for JAML localization at the cell membrane. Finally, exogenous expression of JAML in myeloid leukemia cells resulted in enhanced cell adhesion to endothelial cells. Altogether, our results point to JAML as a novel member of the JAM family expressed on leukocytes with a possible role in leukocyte transmigration.
Introduction
Treatment of acute promyelocytic leukemia (APL) with retinoic acid (RA) is a seminal example of a therapeutic strategy aimed at targeting the activities of a specific oncoprotein with a cell differentiation agent.1-3 The induction of remission is associated with the terminal differentiation of immature leukemic cells into neutrophils. In 95% of clinical APL cases, a specific translocation t(15;17)4 creates a promyelocytic leukemia–retinoic acid receptor alpha (PML-RARα) fusion between the N-terminal region of the PML locus 5-8 and the C-terminus of the RARα.9,10 Expression of PML-RARα in myeloid cell lines increases RA sensitivity11 and RA-induced differentiation of APL cells is dependent on the presence of PML-RARα. This suggests that differentiation of APL cells by RA is triggered through a PML-RARα–dependent signaling pathway.
When hematopoietic cells leave the circulation, they tether to and roll on the endothelial cell surface, spread, and finally emigrate through the endothelial cells to reach the underlying tissues. Such mechanisms have been demonstrated for hematopoietic stem cell homing and engraftment12 and for leukocytes emigration from the bloodstream in response to changes on the surface of blood vessels that signal injury or infection.13,14 Transient opening of interendothelial contacts allows leukocytes to emigrate through the endothelial cells.15 Although a key role has been demonstrated for platelet endothelial cell adhesion molecule (PECAM)16-18 and for junctional ahesion molecule 1 (JAM-1),19 the molecular mechanisms underlying the diapedesis of leukocytes through the endothelial monolayer are not as yet fully understood. PECAM and JAM share stuctural and functional features. Both belong to the immunoglobulin superfamily (IgSF) and engage in homo- and heterotypic interactions.17,19 Two other members of the JAM family, JAM-220,21 (also known as VE-JAM22 ) and JAM-3,23,24 have been identified as components of interendothelial junctional complexes. On leukocytes, a JAM counter-receptor was postulated19 although it is still poorly characterized. JAM-3, which is up-regulated following T-cell activation, is such a JAM-2 counter-receptor.23 More recently, JAM proteins were shown to interact with integrins. JAM-1 was shown to be a ligand of the β2 integrin LFA-1 and to contribute to LFA-1–dependent transendothelial migration of T cells and neutrophils.25 JAM-2 interacts with α4 β2 integrins in T cells26 and JAM-3 interacts with the β2 integrin Mac-1 in platelets.27
In order to identify RA-target genes, which are critical to the process of differentiation, we have used a differential screening strategy with the NB4 APL cell line, which harbors the t(15;17) translocation encoding PML-RARα. These cells undergo granulocytic differentiation when treated with all-trans retinoic acid (ATRA).28 Among 4 novel genes that we have cloned as induced by RA in NB4 cells, we have identified a junctional adhesion molecule–like gene here named JAML (junctional adhesion molecule–like). The predicted JAML protein contains a signal peptide, 2 IgSF domains, a transmembrane segment and an intracellular tail. JAML shares structural homologies with molecules involved in adhesion, such as JAM. Here we show that JAML is a novel JAM protein that is expressed in mature hematopoietic cells. Furthermore, JAML protein is localized at the cell plasma membrane in the areas of cell-cell contacts whereas it was not detected at free cell borders, suggesting that JAML is engaged in homophilic interactions. Finally, JAML expression enhances myeloid leukemia cell adhesion to endothelial cells.
Materials and methods
Cell lines, human cells, culture conditions, induction, and measurements of differentiation
PLB-985,29 NB4,28 NB4.306,30 and HL-60 cells31 were grown and used as described.32,33 HT-29 cells were grown on 9-cm Petri dishes in Dulbecco modified Eagle medium (DMEM) containing 4.5 g/L glucose and 5% fetal bovine serum (Invitrogen, Cergy Pontoise, France). COS-7 cells were grown in 10-cm Petri dishes in DMEM containing 1 g/L glucose, 5% fetal bovine serum, and 1X nonessential amino acids (Invitrogen). U937 and MDCK cells were grown in DMEM containing 10% fetal bovine serum. Human bone marrow endothelial cells (HBMECs34 ; kindly provided by Dr B. Weksler, Weill Medical College of Cornell University, NY), were cultured in DMEM Glutamax supplemented with 10% heat inactivated fetal bovine serum, 7.5 μg/mL endothelial cell growth supplement (Sigma-Aldrich, St Louis, MO), 7 IU heparin (Sigma-Aldrich) and 10 mM HEPES. Cell viability was estimated using standard trypan blue dye exclusion assay. ATRA (Sigma-Aldrich) was dissolved in ethanol. Me2SO (Sigma-Aldrich) was used at the final concentration of 1.25%. Phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) in Me2SO was used at a final concentration of 107 nM. The final concentration of Me2SO was 0.002% and had no effect on differentiation. Differentiation was assessed by the percentage of cells with cell-associated nitro blue tetrazolium (NBT; Sigma-Aldrich), and cell morphology as examined under light microscopy on cytospin slides stained with May-Grünwald-Giemsa.
cDNA library construction and screening
RNAs from NB4 cells before and after 24-hour treatment with 9-cis RA, and HT-29 cells, were extracted using the guanidinium isothiocyanate lysis buffer and purified by centrifugation through cesium chloride cushion. Poly(A)+ RNA was purified using the Oligotex affinity resin (Qiagen, Hilden, Germany). An RA-treated NB4 cDNA library was constructed in the Uni-ZAP XR vector (Stratagene, La Jolla, CA) using the manufacturer's protocol. Using 5 μg poly(A)+ RNA, 3 million recombinant phages were obtained and 120 000 of them were directly plated and transfered onto nylon filter replicas (Hybond NX; Amersham Biosciences, Freiberg, Germany). Subtracted probes were prepared as previously described.36 Briefly, the untreated NB4-specific probe (minus probe) was obtained from 2 μg NB4 poly(A)+ RNA that was reverse transcribed into 320 ng single-stranded cDNA and hybridized to 9.6 μg (30× excess) of HT-29 poly(A)+ RNA. The RA-treated NB4-specific probe (plus probe) was obtained from 3 μg RA-treated NB4 poly(A)+ RNA that was converted into 400 ng single-stranded cDNA and hybridized to 12 μg NB4 poly(A)+ RNA (30× excess). After hydroxylapatite chromatography, 10% and 8% of the cDNA remained single stranded for minus and plus probes, respectively. Both probes (6 ng) were labeled with α-[32P] deoxycytidine triphosphate (dCTP) (111 TBq/mmol) using a random primer labeling kit (Rediprime; Amersham Biosciences) to obtain 4.8 × 107 and 7.6 × 107 cpm, respectively. Nylon replicas were first hybridized with the minus probe (0.8 × 106 cpm/mL) at 42°C in 50% formamide, 5 × saline sodium citrate (SSC), 0.5% sodium dodecyl sulfate (SDS), 0.04% polyvinylpyrrolidone, 0.04% Ficoll, 20 mM sodium pyrophosphate pH 6.5, 10% dextran sulfate, and 100 μg/mL denatured salmon sperm DNA for 36 hours. Stringent washes were performed at 60°C in 0.2 × SSC and 0.1% SDS. Filters were autoradiographed at -80°C for 12 hours and 144 hours using BioMax MS films (Kodak, Rochester, NY). The minus probe was removed using alkali procedure, and filters were hybridized with the plus probe (1.3 × 106 cpm/mL) for 36 hours, stringently washed, and autoradiographed as described above. A secondary screening was performed using the same protocol on 885 plaques giving differential signals with the plus and minus probes.
Plasmid recovery and analysis
Pure plaques were directly recovered as bacterial colonies using the pBluescript/Lambda ZAPII in vivo excision system (Stratagene). Plasmid minipreparations were performed using the Biomek 2000 robot (Beckman Coulter, Brea, CA), digested with EcoRI and XhoI, loaded and electrophoresed into 2 parallel 1% agarose gels. Gels were then blotted onto Hybond-N nylon membranes (Amersham Biosciences), which were hybridized to the plus and minus probes. Inserts from differential clones were purified from agarose gels using spin-X columns (Corning, Corning, NY) and 32P-labeled by random priming. These probes were hybridized to both replicas of the library and dot blots containing previously prepared plasmids in order to perform cross-hybridization analysis.
Sequencing and computer analysis
Plasmid DNA was purified through Nucleobond columns (Macherey-Nagel, Düren, Germany) and double-stranded DNA templates were sequenced using pBluescript-specific and internal primers. Sequence analysis and alignments were carried out with the GCG programs (Wisconsin Package, version 12.0-UNIX; Accelrys, Paris, France). Sequence homologies were identified using the FastA and BLAST programs by searching the GenBank (release 130.0) and EMBL (release 71.0) data banks and in the case of translated sequences by searching the SWISS-PROT (release 40.0), PIR/NBRF (release 73.0), and SPTREMBL (release 21.0) data banks. The bos taurus JAML protein was deduced by translation of accession number BG688339 in comparison to the human JAML sequence.
To determine a dendogram with various IgSF family members containing 2 Ig domains, the alignment was performed using CLUSTALW.37 The phylogenetic tree was constructed using the Neighbor-Joining/UPGMA method (version 3.6a3). The PROTDIST program (version 3.6) was used to compute distance matrix under the Jones, Taylor, and Thornton model of amino acids change. The phylogenetic tree was plotted using NJPlot.38
JAML cDNA cloning
The JAML cDNA fragment isolated in the differential screening contained 1484 base pair (bp) corresponding to the 3′ end of JAML cDNA. A remaining 5′ cDNA sequence was amplified by rapid amplification of complementary DNA ends–polymerase chain reaction (RACE-PCR) from RA-treated NB4 RNA (with 5 × 10-7 M for 48 hours) using the Marathon cDNA amplification kit (Clontech, Palo Alto, CA) with the following primer: 5′-TGGTGACAGAGTCCAGTCTATCTTG-3′. Using the same kit, a contiguous cDNA was generated by reverse transcriptase (RT)–PCR amplification of JAML from RA-treated NB4 RNA. To determine the chromosomal localization and the intron/exon structure of the JAML gene, its complete cDNA was used to retrieve genomic data from databases. Structures of the JAM-1, JAM-2, and JAM-3 have been previously described.23
Eukaryotic expression vectors, in vivo expression, and protein extracts
The JAML coding sequence tagged with the hemagglutinin (HA) epitope at its C-terminus was subcloned into the p513 vector, a derivative of the pSG5 expression vector,39 resulting in the p513-JAML-HA vector; and into the pMTCB6+-derived expression vector40 under the control of the zinc-inducible sheep metallothioneine promoter resulting in the pMT-JAML-HA vector. Construction of the pMT-JAMLK54D-HA vector was achieved by using the QuickChange site-directed mutagenesis kit (Stratagene). The forward oligonucleotide sequence was used (mutated base pairs in bold): 5′-ACAGAAGACAAATGTATATTCGACATAGACTGGACTCTGTCAC-3′. COS-7 cells were transfected using calcium phosphate coprecipitation41 of 0.5 μg DNA vectors (adjusted to 14 μg per 10-cm Petri dish with pBluescript carrier DNA). Medium was changed after 16 hours. Thirty-six hours after transfection, COS-7 cells were harvested, washed once in phosphate-buffered saline (PBS), and resuspended in Laemmli buffer. MDCK and U937 cells were washed twice in serum-free medium, once in Opti-MEM (Invitrogen), resuspended in the same medium at 15 × 106 cells in 0.5 mL, and electroporated (Gene Pulser; Biorad, Hercules, CA) at 300 V, 960 μF with 20 μg of the empty pMT, pMT-JAML-HA, and pMT-JAMLK54D-HA vectors. Cells were then cultured for 48 hours prior to selection with 0.4 μg/mL G418 (Invitrogen).
Northern blot analysis
Total RNA extraction and hybridization were as described.32 Human RNA master blot and human immune system multiple tissue Northern blot II were obtained from Clontech. The JAML probe corresponded to the 3′ end (1484 bp) of the JAML cDNA. Probes were labeled with α-[32P] dCTP (111 TBq/mmol) using a random primer labeling kit (Amersham Bioscience). Radioactivity was detected and quantified using a Storm 860 Phosphor Imager (Amersham Bioscience).
Antibodies and Western blot analysis
A polyclonal rabbit antiserum was raised against the JAML coding sequence lacking N-terminal amino acids (1-19) fused with the glutathione S-transferase (GST) sequence generated by cloning the corresponding JAML sequence into a derivative of pGEX-3X (Amersham Biosciences). After 5 minutes boiling in Laemmli buffer, samples were resolved by SDS–polyacrylamide gel electrophoresis (PAGE), and transferred onto polyvinylidenefluoride (PVDF) membranes (Perkin-Elmer Life Sciences, Boston, MA). Background immunoreactivity was reduced by preincubating membranes with PBS containing 5% nonfat dry milk and 0.1% Tween 20 for 1 hour. Western analysis was carried out with anti-JAML diluted 1:2000 in PBS containing 0.1% Tween 20 or anti-HA monoclonal antibody (Cell Signaling Technology, Beverly, MA) diluted 1:10 000 in PBS containing 0.5% Tween 20. Detection of antibody binding was achieved with horseradish peroxidase–conjugated secondary antibodies (Jackson Laboratories, West Grove, PA). Enzymatic activity was detected using the chemiluminescence reagent plus kit (Perkin-Elmer Life Sciences) and autoradiography. Anti–β-actin antibody was purchased from Santa Cruz Biotechnology, Santa Cruz, CA.
In vitro transcription/translation
In vitro–translated proteins were synthetized with the p513-JAML-HA vector in the presence of [35S]-methionine using the TNT Coupled Reticulocyte Lysate System (Promega, Madison, WI). Glycosylations were achieved using the canine microsomal membrane system as recommended by the manufacturer (Promega). Proteins were visualized by autoradiography after separation by SDS-PAGE.
Immunofluorescence
Transfected MDCK cells were grown on glass slides for 8 hours, and for zinc-induced expression of JAML, 125 μM ZnSO4 was added to the medium. Forty hours after zinc treatment, cells were washed and fixed for 5 minutes with 2% paraformaldehyde in PBS. Cells were permeabilized with PBS, 0.1% Triton X100 for 5 minutes. After a 30-minute treatment with blocking solution (PBS, 1% bovine serum albumin [BSA]), cells were stained for 1 hour with a 1:10 000 dilution of anti-HA monoclonal antibody. Goat anti–mouse-Cy3 (Jackson Laboratories) at a dilution of 1:400 was used as a secondary antibody. DNA was counterstained with Hoechst 33258 dye, and preparations were mounted in mowiol (Calbiochem, Novabiochem, San Diego, CA). Fluorescence was viewed using an Olympus microscope equipped with appropriate optics and filter module for Cy3 detection.
Adhesion assay
When confluent, HBMEC monolayers were either left untreated or pretreated with 100 U/mL recombinant tumor necrosis factor α (TNFα) for 24 hours in DMEM supplemented with 0.1% BSA. U937 cells were stably transfected with the pMT, pMT-JAML-HA, or pMT-JAMLK54D-HA expression vectors and either induced by addition of 100 μM ZnSO4 for 18 hours or left uninduced. U937 cell populations were fluorescently labeled by incubation for 1 hour in RPMI medium containing 10 μL green CFMDA (Molecular Probes, Eugene, OR). After 3 washes in PBS, labeled U937 cells were resuspended in DMEM supplemented with 0.1% BSA and 106 cells were added into each well of 24-well -plates containing confluent HBMEC monolayers. After 30 minutes of incubation at 37°C, nonadherent U937 cells were removed by 3 consecutive washes. Adherent U937 cells were then lysed with 1% Triton X100 and released fluorescence was quantified using a fluorometer (Fusion; Perkin-Elmer). The percentage of adherent U937 cells was calculated in each condition as the ratio between the mean fluorescence recovered in 4 replicate wells and the total amount of fluorescence corresponding to the labeled U937 cells incubated in each well.
Results
JAML mRNA is up-regulated in ATRA-treated APL cells
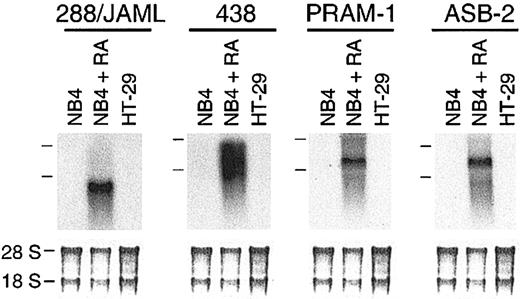
Using subtractive hybridization, we have identified 23 genes induced in RA-treated NB4 cells. Among 4 novel genes (288/JAML,438, PRAM-1, and ASB-2; Figure 1), PRAM-1 encodes an adaptor protein functionally linked to an RA-dependent signaling pathway33 and ASB-2, a protein involved in growth inhibition and commitment in myeloid leukemia cells.42 Nineteen other genes corresponded to known sequences in gene databases (Table 1). Most of the known sequences were characteristics of the myeloid lineage; 2 (CAMPATH-1 and GLUT5) were not known to be induced by RA.
Identification of 4 novel genes induced in RA-treated NB4 cells. (A) Autoradiogram of 288/JAML, 438, 809, and 813 mRNA expression in NB4 cells either untreated or treated for 24 hours with 5 × 10-7 M 9-cis RA, and in HT-29 cells as a nonhematopoietic negative control. Positions of the 28S and 18S rRNAs are indicated on the left side. The lower part is methylene blue–stained 28S and 18S rRNAs on membranes after transfer as assessment of RNA quantities in each lane (5 μg total RNA).
Identification of 4 novel genes induced in RA-treated NB4 cells. (A) Autoradiogram of 288/JAML, 438, 809, and 813 mRNA expression in NB4 cells either untreated or treated for 24 hours with 5 × 10-7 M 9-cis RA, and in HT-29 cells as a nonhematopoietic negative control. Positions of the 28S and 18S rRNAs are indicated on the left side. The lower part is methylene blue–stained 28S and 18S rRNAs on membranes after transfer as assessment of RNA quantities in each lane (5 μg total RNA).
Characteristics of the differential cDNAs identified
Clone . | Description . | GenBank accession no. . |
|---|---|---|
| 3 | 5-lipoxygenase activating protein (FLAP) | X52195 |
| 13 | Monocyte chemoattractant protein 1 (MCP-1) | X14768 |
| 16 | Neutrophil cytosol factor 1 (p47phox) | M25665 |
| 20 | G0S2 | M69199 |
| 26 | Cellular transglutaminase homologue (TGase H) | M98479 |
| 301 | CD11b | J03925 |
| 617 | Plasminogen activator inhibitor-2 (PAI-2) | J03603 |
| 685 | Lymph node homing receptor (L-selectin) | M25280, X16070 |
| 685 | Leu-8 | X17519 |
| 685 | LAM-1 | X16150 |
| 732 | Interleukin-8 (IL-8) | M17017 |
| 748 | Bfl-1/A1 | U27467, U29680 |
| 752 | Defensin 1 | M26602, M21130 |
| 756 | Corticostatin HP-4 (defensin 4) | X65977 |
| 781 | Delta-aminolevulinate synthase (ALAS1) | X56351 |
| 796 | CAMPATH-1 (CD52) | X62466 |
| 806 | Coactosin-like protein (CLP) | L54057 |
| 808 | ICAM-1 | J03132 |
| 820 | c-fgr | M19722 |
| 832 | GLUT5 glucose transporter | M55531 |
| 850 | Src-like adapter protein (SLAP) | D89077 |
Clone . | Description . | GenBank accession no. . |
|---|---|---|
| 3 | 5-lipoxygenase activating protein (FLAP) | X52195 |
| 13 | Monocyte chemoattractant protein 1 (MCP-1) | X14768 |
| 16 | Neutrophil cytosol factor 1 (p47phox) | M25665 |
| 20 | G0S2 | M69199 |
| 26 | Cellular transglutaminase homologue (TGase H) | M98479 |
| 301 | CD11b | J03925 |
| 617 | Plasminogen activator inhibitor-2 (PAI-2) | J03603 |
| 685 | Lymph node homing receptor (L-selectin) | M25280, X16070 |
| 685 | Leu-8 | X17519 |
| 685 | LAM-1 | X16150 |
| 732 | Interleukin-8 (IL-8) | M17017 |
| 748 | Bfl-1/A1 | U27467, U29680 |
| 752 | Defensin 1 | M26602, M21130 |
| 756 | Corticostatin HP-4 (defensin 4) | X65977 |
| 781 | Delta-aminolevulinate synthase (ALAS1) | X56351 |
| 796 | CAMPATH-1 (CD52) | X62466 |
| 806 | Coactosin-like protein (CLP) | L54057 |
| 808 | ICAM-1 | J03132 |
| 820 | c-fgr | M19722 |
| 832 | GLUT5 glucose transporter | M55531 |
| 850 | Src-like adapter protein (SLAP) | D89077 |
Leu-8 indicates leukocyte surface antigen Leu-8; LAM-1, leukocyte adhesion molecule-1; Bfl-1/A1, Bcl-2—related gene expression in fetal liver-1/Bcl-2—related protein A1; ICAM-1, intercellular adhesion molecule-1; and c-fgr, Gardner-Rasheed feline sarcoma viral proto-oncogene.
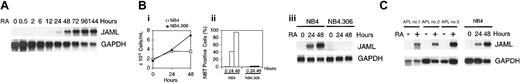
ATRA-induced granulocytic differentiation of NB4 cells was accompanied by progressive up-regulation of the JAML mRNA, which was detected 12 hours after treatment (Figure 2A). As expected, ATRA did not affect growth and differentiation (Figure 2Bi-ii, respectively) of NB4.306, an ATRA-resistant NB4 subclone, which has lost the capacity to encode an intact ATRA-binding PML-RARα fusion protein.30 In these cells, no induction of JAML mRNA was observed (Figure 2Biii). In contrast, expression of JAML mRNA was strongly up-regulated in NB4 cells induced to growth arrest and differentiation by ATRA (Figure 2B). JAML mRNA was also highly induced by ATRA in blasts freshly obtained from peripheral blood of newly diagnosed patients with APL (Figure 2C) and expressing the PML-RARα gene. This induction correlated with ATRA-induced differentiation of these cells as assessed morphologically and by NBT reduction assay (data not shown). Altogether, our data strongly suggested that increased JAML mRNA expression was associated with the capacity of APL cells to undergo granulocytic differentiation when treated with ATRA.
Induction of JAML mRNA in RA-treated cells. (A) Time-response to ATRA in NB4 cells. Cells were treated for different times using 5 × 10-7 M ATRA. (B) JAML expression correlates with the capacity of NB4 cells to differentiate. NB4 and NB4.306 cells were cultured with 10-6 M ATRA and harvested after 0, 24, and 48 hours. Viable cells (i) and percent of NBT-positive cells (ii) were counted each day and JAML mRNA expression was analyzed (iii). (C) JAML mRNA is induced in ATRA-treated primary APL cells. Cells were purified from 3 untreated patients cultivated in the absence (–) or presence (+) of 10-7 M ATRA for 5 (APL no. 2)or6(APL no. 1 and APL no. 3) days. Northern blots were performed using 8 μg (A,Biii) or 2 μg (C) total RNA. Hybridization with a glyceraldehyde phosphate dehydrogenase (GAPDH) probe controlled for RNA quantities in each lane.
Induction of JAML mRNA in RA-treated cells. (A) Time-response to ATRA in NB4 cells. Cells were treated for different times using 5 × 10-7 M ATRA. (B) JAML expression correlates with the capacity of NB4 cells to differentiate. NB4 and NB4.306 cells were cultured with 10-6 M ATRA and harvested after 0, 24, and 48 hours. Viable cells (i) and percent of NBT-positive cells (ii) were counted each day and JAML mRNA expression was analyzed (iii). (C) JAML mRNA is induced in ATRA-treated primary APL cells. Cells were purified from 3 untreated patients cultivated in the absence (–) or presence (+) of 10-7 M ATRA for 5 (APL no. 2)or6(APL no. 1 and APL no. 3) days. Northern blots were performed using 8 μg (A,Biii) or 2 μg (C) total RNA. Hybridization with a glyceraldehyde phosphate dehydrogenase (GAPDH) probe controlled for RNA quantities in each lane.
The JAML protein is a novel member of the IgSF localized at areas of cell-cell contacts
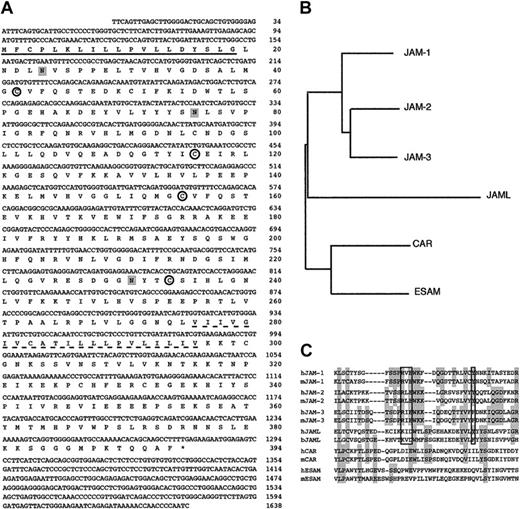
A 1638-bp JAML cDNA (GenBank Accession no. AJ515553) obtained from ATRA-treated NB4 cells (Figure 3A) contains a complete open reading frame predicted to encode a protein of 394 amino acids followed by a 3′-untranslated region and a poly(A) tail. A 19 amino acids signal peptide was predicted using SignalP.43 Overall, the cDNA is predicted to encode a type I integral transmembrane protein with an extracellular portion of 275 amino acids, a transmembrane segment of 21 amino acids, and a cytoplasmic tail of 98 amino acids. The extracellular region of JAML comprises 2 Ig domains resembling surface antigens such as epithelial v-like antigen (EVA),44 human Coxsackie and adenovirus receptor (HCAR),45,46 and JAM proteins.23 The ectodomain possesses 2 Ig-like folds, the N-terminal domain is a C2-type whereas the membrane proximal domain is a V-type. When compared with other IgSF members, JAML appears to be related to the JAM family (Figure 3B). A motif (KID58 flanked by Y72 in human, KVD58 flanked by F72 in bos taurus) similar to the dimerization motif R(V, I, L)E flanked by an aromatic amino acid Tyr/Phe which was shown to be essential for JAM dimers formation in solution47 was found in JAML proteins (Figure 3C). By using information derived from the chromosome 11 sequence, we mapped the human JAML gene to the 11q23 region. A remarkable conservation of gene structure and phase of introns within the members of the JAM family points to their evolutionary relationship (Table 2). Divergence between JAML and JAM-1, JAM-2, and JAM-3 occurs in the 5′ noncoding region of JAML, which is encoded by a separate exon, and in the first IgSF domain, which is encoded by 2 exons. Altogether, this indicates that we have identified a novel protein exhibiting homologies with members of the JAM family.
JAML is related to JAM proteins. (A) cDNA and deduced amino acid sequences of the human JAML. Nucleotide residues are numbered in the 5′ to 3′ orientation, and amino acids in the reading frame are designated by the one-letter code. Asterisk indicates the stop codon. The putative hydrophobic signal peptide (underlined) and transmembrane sequences (dotted underline) are marked. Potential N-linked glycosylation sites are shaded. Cysteines likely to form disulfide bonds in the 2 Ig domains are circled. (B) Dendogram of various IgSF family members containing 2 Ig domains (JAM-1, JAM-2, JAM-3, JAML, Coxsackie and adenovirus receptor (CAR), and endothelial cell-selective adhesion molecule (ESAM)). (C) Alignment of the amino-terminal region of the membrane distal Ig domains of these proteins and their mouse (JAM-1, JAM-2, JAM-3, CAR, ESAM) or bos taurus (JAML) homologs. Shaded regions represent residues identical between at least 4 proteins. The dimerization motif is boxed.
JAML is related to JAM proteins. (A) cDNA and deduced amino acid sequences of the human JAML. Nucleotide residues are numbered in the 5′ to 3′ orientation, and amino acids in the reading frame are designated by the one-letter code. Asterisk indicates the stop codon. The putative hydrophobic signal peptide (underlined) and transmembrane sequences (dotted underline) are marked. Potential N-linked glycosylation sites are shaded. Cysteines likely to form disulfide bonds in the 2 Ig domains are circled. (B) Dendogram of various IgSF family members containing 2 Ig domains (JAM-1, JAM-2, JAM-3, JAML, Coxsackie and adenovirus receptor (CAR), and endothelial cell-selective adhesion molecule (ESAM)). (C) Alignment of the amino-terminal region of the membrane distal Ig domains of these proteins and their mouse (JAM-1, JAM-2, JAM-3, CAR, ESAM) or bos taurus (JAML) homologs. Shaded regions represent residues identical between at least 4 proteins. The dimerization motif is boxed.
Comparison of gene structure between JAM family members
HGNC exon . | No. . | JAM-1 1q21.2-21.3 . | i . | JAM-2 21q21.2 . | i . | JAM-3 11q25 . | i . | JAML 11q23.3 . | i . |
|---|---|---|---|---|---|---|---|---|---|
| 5′ | 0 | — | — | — | — | — | — | >73 | — |
| Intron | 0-1 | — | — | — | — | — | — | 10 054 | — |
| ATG/signal sequence | 1 | 103 | — | >301 | — | >76 | — | 64 | — |
| Intron | 1-2 | 44 222 | 1 | 43 994 | 1 | 79 105 | 1 | 2262 | 1 |
| EC, Ig-fold 1 | 2 | 59 | — | 66 | — | 66 | — | — | — |
| Intron | 2-3 | 167 | 1 | 5916 | 1 | 740 | 1 | — | — |
| EC, Ig-fold 1 | 3 | 98 | — | 108 | — | 114 | — | 153 | — |
| Intron | 3-4 | 252 | 1 | 3782 | 1 | 3470 | 1 | 1694 | 1 |
| EC, Ig-fold 1 | 4 | 137 | — | 153 | — | 153 | — | 228 | — |
| Intron | 4-5 | 282 | 1 | 4768 | 1 | 398 | 1 | 4493 | 1 |
| EC, Ig-fold 2 | 5 | 203 | — | 203 | — | 203 | — | 110 | — |
| Intron | 5-6 | 167 | 0 | 3290 | 0 | 951 | 0 | 2217 | 0 |
| EC, Ig-fold 2 | 6 | 103 | — | 100 | — | 100 | — | 238 | — |
| Intron | 6-7 | 128 | 1 | 3709 | 1 | 2511 | 1 | 2815 | 1 |
| Transmembrane | 7 | 108 | — | 108 | — | 130 | — | 137 | — |
| Intron | 7-8 | 231 | 1 | 3347 | 1 | — | — | 2384 | 1 |
| IC | 8 | 13 | — | 16 | — | — | — | 95 | — |
| Intron | 8-9 | 304 | 2 | 2890 | 2 | 87 | 2 | 1176 | 2 |
| IC | 9 | 49 | — | 43 | — | 55 | — | 88 | — |
| Intron | 9-10 | 136 | 0 | 2257 | 0 | 321 | 0 | 2318 | 0 |
| IC/STOP | 10 | >937 | — | >283 | — | >36 | — | >452 | — |
HGNC exon . | No. . | JAM-1 1q21.2-21.3 . | i . | JAM-2 21q21.2 . | i . | JAM-3 11q25 . | i . | JAML 11q23.3 . | i . |
|---|---|---|---|---|---|---|---|---|---|
| 5′ | 0 | — | — | — | — | — | — | >73 | — |
| Intron | 0-1 | — | — | — | — | — | — | 10 054 | — |
| ATG/signal sequence | 1 | 103 | — | >301 | — | >76 | — | 64 | — |
| Intron | 1-2 | 44 222 | 1 | 43 994 | 1 | 79 105 | 1 | 2262 | 1 |
| EC, Ig-fold 1 | 2 | 59 | — | 66 | — | 66 | — | — | — |
| Intron | 2-3 | 167 | 1 | 5916 | 1 | 740 | 1 | — | — |
| EC, Ig-fold 1 | 3 | 98 | — | 108 | — | 114 | — | 153 | — |
| Intron | 3-4 | 252 | 1 | 3782 | 1 | 3470 | 1 | 1694 | 1 |
| EC, Ig-fold 1 | 4 | 137 | — | 153 | — | 153 | — | 228 | — |
| Intron | 4-5 | 282 | 1 | 4768 | 1 | 398 | 1 | 4493 | 1 |
| EC, Ig-fold 2 | 5 | 203 | — | 203 | — | 203 | — | 110 | — |
| Intron | 5-6 | 167 | 0 | 3290 | 0 | 951 | 0 | 2217 | 0 |
| EC, Ig-fold 2 | 6 | 103 | — | 100 | — | 100 | — | 238 | — |
| Intron | 6-7 | 128 | 1 | 3709 | 1 | 2511 | 1 | 2815 | 1 |
| Transmembrane | 7 | 108 | — | 108 | — | 130 | — | 137 | — |
| Intron | 7-8 | 231 | 1 | 3347 | 1 | — | — | 2384 | 1 |
| IC | 8 | 13 | — | 16 | — | — | — | 95 | — |
| Intron | 8-9 | 304 | 2 | 2890 | 2 | 87 | 2 | 1176 | 2 |
| IC | 9 | 49 | — | 43 | — | 55 | — | 88 | — |
| Intron | 9-10 | 136 | 0 | 2257 | 0 | 321 | 0 | 2318 | 0 |
| IC/STOP | 10 | >937 | — | >283 | — | >36 | — | >452 | — |
HGNC indicates HUGO Nomenclature Committee; i, phase of introns; EC, extracellular domain; IC, intracellular domain; and Ig-fold, immunoglobulin-like fold.
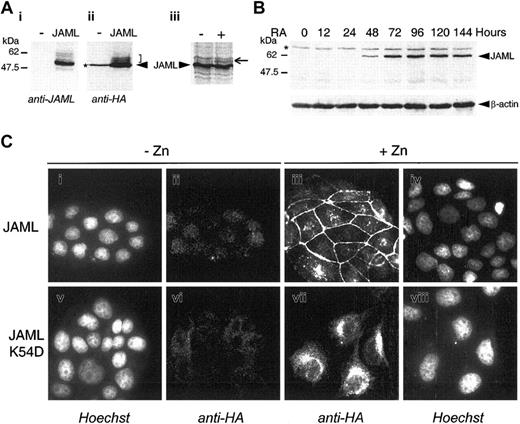
In COS cells transfected with JAML in frame with a carboxy-terminal HA epitope tag, both anti-JAML polyclonal serum and anti-HA monoclonal antibodies detected 3 bands with molecular weights of 60 kDa, 58 kDa, and 50 kDa. We interpreted these bands as corresponding to different glycoforms of the protein (Figure 4Ai-ii). Indeed, when microsomal membranes were added to an in vitro–coupled transcription/translation reaction, a higher-molecular-weight species was detected corresponding to a glycosylated JAML protein (Figure 4Aiii). In ATRA-treated NB4 cells, anti-JAML polyclonal antibodies detected a specific 60 kDa protein 48 hours after treatment (Figure 4B). To investigate the cellular distribution of the JAML protein, a JAML expression vector (pMT-JAML-HA) under the control of the zinc-inducible metallothionein promoter was stably transfected into MDCK cells. As shown in Figure 4Ciii, the JAML protein was present at the cell plasma membrane only in areas of cell-cell contacts and remained diffuse or absent at free cell borders. Mutation of the dimerization motif by substitution of lysine 54 by aspartic acid abolished membrane localization of the JAMLK54D mutated protein (Figure 4Cvii). Altogether, our results indicated that the JAML protein was localized at the cell plasma membrane and suggested that it may engage in homophilic interactions via its dimerization interface.
JAML is a transmembrane protein induced upon ATRA-treatment in NB4 cells. (A) COS-7 cells were mock-transfected (-) or transfected with the p513-JAML-HA vector (JAML). Cell lysates were separated by SDS-PAGE and subjected to immunoblotting with anti-JAML (i) or anti-HA (ii) antibodies. The arrowhead indicates the major specific band. Brackets indicate minor specific bands likely corresponding to glycosylated JAML protein. (iii) JAML protein was in vitro–translated in the absence (–) or presence (+) of canine microsomal membranes. Arrow indicates a glycosylated JAML protein. (B) JAML protein expression during ATRA-induced differentiation of NB4 cells. NB4 cells were treated with 5 × 10-7 M ATRA for different times. Protein lysates corresponding to 105 cells were analyzed by Western blot, using anti-JAML or anti–β-actin polyclonal antibodies. In panels A and B, asterisks indicate nonspecific bands. (C) Cellular localization of JAML. MDCK cells stably transfected with the pMT-JAML-HA or pMT-JAMLK54D-HA were either untreated (-Zn) or treated (+Zn) with 125 μM ZnSO4 for 40 hours. Cells were treated with anti-HA monoclonal antibody and stained with Cy3-conjugated goat anti-mouse antibody (ii,iii,vi, vii). In panels i, iv, v, and viii, the same cells as in panels ii, iii, vi, and vii, respectively, were counterstained with Hoechst 33258 reagent.
JAML is a transmembrane protein induced upon ATRA-treatment in NB4 cells. (A) COS-7 cells were mock-transfected (-) or transfected with the p513-JAML-HA vector (JAML). Cell lysates were separated by SDS-PAGE and subjected to immunoblotting with anti-JAML (i) or anti-HA (ii) antibodies. The arrowhead indicates the major specific band. Brackets indicate minor specific bands likely corresponding to glycosylated JAML protein. (iii) JAML protein was in vitro–translated in the absence (–) or presence (+) of canine microsomal membranes. Arrow indicates a glycosylated JAML protein. (B) JAML protein expression during ATRA-induced differentiation of NB4 cells. NB4 cells were treated with 5 × 10-7 M ATRA for different times. Protein lysates corresponding to 105 cells were analyzed by Western blot, using anti-JAML or anti–β-actin polyclonal antibodies. In panels A and B, asterisks indicate nonspecific bands. (C) Cellular localization of JAML. MDCK cells stably transfected with the pMT-JAML-HA or pMT-JAMLK54D-HA were either untreated (-Zn) or treated (+Zn) with 125 μM ZnSO4 for 40 hours. Cells were treated with anti-HA monoclonal antibody and stained with Cy3-conjugated goat anti-mouse antibody (ii,iii,vi, vii). In panels i, iv, v, and viii, the same cells as in panels ii, iii, vi, and vii, respectively, were counterstained with Hoechst 33258 reagent.
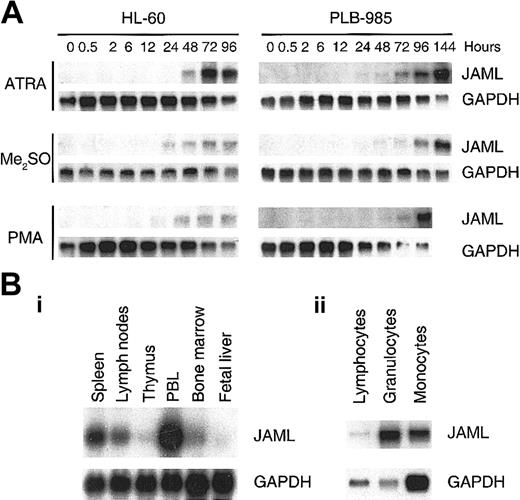
JAML mRNA is up-regulated during induced differentiation of myeloid leukemia cells and is expressed in normal hematopoietic tissues
In HL-60 and PLB-985 cells, JAML mRNA was progressively expressed as cells were induced to differentiation toward the granulocytic pathway with RA or Me2SO, and toward the monocytic pathway with PMA (Figure 5A). JAML mRNA was mainly expressed in peripheral blood leukocytes and to a lower extent in bone marrow, spleen, and lymph nodes (Figure 5Bi). In freshly purified mononulear cells, JAML mRNA was expressed in human granulocytes as well as, albeit to a much lesser extent, in monocytes and lymphocytes (Figure 5Bii). Using the Human RNA Master Blot, JAML mRNA was also expressed in fetal liver, fetal spleen, and fetal thymus but was not detected in nonhematopoietic tissues, except lung which is known to contain a number of myelomonocytic cells (data not shown). Altogether, our results indicated that JAML mRNA was expressed during normal myelopoiesis as well as in leukemia cells induced to myeloid differentiation.
JAML mRNA is up-regulated during induced differentiation of myeloid leukemia cells and is expressed in normal hematopoietic tissues. (A) Up-regulation of JAML mRNA during induced differentiation of myeloid leukemia cells. Autoradiograms of JAML mRNA expression before and after treatment of HL-60 and PLB-985 cells with different inducers of differentiation. Northern blots were performed using 5 μg total RNA. Hybridization with a GAPDH probe controlled for RNA quantities in each lane. (B) Autoradiograms of JAML mRNA expression in immune tissues (i) and peripheral blood cells (ii). Northern blots were either obtained from Clontech (i) or were performed using 5 μg total RNA from granulocytes, monocytes, and lymphocytes (ii). In panels A and B, GAPDH was used as a probe for assessment of RNA quantities in each lane.
U937/MT, U937/MT-JAML-HA, and U937/MT-JAMLK54D-HA cells stably transfected with the pMT, the pMT-JAML-HA, or pMT-JAMLK54D-HA respectively were untreated (-Zn) or treated (+Zn) with ZnSO4 for 18 hours prior to fluorescent labeling and subjected to a cell adhesion assay using human bone marrow endothelial cells either untreated (HBMECs) or treated (HBMECs + TNFα) as a target layer. Results are expressed in each condition as the amount of released fluorescence from adherent cells, normalized to unity for the corresponding -Zn/-TNFα condition. The adhesion level of U937/MT-JAML-HA cells onto untreated or TNFα-pretreated HBMECs, in the absence of Zn treatment (-Zn), was measured as 2.3% and 15.5% respectively, of the total number of incubated cells. Values shown are ± SD.
JAML mRNA is up-regulated during induced differentiation of myeloid leukemia cells and is expressed in normal hematopoietic tissues. (A) Up-regulation of JAML mRNA during induced differentiation of myeloid leukemia cells. Autoradiograms of JAML mRNA expression before and after treatment of HL-60 and PLB-985 cells with different inducers of differentiation. Northern blots were performed using 5 μg total RNA. Hybridization with a GAPDH probe controlled for RNA quantities in each lane. (B) Autoradiograms of JAML mRNA expression in immune tissues (i) and peripheral blood cells (ii). Northern blots were either obtained from Clontech (i) or were performed using 5 μg total RNA from granulocytes, monocytes, and lymphocytes (ii). In panels A and B, GAPDH was used as a probe for assessment of RNA quantities in each lane.
U937/MT, U937/MT-JAML-HA, and U937/MT-JAMLK54D-HA cells stably transfected with the pMT, the pMT-JAML-HA, or pMT-JAMLK54D-HA respectively were untreated (-Zn) or treated (+Zn) with ZnSO4 for 18 hours prior to fluorescent labeling and subjected to a cell adhesion assay using human bone marrow endothelial cells either untreated (HBMECs) or treated (HBMECs + TNFα) as a target layer. Results are expressed in each condition as the amount of released fluorescence from adherent cells, normalized to unity for the corresponding -Zn/-TNFα condition. The adhesion level of U937/MT-JAML-HA cells onto untreated or TNFα-pretreated HBMECs, in the absence of Zn treatment (-Zn), was measured as 2.3% and 15.5% respectively, of the total number of incubated cells. Values shown are ± SD.
JAML facilitates leukocyte adhesion to endothelial cells
To establish whether JAML can facilitate the interaction between leukocytes and endothelial cells, U937 cells were stably transfected with the pMT-JAML-HA or pMT-JAMLK54D-HA constructs, or with the empty vector as control. These nonclonal cell lines were fluorescently labeled and subjected to a cell adhesion assay using HBMECs as a target layer, either treated by TNFα or not treated. As expected, only a low level of control U937 cell adhesion to endothelial cells was observed and this adhesion was largely enhanced by treatment of the endothelial cell monolayer with TNFα (Table 3). We observed that JAML expression induced significantly both basal and cytokine-induced U937 cell adhesion to endothelial cell monolayers, whereas the expression of JAMLK54D mutated protein had no effect (Table 3). These results strongly suggest that JAML expression enhances myeloid leukemia cell adhesion to endothelial cells.
JAML expression enhances U937 cell adhesion to HBMECs
. | HBMECs . | HBMECs + TNFα . |
|---|---|---|
| U937/MT | ||
| - Zn | 1.00 ± 0.36 | 5.29 ± 0.17 |
| + Zn | 1.07 ± 0.33 | 4.79 ± 0.29 |
| U937/MT-JAML | ||
| - Zn | 1.00 ± 0.30 | 6.74 ± 0.52 |
| + Zn | 2.65 ± 0.17 | 11.04 ± 1.39 |
| U937/MT-JAMLK54D | ||
| - Zn | 1.00 ± 0.13 | 4.83 ± 0.30 |
| + Zn | 1.38 ± 0.47 | 4.23 ± 0.47 |
. | HBMECs . | HBMECs + TNFα . |
|---|---|---|
| U937/MT | ||
| - Zn | 1.00 ± 0.36 | 5.29 ± 0.17 |
| + Zn | 1.07 ± 0.33 | 4.79 ± 0.29 |
| U937/MT-JAML | ||
| - Zn | 1.00 ± 0.30 | 6.74 ± 0.52 |
| + Zn | 2.65 ± 0.17 | 11.04 ± 1.39 |
| U937/MT-JAMLK54D | ||
| - Zn | 1.00 ± 0.13 | 4.83 ± 0.30 |
| + Zn | 1.38 ± 0.47 | 4.23 ± 0.47 |
U937/MT, U937/MT-JAML-HA, and U937/MT-JAMLK54D-HA cells stably transfected with the pMT, the pMT-JAML-HA, or pMT-JAMLK54D-HA respectively were untreated (-Zn) or treated (+Zn) with ZnSO4 for 18 hours prior to fluorescent labeling and subjected to a cell adhesion assay using human bone marrow endothelial cells either untreated (HBMECs) or treated (HBMECs + TNFα) as a target layer. Results are expressed in each condition as the amount of released fluorescence from adherent cells, normalized to unity for the corresponding -Zn/-TNFα condition. The adhesion level of U937/MT-JAML-HA cells onto untreated or TNFα-pretreated HBMECs, in the absence of Zn treatment (-Zn), was measured as 2.3% and 15.5% respectively, of the total number of incubated cells. Values shown are ± SD.
Discussion
We have identified a novel protein, JAML, exhibiting homologies with members of the JAM family. JAML was induced by ATRA in human myeloid leukemia promyelocytes. JAML mRNA was up-regulated in myeloid leukemia cells induced to differentiate through the granulocytic and monocytic pathways and was expressed in normal hematopoietic cells. JAML is structurally similar to transmembrane proteins involved in cell-cell contacts and was localized to cell-cell borders. Induced expression of an exogenous JAML in myeloid leukemia cells resulted in enhanced cell adhesion to endothelial cells.
JAML is predicted to encode a type I integral transmembrane protein with 2 extracellular IgSF domains characteristic of proteins implicated as mediators of cell-cell interactions, generally through heterophilic interactions with other IgSF members. Searches of DNA and protein databases revealed that the JAML sequence has homologies with Ig domains of HCAR,45,46 EVA,44 and myelin P0.48 Furthermore, JAML shares structural features with members of the JAM family.19-24 We mapped the human JAML gene to the 11q23 region; whereas JAM-1 has been found at 1q21 and JAM-2 at 21q21. Although, the JAM genes are not clustered, the JAML gene lies upstream of the ESAM (endothelium cell-selective adhesion molecule, 11q24), CTH genes (11q24) and JAM-3 (11q25).23 Interestingly, 2 salt bridges formed in a complementary manner by a novel dimerization motif were shown to be essential for JAM dimers formation in solution.47 A similar motif in the JAML protein was shown to be required for JAML localization at the plasma membrane, suggesting that this protein may engage in homophilic interactions via this dimerization interface. This is further emphasized by the fact that JAML protein is localized at the cell plasma membrane in the areas of cell-cell contacts, whereas it was not detected at free cell borders.
We have found that the drastic increase in JAML mRNA expression induced by RA in NB4 cells correlated with growth arrest and differentiation of these cells. Cycloheximide treatment did block RA induction of JAML mRNA (data not shown), suggesting that its expression was not regulated at the transcriptional level. JAML mRNA increased during RA- or Me2SO-induced granulocytic differentiation and by PMA, which induced monocytic differentiation of both HL-60 and PLB-985 cells. Consistent with JAML expression in myeloid leukemia cells induced to differentiate, JAML mRNA was expressed in normal granulocytes and monocytes. Altogether, our results point to JAML as a novel member of the JAM family expressed on leukocytes. JAM family members are modulators of leukocyte adhesion to and transmigration through endothelial monolayers. Indeed, we have demonstrated here that JAML can facilitate the adhesion of U937 cells to endothelial cells. Furthermore, the observation that a JAML dimerization mutant protein has no effect strongly suggests that JAML may support adhesion of leukocytes to endothelial cells by engaging either in homotypic or heterotypic interactions with endothelial cell adhesion partners. Whether JAML supports leukocyte transmigration and possibly interacts with viral protein to mediate viral internalization as described for JAM-149 and HCAR45,46 will be further investigated.
Prepublished online as Blood First Edition Paper, July 17, 2003; DOI 10.1182/blood-2002-11-3462.
Supported by INSERM, CNRS, and grants from the Association Pour la Recherche sur le Cancer, the Fondation de France, the Comité de Paris de la Ligue Contre le Cancer, the Lady Tata Memorial Trust, and the Leukemia Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs A. Benecke and R. M. Mège for helpful comments and suggestions and Dr B. Weksler for kindly providing us with the HBMECs. Sequencing was conducted by C. Cruaud at the Centre National de Séquençage (Evry, France).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal