Abstract
Previous studies suggested that the SH2-containing inositol-5-phosphatase (SHIP) may play a tumor suppressor-like function in BCR-ABL–mediated leukemogenesis. To investigate this possibility, we first developed a new assay for quantitating transplantable multilineage leukemia-initiating cells (L-ICs) in hematopoietic stem cell (HSC)–enriched mouse bone marrow (BM) cells transduced with a BCR-ABL–GFP (green fluorescent protein) retrovirus. The frequency of L-ICs (1 of 430 Sca-1+lin– cells) was 7-fold lower than the frequency of HSCs in the Sca-1+lin– subset transduced with a control virus (1 of 65 cells). Forced BCRABL expression was also accompanied by a loss of regular HSC activity consistent with the acquisition of an increased probability of differentiation. Interestingly, the frequency and in vivo behavior of wild-type (+/+) and SHIP–/– L-ICs were indistinguishable, and in vitro, Sca-1+lin– BCR-ABL–transduced SHIP–/– cells showed a modestly reduced factor independence. Comparison of different populations of cells from patients with chronic myeloid leukemia (CML) in chronic phase and normal human BM showed that the reduced expression of full-length SHIP proteins seen in the more mature (CD34–lin+) leukemic cells is not mirrored in the more primitive (CD34+lin–) leukemic cells. Thus, SHIP expression appears to be differently altered in the early and late stages of differentiation of BCR-ABL–transformed cells, underscoring the importance of the cellular context in which its mechanistic effects are analyzed.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder characterized in the initial chronic phase of the disease by an excessive production of granulocytes and the presence of a unique rearrangement of the BCR gene on chromosome 22 and the ABL gene on chromosome 9.1 The 210-kDa product of the BCR-ABL fusion gene has increased tyrosine kinase activity2 that is important to its leukemogenic activity.3 Cells expressing the BCR-ABL gene, including primary CML cells, show alterations in many intracellular signaling pathways, including the Ras, PI3 (phosphatidylinositol 3) kinase, and JAK/STAT (Janus kinase/signal transducers and activators of transcription) pathways.4,5 Some of these perturbations may be direct through interactions with the multiple protein-binding domains of p210 BCR-ABL. However, others are likely to represent the downstream consequences of these events, which include the activation of an unusual autocrine mechanism involving interleukin-3 (IL-3) and granulocyte colony-stimulating factor (G-CSF).6,7 This complexity of intracellular effects caused directly or indirectly by the BCR-ABL oncoprotein, coupled with the relatively small numbers of CML stem cells present in patients with chronic phase disease, has made it very difficult to identify the molecular basis of the perturbed biology of these particular cells.1,8
SH2-containing inositol-5-phosphatase (SHIP) is a 145-kDa protein first identified by its ability to interact with SHC (Src homologous and collagen) following its transient tyrosine phosphorylation in growth factor–activated hematopoietic cells.9,10 Subsequent studies revealed SHIP to be an SH2-containing inositol phosphatase that hydrolyzes the 5′-phosphate from inositol-1,3,4,5-tetraphosphate and phosphatidylinositol (PI)–3,4,5-triphosphate. The latter appears to play a key role in modulating the ability of certain proteins containing pleckstrin homology domains (eg, Btk, PKB/Akt, PDK1, GRP1, Vav, and PLCγ1) or SH2 domains (eg, PI3 kinase and PLCγ1) to be translocated to the plasma membrane.10 SHIP expression is confined to hematopoietic cells (reviewed in Rohrschneider et al11 ), and targeted disruption of the SHIP gene confers on hematopoietic progenitor cells a hypersensitivity to many growth factors, in vivo expansion of granulocyte-macrophage progenitor numbers, and the development of a myeloproliferative syndrome.12 In cells expressing BCR-ABL, SHIP is constitutively phosphorylated and coprecipitates with p210BCR-ABL,13-15 although the level of SHIP expression is markedly reduced in some of these cells.16 A related protein, SHIP2, was also found to be constitutively tyrosine phosphorylated and bound to BCR-ABL in CML cells.17 These findings suggested that SHIP might play a tumor suppressor-like role in BCR-ABL–mediated leukemia.
To investigate this possibility, we initially compared the frequency and biologic properties of leukemia-initiating cells (L-ICs) in the Sca-1+ lin– subset of BCR-ABL–transduced bone marrow (BM) cells from 5-fluorouracil (FU)–treated wild-type (+/+) and SHIP–/– mice. Contrary to the results anticipated, the frequency of primitive SHIP–/– hematopoietic cells that acquired leukemogenic activity in vivo was unchanged, and their autonomous growth activity in vitro was reduced rather than being enhanced. Further examination of SHIP expression in primary BCR-ABL–transduced murine BM and in primitive leukemic cells from patients with CML in chronic phase revealed the levels of full-length SHIP proteins to be elevated with marked down-regulation of SHIP being restricted to the terminal stages of CML cell differentiation.
Materials and methods
Virus
The MSCV-BCR-ABL-IRES-GFP (BCR-ABL) and control MSCV-IRESGFP (MIG) vectors have been described previously.7 Ecotropic Phoenix packaging cells18 were transfected using calcium phosphate19 with 10 to 30 μg retroviral plasmid DNA isolated by Qiagen plasmid purification (Qiagen, Hilden, Germany), and the medium overlaying the transfected cells was changed 24 hours later. Virus-containing supernatants (VCSs) were harvested after another 24 hours and then passed through a 0.45-μm filter and frozen at –80°C. Viral titers (4 to 8 × 105/mL for the BCR-ABL virus and 5 to 10 × 105/mL for the MIG virus) were determined by assessment of GFP+ BaF3 cells 2 days after a single exposure to different dilutions of VCSs.
Animals
SHIP–/– mice12 were backcrossed for 17 generations onto a C57BL/6-Ly5.2 background and along with C57BL/6-+/+-Ly5.2 and C57BL/6-+/+-Peb3b-Ly5.1 mice were bred and maintained under microisolation conditions. Mice to receive transplants were irradiated at 8 to 10 weeks of age with 900 cGy 137Cs γ-rays and received acidified water thereafter. Cells were injected into mice intravenously a few hours after they had been irradiated. Mice were subsequently monitored daily for signs of weight loss or lethargy. Peripheral blood (PB) samples were obtained every 1 to 2 weeks by nicking the tail vein, and the white blood cells (WBCs) were then counted and analyzed by fluorescence-activated cell sorter (FACS) as described in “Cell sorting and flow cytometry” after lysis of the red blood cells (RBCs) with ammonium chloride.
Transduction of mouse BM cells
BM cells from 6- to 10-week-old C57BL/6 and SHIP–/– mice were harvested 4 days after injection of 150 mg/kg 5-FU. The cells were then washed in phosphate-buffered saline (PBS) containing 2% fetal calf serum (FCS), and low-density cells were isolated by centrifugation on a 1.077 g/mL Ficoll-Hypaque solution.20 Low-density cells were then cultured for 48 hours at 3 to 5 × 105/mL in serum-free medium (SFM), ie, Iscove medium supplemented with BIT (StemCell Technologies, Vancouver, BC, Canada) and 40 μg/mL low-density lipoproteins (Sigma Chemical, St Louis, MO), to which 100 ng/mL murine Steel factor (SF, purified from cDNA-transfected COS cell supernatants), 10 ng/mL murine IL-3 (Stem Cell Technologies), and 10 ng/mL human IL-6 (Cangene, Mississauga, ON, Canada) were also added. The cells were then collected and resuspended at 5 × 105 cells/mL in VCS diluted 1:3 in Dulbecco modified Eagle medium (DMEM) containing 15% FCS, protamine sulfate (5 μg/mL), and SF, IL-3, and IL-6 at the same concentrations as previous. The cells were then transferred to Petri dishes that had been precoated with 3 μg/cm2 fibronectin (Sigma) and preloaded with virus for 30 minutes at room temperature.21 This transduction was repeated once 4 hours later. The cells were then incubated at 37°C for a further 24 to 48 hours before additional studies as indicated.
Human cells
Heparin-treated blood and BM cells were obtained from 8 patients with untreated BCR-ABL+ chronic phase CML, and samples of normal adult human BM cells were either from harvests taken for local allogeneic transplants or were from cadaveric donors (Northwest Tissue Center, Seattle, WA). In all cases, informed consent was obtained, and the procedures used, including the Declaration of Helsinki protocol, were approved by the Research Ethics Board of the University of British Columbia (Vancouver). All samples were enriched for CD34+ cells by immunomagnetic removal of lineage marker+ (lin+) cells using StemSep columns (StemCell). CD34–lin+ cells were purified from low-density cells as described.6 For these studies, CML samples were from patients with exclusively leukemic cells in all subpopulations studied, determined in previous analyses as described previously.6
Cell sorting and flow cytometry
Transduced mouse cells were washed and suspended in Hanks solution with 2% FCS (HF) at 5 to 10 × 106 cells/mL and then incubated first with 6 μg/mL 2.4G2 (anti-Fc receptor antibody)22 followed by a mixture of biotinylated lineage-specific rat monoclonal antibodies (anti-B220 [RA3-6B2], anti–Gr-1 [RB6-8C5], anti–Ly-1 [53-7.3], Ter119 [anti-Ly-76]), and finally with phycoerythrin (PE)–labeled anti–Sca-1 (anti–Ly-6A/E; E13-161.7), each time for 30 minutes at 4°C as described.23 Anti–Mac-1 was omitted from this cocktail of antilineage (lin) marker antibodies because expression of Mac-1 is up-regulated on activated stem cells.24 Antibodylabeled cells were then washed once, incubated with allophycocyanin (APC)–labeled streptavidin (SA-APC) for 30 minutes at 4°C, washed twice again, and finally resuspended in 1 μg/mL propidium iodide (PI; Sigma). A FACStar Plus (Becton Dickinson Immunocytometry Systems, San Jose, CA) was used to isolate populations of more than 99% pure, viable GFP+ Sca-1+lin– cells. Bulk cells were collected into Eppendorf tubes containing DMEM with 10% FCS or PBS. Single cells were sorted directly into the individual wells of 96-well microtiter plates preloaded with 100 μL DMEM plus 10% FCS using the FACS single-cell deposition unit. Subsequent surface expression on cultured cells of Gr-1, Mac-1, B220, CD4, CD8, Ter119, Sca-1, c-kit, and Ly5-2 was evaluated using directly conjugated PE- or APC-conjugated monoclonal antibodies against these antigens after staining as described earlier.
Isolation of lin–CD34+, lin–CD34+CD38+, lin–CD34+CD38–, and lin+CD34– cells from samples of previously cryopreserved normal BM and patients with chronic phase CML was as described previously.25 Data acquisition using the FACScan and FACScalibur and analysis was performed with CellQuest software (Becton Dickinson).
Cell culture
Sorted cells were cultured at 1 to 5 × 105 cells/mL in DMEM plus 10% FCS with or without 100 ng/mL SF, 10 ng/mL IL-3, and 10 ng/mL IL-6 at 37°C. Viable cell numbers were determined from hematocytometer counts of trypan blue–excluding cells. Subsequent clone formation (presence of ≥ 2 refractile cells) and clone size (total number of refractile cells) 7 days after seeding wells with single viable (PI–) cells (using the FACS cloning device) was assessed by direct visual inspection using an inverted microscope.
Progenitor assays
Murine colony-forming cell (CFC) and long-term culture-initiating cell (LTC-IC) assays were performed using culture media from StemCell Technologies. CFC assays were performed in FCS-containing methylcellulose cultures with 10 ng/mL human IL-6, 10 ng/mL murine IL-3, 50 ng/mL murine SF, and 3 U/mL human erythropoietin (EPO; StemCell Technologies). Colony counts were performed after 12 to 14 days of incubation at 37°C, using standard scoring criteria.23 LTCs were set up on pre-established irradiated mouse marrow LTC feeder layers and maintained at 33°C with weekly half medium changes and the addition of fresh hydrocortisone (10–6 mM) prior to performing CFC assays on the cells harvested after 4 weeks as previously described.23
Expanded populations of BCR-ABL–transduced +/+ and SHIP–/– cells were generated from single colonies generated in methylcellulose cultures or clones initially generated in liquid cultures of single Sca-1+lin–GFP+ cells deposited into 96-well plates using the FACS. Initial clones were then expanded in DMEM containing 10% FCS, 100 ng/mL SF, 10 ng/mL IL-3, and 10 ng/mL IL-6, or 5% pokeweed mitogen (PWM)–stimulated murine spleen cell–conditioned medium (StemCell Technologies). In some experiments, 0.1 to 10 μmol STI571 (Novartis, Basel, Switzerland) was added to some of the cultures.
Quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR) analyses
Total RNA was extracted from aliquots of cells using the Absolutely RNA Microprep kit (Stratagene, La Jolla, CA) or TRIzol (Invitrogen, Burlington, ON, Canada). The RT reaction was performed in 20 μL with superscript II reverse transcriptase (Invitrogen) using random hexamer oligonucleotides (Amersham Pharmacia, Piscataway, NJ) as described.7 Real-time PCR was performed using 25 μL 2 × SYBY Green PCR Master Mix (Applied Biosystems, Foster City, CA), 1 μL of 20 pM specific primers, 1 to 2 μL cDNA, and water to a final volume of 50 μL. Specific forward and reverse primers to produce approximately 100-bp amplicons for optimal amplification in real-time PCR of reverse transcribed cDNA for murine SHIP were 5′-GCCTCCCAGAAACATTCCTATG-3′ and 5′-GGTTCTCTGTTGCAAGAGGAAG-3′, for human SHIP were 5′-GACACAGAAAGTGTCGTGTCTC-3′ and 5′-GGAACCTCCTTGGCCTCACAG-3′, and for both human and murine glyceraldehyde-3-phosphtae dehydrogenase (GAPDH) were 5′-CCCATCACCATCTTCCAGGAG-3′ and 5′-CTTCTCCATGGTGGTGAAGACG-3′. Real-time PCR and data analysis were performed on an iCycler iQ system, using iCycler iQ Real-time Detection Software (BIO-RAD, Hercules, CA). Optimal reaction conditions for amplification of human and murine SHIP and GAPDH were as follows: 40 cycles of 3-step PCR (94°C for 15 seconds, 60°C for 20 seconds, 72°C for 30 seconds with a single fluorescence measurement) after initial denaturation (94°C for 5 minutes). The relative quantification data of human and murine SHIP in comparison to a reference gene (GAPDH) was generated on the basis of a mathematical model for relative quantification in real-time RT-PCR as described.26,27
Southern analyses
High molecular weight DNA was isolated using DNAZol (GIBCO/BRL). DNA (15 μg) was digested with EcoR1, electrophoresed, and transferred to a nylon membrane.6 The blot was then hybridized with a GFP probe derived from the retroviral vector, and a BCR-ABL cDNA was used to confirm the presence of an intact BCR-ABL gene.
Western analyses
Cells (105) were lysed in phosphorylation solubilization buffer (PSB; 50 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer, 100 mM NaF, 10 nM sodium pyrophosphate, 2 mM Na3VO4, 4 mM EDTA (ethylenediaminetetraacetic acid) containing 0.5% Nonidet P-40.15 Western analyses were performed using a monoclonal anti-ABL antibody (8E9; Pharmingen, Mississauga, ON, Canada), an antiphosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY), anti-STAT5A antibodies (Upstate Biotechnology), a monoclonal anti-SHIP antibody (P1C1; Santa Cruz Biotechnology, Santa Cruz, CA), 2 polyclonal anti-SHIP antibodies against the SH2 domain and c-terminal region of SHIP (α-SHIP166), an anti-SHIP2 antibody,17 and a monoclonal human antiactin antibody (Sigma).
Transplantation experiments
Hematopoietic stem cells (HSCs) were enumerated using a limiting dilution transplantation assay for cells with competitive, long-term, lymphomyeloid repopulating activity as described previously.23,28 Briefly, different numbers of MIG-transduced Sca-1+lin–GFP+ cells were injected into groups of irradiated Ly5 congenic recipients together with 105 normal BM cells of the same strain as the recipient. The blood of these mice was then analyzed by FACS 3, 6, 12, 20, and 32 weeks later. Only mice with more than 1% donor-derived (GFP+) cells, including both lymphoid (B220+) and myeloid (Gr-1+ and/or Mac-1+) subpopulations at 20 weeks, were considered to be repopulated with transduced HSCs. L-IC frequencies were similarly determined after injection of different numbers of BCR-ABL–transduced Sca-1+lin–GFP+ cells together with 105 recipient strain normal BM cells into groups of Ly5 congenic recipients from the frequency of these mice scored as positive for leukemia. The endpoint for leukemia used was an increased WBC count, splenomegaly, and/or hepatomegaly and the accompanying presence of GFP+ cells. The clonality of the leukemias seen in mice injected with limiting numbers of L-ICs was confirmed by Southern blot analysis of DNA from terminally ill mice. Normal HSC and L-IC frequencies were calculated from the proportion of negative recipients in groups given different dilutions of cells using Poisson statistics and the method of maximum likelihood provided in L-calc software (StemCell Technologies).
Results
Development of a new assay for quantitating the frequency of leukemic stem cells in BCR-ABL–transduced mouse BM populations
Multiple types of mouse BM cells, including relatively mature cells, can generate rapidly lethal malignant populations after being transduced with BCR-ABL vectors that cause high-level BCRABL expression.29-32 Even when cells are obtained from donors pretreated with 5-FU to obtain a target population that is enriched in more primitive cells, fatal lymphoid or myeloid leukemias are common in mice injected with large numbers of cells indicative of disease generation from lineage-restricted progenitors.33,34 To create a model of BCR-ABL–transduced murine leukemia that consistently resembles human CML more closely in its origin from cells with multilineage differentiation potential, we, therefore, sought to develop a method that would enrich for HSCs in the immediately transduced population and then assess the leukemogenic activity of these more highly purified cells.
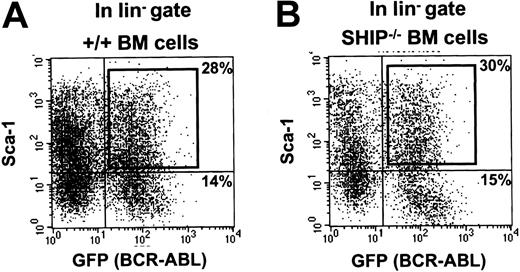
Previous quantitative studies have shown that the frequency of HSCs in 5-FU–treated BM cells is approximately 1/2 × 103,28 and this frequency decreases several-fold within the first few days in vitro under the transduction conditions used here.35 We, therefore, undertook a first set of experiments to determine whether a higher frequency of HSCs could be obtained by isolation of the Sca-1+lin– subset of BM cells harvested 24 hours after being infected with a control GFP virus (MIG) as described in “Methods and materials.” Sca-1+lin– cells represented 28% ± 4% of the total cells present at this time, and the proportion of lin– cells that were Sca-1+ was not different between the GFP+ and GFP– fractions (n = 6). The HSC frequency determined for the Sca-1+lin– MIG-transduced BM cells by limiting dilution transplantation assays was 1 of 65 cells (Table 1). This value is comparable to that previously reported for freshly isolated Sca-1+lin– BM cells from normal mice.23
Stem cell frequency (CRU) of MIG-transduced Sca-1+lin–GFP+ BM cells from +/+ and SHIP–/– mice
Phenotype of cells injected per mouse . | No. of mice positive for GFP+ lymphoid-myeloid cells/no. of mice receiving transplants . |
|---|---|
| +/+ Sca-1+lin-GFP+ | |
| 10 | 0/3 |
| 100 | 5/6 |
| 1000 | 6/6 |
| 3000 | 3/3 |
| 5000 | 3/3 |
| 10 000 | 3/3 |
| SHIP-/- Sca-1+lin-GFP+ | |
| 10 | 0/3 |
| 100 | 4/5 |
| 1000 | 5/5 |
| 3000 | 3/3 |
| 5000 | 3/3 |
| 10 000 | 3/3 |
Phenotype of cells injected per mouse . | No. of mice positive for GFP+ lymphoid-myeloid cells/no. of mice receiving transplants . |
|---|---|
| +/+ Sca-1+lin-GFP+ | |
| 10 | 0/3 |
| 100 | 5/6 |
| 1000 | 6/6 |
| 3000 | 3/3 |
| 5000 | 3/3 |
| 10 000 | 3/3 |
| SHIP-/- Sca-1+lin-GFP+ | |
| 10 | 0/3 |
| 100 | 4/5 |
| 1000 | 5/5 |
| 3000 | 3/3 |
| 5000 | 3/3 |
| 10 000 | 3/3 |
Stem cell frequency is 1 in 65 (95% confidence interval [CI], 39-100) for +/+ Sca-1+lin-GFP+ cells and 1 in 70 (95% CI, 42-120) for SHIP-/- Sca-1+lin-GFP+ cells in vivo.
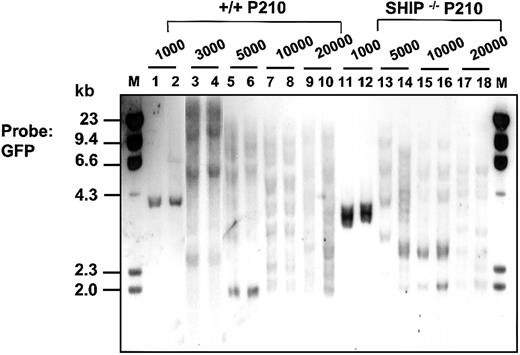
Similar assays were then applied to BCR-ABL–transduced BM cells from 5-FU–treated mice. One day after completion of the infection protocol, the fraction of cells that were Sca-1+lin– (22% ± 3% of the total, n = 6, Figure 1A) was similar to that seen in the MIG-infected populations noted earlier. However, the proportion of GFP+ cells in all subpopulations after exposure to the BCR-ABL virus was lower, as expected from the lower titer of the BCR-ABL virus preparation. The Sca-1+lin– GFP+ cells isolated from the BCR-ABL–infected cells were then assessed for their ability to generate leukemic clones in lethally irradiated recipients by injecting these mice with decreasing numbers of the sorted transduced cells. To protect the recipients against the lethal effects of the irradiation they were co-injected with 105 normal BM cells of the same genotype as the host.36 The frequency of cells with leukemia-initiating activity (L-ICs) calculated from the proportions of mice that did (or did not) develop leukemia in these experiments was 1 of 430 Sca-1+lin–GFP+ (BCR-ABL+) cells (Table 2). The clonality of the diseases produced by limiting numbers of L-ICs was confirmed by Southern blot analysis of DNA isolated from the spleen and liver of terminally ill mice and hybridized with a GFP probe (Figure 2). Rehybridization with a BCR-ABL cDNA probe confirmed the presence of the full-length 7.3-kb BCR-ABL gene in each case (data not shown).
FACS profiles of BCR-ABL–transduced +/+ and SHIP–/– BM cells. Dot plots showing gates used to isolate the Sca-1+ subset of GFP+ cells from the lin– cells after exposure of +/+ (A) and SHIP–/– (B) BM cells to the BCR-ABL-GFP virus.
FACS profiles of BCR-ABL–transduced +/+ and SHIP–/– BM cells. Dot plots showing gates used to isolate the Sca-1+ subset of GFP+ cells from the lin– cells after exposure of +/+ (A) and SHIP–/– (B) BM cells to the BCR-ABL-GFP virus.
Leukemia-initiating cell frequency (L-IC) of BCR-ABL–transduced Sca-1+lin–GFP+ cells from +/+ and SHIP–/– mice
Phenotype of cells injected per mouse . | No. of leukemic mice/no. of mice receiving transplants . | Maximum WBC count, × 106/mL . |
|---|---|---|
| +/+ Sca-1+lin-GFP+ | ||
| 10 | 0/8 | 6 |
| 100 | 0/8 | 8 |
| 1000 | 8/8 | 26 |
| 3000 | 5/5 | 28 |
| 5000 | 5/5 | 36 |
| 10 000 | 5/5 | 40 |
| 20 000 | 5/5 | 60 |
| SHIP-/- Sca-1+lin-GFP+ | ||
| 10 | 0/8 | 7 |
| 100 | 0/8 | 8 |
| 1000 | 7/7 | 24 |
| 3000 | 6/6 | 27 |
| 5000 | 5/5 | 38 |
| 10 000 | 5/5 | 42 |
| 20 000 | 5/5 | 58 |
Phenotype of cells injected per mouse . | No. of leukemic mice/no. of mice receiving transplants . | Maximum WBC count, × 106/mL . |
|---|---|---|
| +/+ Sca-1+lin-GFP+ | ||
| 10 | 0/8 | 6 |
| 100 | 0/8 | 8 |
| 1000 | 8/8 | 26 |
| 3000 | 5/5 | 28 |
| 5000 | 5/5 | 36 |
| 10 000 | 5/5 | 40 |
| 20 000 | 5/5 | 60 |
| SHIP-/- Sca-1+lin-GFP+ | ||
| 10 | 0/8 | 7 |
| 100 | 0/8 | 8 |
| 1000 | 7/7 | 24 |
| 3000 | 6/6 | 27 |
| 5000 | 5/5 | 38 |
| 10 000 | 5/5 | 42 |
| 20 000 | 5/5 | 58 |
L-IC frequency is 1 in 430 (95% CI, 202-900) for +/+ Sca-1+lin-GFP+ cells and 1 in 450 (95% CI, 207-984) for SHIP-/- Sca-1+lin-GFP+ cells in vivo.
Southern blot analysis showing clonal repopulation of mice receiving transplants with limiting numbers of Sca1+lin– BCR-ABL–transduced (GFP+)+/+ and SHIP–/– cells. Genomic DNA was digested with EcoR1 and probed for GFP. EcoR1 cleaves once within the retroviral sequence; therefore, each fragment represents a unique site of retroviral integration. Lanes 1-10 and 11-18 show the results for DNA isolated from the spleen or liver of sick mice injected with 103 to 2 × 104 Sca-1+lin–GFP+ BCR-ABL–transduced cells from +/+ cells and SHIP–/– mice, respectively. Molecular size markers are also indicated. M indicates marker.
Southern blot analysis showing clonal repopulation of mice receiving transplants with limiting numbers of Sca1+lin– BCR-ABL–transduced (GFP+)+/+ and SHIP–/– cells. Genomic DNA was digested with EcoR1 and probed for GFP. EcoR1 cleaves once within the retroviral sequence; therefore, each fragment represents a unique site of retroviral integration. Lanes 1-10 and 11-18 show the results for DNA isolated from the spleen or liver of sick mice injected with 103 to 2 × 104 Sca-1+lin–GFP+ BCR-ABL–transduced cells from +/+ cells and SHIP–/– mice, respectively. Molecular size markers are also indicated. M indicates marker.
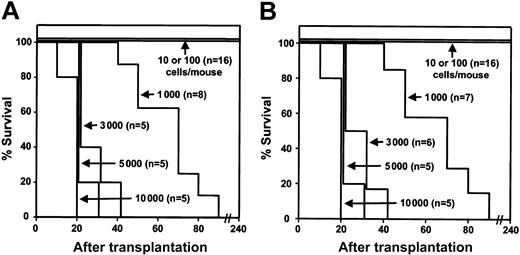
Figure 3A shows the survival curves obtained, indicating the cell-dose dependent latency of the leukemias that developed in these experiments. From the results shown in Table 2, it can be seen that significant effects on survival were restricted to mice receiving transplants with very small numbers (1-2) of L-ICs. Interestingly, in spite of 7-fold lower frequency of L-ICs in the BCR-ABL–transduced Sca-1+lin– cells than the frequency of HSCs measured in the matched MIG-transduced cells, mice receiving transplants with less than one L-IC (ie, 102 Sca-1+lin–GFP+ cells) showed only transient engraftment with GFP+ WBCs which disappeared after the first 3 weeks. These findings indicate that following BCR-ABL transduction there was a loss of the expected residual numbers of cells with “normal” HSC activity.
Survival curves for mice receiving transplants with different doses of Sca-1+lin– BCR-ABL–transduced +/+ and SHIP–/– cells are indistinguishable. Survival curves of C57BL/6J Pep3b mice injected with different numbers of FACS-sorted Sca-1+lin–GFP+ BM cells from BCR-ABL–transduced +/+ B6 mice (A) or B6-SHIP–/– mice (B).
Survival curves for mice receiving transplants with different doses of Sca-1+lin– BCR-ABL–transduced +/+ and SHIP–/– cells are indistinguishable. Survival curves of C57BL/6J Pep3b mice injected with different numbers of FACS-sorted Sca-1+lin–GFP+ BM cells from BCR-ABL–transduced +/+ B6 mice (A) or B6-SHIP–/– mice (B).
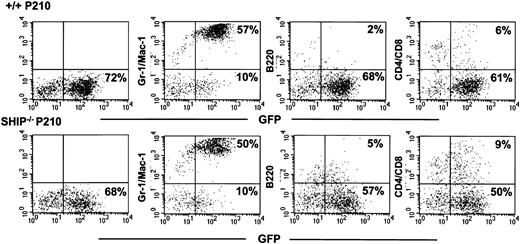
More than 80% of all the circulating leukocytes in the leukemic mice had the morphology of neutrophils, and Gr-1+ and/or Mac-1+ (myeloid) GFP+ cells constituted a predominant fraction of the BM cells removed from moribund animals (Figure 4). However, GFP+ B220+ (B-lineage), CD4+ and/or CD8+ (T lineage) cells (Figure 4) as well as Ter119+ (erythroid) cells (data not shown) were also consistently detected in these BM cell suspensions, including harvests from clonally repopulated mice, indicating the reproducible multilineage character of the clones generated. Splenomegaly and hepatomegaly were also frequently seen in these mice. Transplantation of BM or spleen cells from primary diseased mice produced a similar multilineage lethal disease when transplanted into secondary recipients (n = 6, data not shown).
Multiple lineages of GFP+ cells in clonally repopulated leukemic mice. Shown are a series of FACS analyses of BM cells from representative sick mice injected with 103 Sca-1+lin–GFP+ BCR-ABL+ +/+ cells (top row) and SHIP–/– cells (bottom row). The presence of GFP+ cells in the compartments of Gr-1/Mac-1+, B220+, and CD4/CD8+ cells can be seen in recipients of both genotypes of BCR-ABL–transduced cells.
Multiple lineages of GFP+ cells in clonally repopulated leukemic mice. Shown are a series of FACS analyses of BM cells from representative sick mice injected with 103 Sca-1+lin–GFP+ BCR-ABL+ +/+ cells (top row) and SHIP–/– cells (bottom row). The presence of GFP+ cells in the compartments of Gr-1/Mac-1+, B220+, and CD4/CD8+ cells can be seen in recipients of both genotypes of BCR-ABL–transduced cells.
Absence of SHIP has no effect on BCR-ABL–induced leukemogenesis from primitive transduced murine BM cells
A similar series of experiments were then carried out with BM cells from 5-FU–pretreated SHIP–/– mice. The yield of Sca-1+lin– SHIP–/– cells 24 hours after transduction with control virus was similar to that measured for +/+ cells (23% ± 4% of the total, n = 6, Figure 1B), and the HSC frequency of MIG-transduced Sca-1+lin–GFP+ cells from SHIP–/– mice was also similar to that determined for +/+ cells (Table 1). The leukemogenic properties of the BCR-ABL–transduced Sca-1+lin– SHIP–/– BM cells were then analyzed. As can be seen in Table 2 and Figures 2, 3, 4, the frequency of these SHIP–/– L-ICs and the clonal, multilineage disease they produced with a prolonged latency when injected at limiting dilution were indistinguishable from the results obtained with BCR-ABL–transduced +/+ cells. SHIP–/– leukemias were also similarly transplantable into secondary mice (n = 6, data not shown).
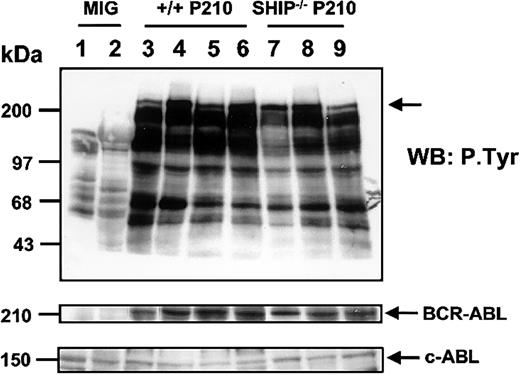
To determine whether differences in p210BCR-ABL expression or general protein tyrosine phosphorylation might, nevertheless, be discerned in the leukemic cells generated in vivo from +/+ and SHIP–/– cells, Western blot analysis was performed on extracts of cells harvested from mice with splenomegaly or hepatomegaly. As illustrated in Figure 5, no significant difference could be seen in any of these parameters between leukemias of either genotype. Interestingly, however, for both genotypes, there was evidence of a higher tyrosine phosphorylation of p210BCR-ABL in the leukemic cells obtained from the liver as compared with the spleen (Figure 5, lanes 4,6,8).
Similar tyrosine phosphorylation of BCR-ABL in BCR-ABL– transduced +/+ and SHIP–/– leukemic murine cells. Shown are Western blot analyses of cell lysates from the spleen and liver of a control (MIG-transduced) mouse (lanes 1-2), the spleen and liver of sick mice initially receiving transplants with 103 and 5 × 103 BCR-ABL–transduced Sca-1+lin– GFP+ cells (lanes 3-6), and cell lysates from the spleen and liver of sick mice injected with similar numbers of Sca-1+lin–GFP+ cells from BCR-ABL–transduced SHIP–/– cells (lanes 7-9). Lysates from equal numbers of cells were separated on 8% polyacrylamide gradient gels. The filter was first probed with an antiphosphotyrosine (4G10) antibody, then stripped and reprobed with an anti-ABL antibody (8E9). The positions of prestained molecular weight markers are indicated on the left side of the blot.
Similar tyrosine phosphorylation of BCR-ABL in BCR-ABL– transduced +/+ and SHIP–/– leukemic murine cells. Shown are Western blot analyses of cell lysates from the spleen and liver of a control (MIG-transduced) mouse (lanes 1-2), the spleen and liver of sick mice initially receiving transplants with 103 and 5 × 103 BCR-ABL–transduced Sca-1+lin– GFP+ cells (lanes 3-6), and cell lysates from the spleen and liver of sick mice injected with similar numbers of Sca-1+lin–GFP+ cells from BCR-ABL–transduced SHIP–/– cells (lanes 7-9). Lysates from equal numbers of cells were separated on 8% polyacrylamide gradient gels. The filter was first probed with an antiphosphotyrosine (4G10) antibody, then stripped and reprobed with an anti-ABL antibody (8E9). The positions of prestained molecular weight markers are indicated on the left side of the blot.
Absence of SHIP reduces the ability of primitive BCR-ABL–transduced BM cells to acquire autonomous growth potential in vitro
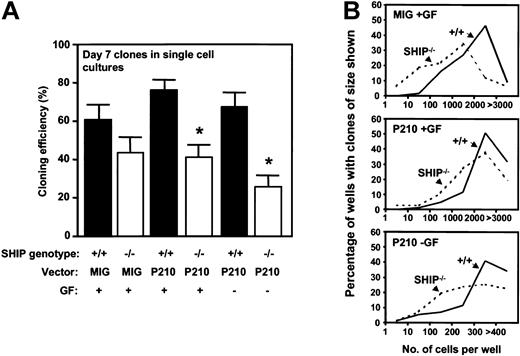
Sca-1+lin–GFP+ cells isolated from BCR-ABL–transduced +/+ and SHIP–/– BM cells showed identical growth rates when plated in bulk liquid suspension culture with or without added growth factors and monitored over the first 2 weeks (data not shown). To determine whether potential differences might be unmasked if the cells were cultured under more stringent conditions, single BCRABL–transduced Sca-1+lin–GFP+ +/+ and SHIP–/– cells were plated in SFM with or without added growth factors, and the frequency and size of clones generated were then assessed. As shown in Figure 6, the BCR-ABL–transduced SHIP–/– cells showed a reduced cloning efficiency and generated smaller clones than the BCR-ABL–transduced +/+ cells, particularly when no exogenous growth factors were added. However, reduced growth of Sca-1+lin– SHIP–/– cells was also observed in the absence of BCR-ABL, suggesting that the reduced growth exhibited by isolated BCR-ABL–transduced SHIP–/– cells may be due to an innately compromised ability of primitive SHIP–/– BM cells to respond to growth factor stimulation.
Reduced factor-independent growth of single BCR-ABL–transduced SHIP–/– Sca1+lin–GFP+ cells in vitro. (A) Percentage of transduced cells generating clones of more than 2 refractile cells after 7 days in the presence or absence of added growth factors. Values shown are the mean ± SEM from 3 experiments with each genotype of BM cells. *Indicates significantly different from BCR-ABL–transduced +/+ cells (P < .05). (B) Clone size distributions obtained from MIG– or BCR-ABL–transduced SHIP–/– cells compared with MIG– or BCR-ABL–transduced+/+ cells after being cultured for 7 days in the presence or absence of growth factors. Data for a representative experiment of 3 performed are shown.
Reduced factor-independent growth of single BCR-ABL–transduced SHIP–/– Sca1+lin–GFP+ cells in vitro. (A) Percentage of transduced cells generating clones of more than 2 refractile cells after 7 days in the presence or absence of added growth factors. Values shown are the mean ± SEM from 3 experiments with each genotype of BM cells. *Indicates significantly different from BCR-ABL–transduced +/+ cells (P < .05). (B) Clone size distributions obtained from MIG– or BCR-ABL–transduced SHIP–/– cells compared with MIG– or BCR-ABL–transduced+/+ cells after being cultured for 7 days in the presence or absence of growth factors. Data for a representative experiment of 3 performed are shown.
To further investigate the effect of SHIP deprivation on the ability of primitive BCR-ABL–transduced cells to proliferate autonomously in vitro, we assessed their ability to form colonies of differentiated cells in semisolid media. As shown previously,7 BCR-ABL transduction increased (approximately 3-fold) the frequency of CFCs in the Sca-1+lin– fraction of +/+ cells and conferred on approximately two thirds of them an autonomous growth potential (Table 3). Although there was no difference in the frequency or factor dependence of the CFCs in the MIG-transduced Sca-1+lin– SHIP–/– and +/+ cells, the frequency of all types of CFCs in the BCR-ABL–transduced (GFP+) Sca-1+lin– SHIP–/– population was lower than in the corresponding +/+ population both with and without added growth factors (P < .05).
Reduced factor-independent colony formation by BCR-ABL–transduced SHIP–/– CFCs in vitro
. | No. of CFCs detected per 103 Sca-1+lin-GFP+ cells . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| CFC assay conditions . | BFU-E . | CFU-GEMM . | CFU-GM . | Total . | |||
| MIG | |||||||
| 3GF | |||||||
| +/+ | 15 ± 3 | 12 ± 4 | 85 ± 10 | 112 ± 12 | |||
| -/- | 12 ± 3 | 10 ± 4 | 95 ± 12 | 118 ± 14 | |||
| EPO | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| No GF | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| p210 | |||||||
| 3GF | |||||||
| +/+ | 50 ± 8 | 30 ± 6 | 215 ± 25 | 295 ± 35 | |||
| -/- | 20 ± 6 | 35 ± 8 | 135 ± 20 | 198 ± 32 | |||
| EPO | |||||||
| +/+ | 20 ± 2 | 23 ± 6 | 128 ± 12 | 172 ± 18 | |||
| -/- | 16 ± 4 | 6 ± 3 | 82 ± 8 | 105 ± 4 | |||
| No GF | |||||||
| +/+ | 14 ± 2 | 20 ± 4 | 90 ± 8 | 124 ± 12 | |||
| -/- | 8 ± 2 | 0 | 68 ± 6 | 76 ± 6 | |||
. | No. of CFCs detected per 103 Sca-1+lin-GFP+ cells . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| CFC assay conditions . | BFU-E . | CFU-GEMM . | CFU-GM . | Total . | |||
| MIG | |||||||
| 3GF | |||||||
| +/+ | 15 ± 3 | 12 ± 4 | 85 ± 10 | 112 ± 12 | |||
| -/- | 12 ± 3 | 10 ± 4 | 95 ± 12 | 118 ± 14 | |||
| EPO | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| No GF | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| p210 | |||||||
| 3GF | |||||||
| +/+ | 50 ± 8 | 30 ± 6 | 215 ± 25 | 295 ± 35 | |||
| -/- | 20 ± 6 | 35 ± 8 | 135 ± 20 | 198 ± 32 | |||
| EPO | |||||||
| +/+ | 20 ± 2 | 23 ± 6 | 128 ± 12 | 172 ± 18 | |||
| -/- | 16 ± 4 | 6 ± 3 | 82 ± 8 | 105 ± 4 | |||
| No GF | |||||||
| +/+ | 14 ± 2 | 20 ± 4 | 90 ± 8 | 124 ± 12 | |||
| -/- | 8 ± 2 | 0 | 68 ± 6 | 76 ± 6 | |||
Each 1-mL CFC assay culture contained 103 Sca-1+lin-GFP+ cells. Data shown are the mean ± SEM of values obtained from 3 independent experiments. BFU-E indicates erythroid burst-forming unit; CFU-GEMM, granulocyte-erythroid-macrophage-megakaryocyte, colony-forming unit; CFU-GM, granulocyte-macrophage colony-forming unit; and GF, growth factor.
To assess effects on a more primitive subset of functionally defined progenitors, LTC-IC assays were also performed on the 2 genotypes of Sca-1+lin–GFP+ cells. As for the HSCs assayed in vivo and the CFCs assayed directly in the Sca-1+lin– fractions, the 4-week LTC output of all types of CFCs from MIG-transduced GFP+ cells of +/+ and SHIP–/– origin was similar. Also as expected,7 these values were increased in the assays of BCR-ABL–transduced +/+ cells (approximately 6-fold, Table 4) and also in the assays of BCR-ABL–transduced SHIP–/– cells. The total number of CFCs generated from the BCR-ABL–transduced SHIP–/– LTC-ICs was slightly, but not significantly, reduced by comparison with the BCR-ABL–transduced +/+ LTC-ICs. However, their autonomous growth potential was significantly reduced (P < .05). In addition, the colonies generated from the BCR-ABL–transduced SHIP–/– LTC-ICs were much smaller than from those produced by the BCR-ABL–transduced +/+ cells.
Reduced factor-independent colony formation in vitro by BCR-ABL–transduced SHIP–/– LTC-IC–derived CFCs
. | No. of LTC-IC—derived CFCs detected per 103 Sca-1+lin-GFP+ cells . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| CFC assay conditions . | BFU-E . | CFU-GEMM . | CFU-GM . | Total . | |||
| MIG | |||||||
| 3GF | |||||||
| +/+ | 29 ± 4 | 28 ± 3 | 205 ± 77 | 262 ± 84 | |||
| -/- | 25 ± 2 | 26 ± 4 | 210 ± 60 | 265 ± 76 | |||
| EPO | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| No GF | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| p210 | |||||||
| 3GF | |||||||
| +/+ | 150 ± 30 | 90 ± 30 | 1545 ± 145 | 1785 ± 205 | |||
| -/- | 115 ± 15 | 40 ± 10 | 1440 ± 150 | 1590 ± 220 | |||
| EPO | |||||||
| +/+ | 135 ± 15 | 60 ± 10 | 980 ± 60 | 1180 ± 110 | |||
| -/- | 38 ± 7 | 20 ± 2 | 720 ± 120 | 780 ± 128 | |||
| No GF | |||||||
| +/+ | 80 ± 30 | 40 ± 10 | 930 ± 90 | 1050 ± 220 | |||
| -/- | 20 ± 4 | 0 | 710 ± 120 | 730 ± 120 | |||
. | No. of LTC-IC—derived CFCs detected per 103 Sca-1+lin-GFP+ cells . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| CFC assay conditions . | BFU-E . | CFU-GEMM . | CFU-GM . | Total . | |||
| MIG | |||||||
| 3GF | |||||||
| +/+ | 29 ± 4 | 28 ± 3 | 205 ± 77 | 262 ± 84 | |||
| -/- | 25 ± 2 | 26 ± 4 | 210 ± 60 | 265 ± 76 | |||
| EPO | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| No GF | |||||||
| +/+ | 0 | 0 | 0 | 0 | |||
| -/- | 0 | 0 | 0 | 0 | |||
| p210 | |||||||
| 3GF | |||||||
| +/+ | 150 ± 30 | 90 ± 30 | 1545 ± 145 | 1785 ± 205 | |||
| -/- | 115 ± 15 | 40 ± 10 | 1440 ± 150 | 1590 ± 220 | |||
| EPO | |||||||
| +/+ | 135 ± 15 | 60 ± 10 | 980 ± 60 | 1180 ± 110 | |||
| -/- | 38 ± 7 | 20 ± 2 | 720 ± 120 | 780 ± 128 | |||
| No GF | |||||||
| +/+ | 80 ± 30 | 40 ± 10 | 930 ± 90 | 1050 ± 220 | |||
| -/- | 20 ± 4 | 0 | 710 ± 120 | 730 ± 120 | |||
Each 1-mL LTC-IC assay was initiated with 103 Sca-1+lin-GFP+ cells. The LTCs were harvested after 4 weeks, and the cells in them were then assayed for CFCs in FCS-containing methylcellulose medium in the presence or absence of growth factors. Data shown are the total CFCs recovered per initial LTC and represent mean ± SEM of values obtained from 3 independent experiments.
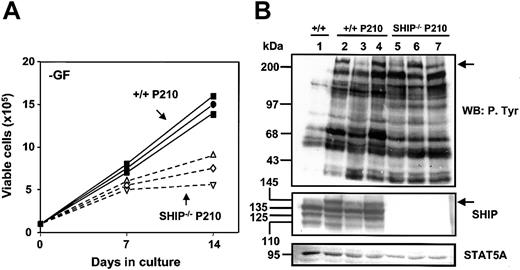
In a final series of experiments to examine the effect of BCR-ABL on the growth of SHIP–/– cells, we expanded single BCR-ABL–transduced +/+ and SHIP–/– cells in the presence of growth factors and then examined the growth kinetics of the cells obtained. As noted previously for +/+ cells,7 the cells in all of these cultures showed a Sca-1+c-kit+lin– immunoglobulin E (IgE) receptor+ phenotype and a mast cell morphology (data not shown). Southern blot analysis showed the presence in each clone of a single complete copy of the BCR-ABL cDNA (data not shown). Although all clones showed some ability to expand in the absence of growth factor, in addition to those derived from BCR-ABL–transduced SHIP–/– cells (n = 6) grew more slowly than their BCR-ABL–transduced +/+ counterparts (n = 6). Results for 3 representative BCR-ABL–transduced +/+ and SHIP–/– clones are shown in Figure 7A.
Reduced growth factor independence of BCR-ABL–transduced SHIP–/– cells and up-regulated expression of the full-length 145-kDa SHIP in BCR-ABL–transduced +/+ cells. (A) Autonomous growth of 3 expanded clones (nos. 1, 3, and 4) of BCR-ABL–transduced +/+ BM cells (solid symbols) and 3 expanded clones (nos. 2-4) of transduced SHIP–/– BM cells (open symbols) in the absence of exogenous growth factors. Viable cell numbers were determined by hematocytometer counts of trypan blue–excluding cells. Results are expressed as the mean ± SEM of data from 3 independent experiments. (B) Western blot analyses of cell lysates from control (MIG-transduced) cells (lane 1); 3 clones of BCR-ABL– transduced +/+ cells (lanes 2-4, clones 1-3); and 3 clones of BCR-ABL–transduced SHIP–/– cells (lanes 5-7, clones 1-3). Lysates of equal numbers of cells were separated on 8% polyacrylamide gradient gels. The filter was first probed with antiphosphotyrosine (4G10) (top panel), then stripped, and reprobed with anti-SHIP antibodies against the SH2 domain and c-terminal region of SHIP and anti-STAT5 (STAT5A) (middle and bottom panels). The positions of prestained molecular weight markers are indicated on the left side of each blot.
Reduced growth factor independence of BCR-ABL–transduced SHIP–/– cells and up-regulated expression of the full-length 145-kDa SHIP in BCR-ABL–transduced +/+ cells. (A) Autonomous growth of 3 expanded clones (nos. 1, 3, and 4) of BCR-ABL–transduced +/+ BM cells (solid symbols) and 3 expanded clones (nos. 2-4) of transduced SHIP–/– BM cells (open symbols) in the absence of exogenous growth factors. Viable cell numbers were determined by hematocytometer counts of trypan blue–excluding cells. Results are expressed as the mean ± SEM of data from 3 independent experiments. (B) Western blot analyses of cell lysates from control (MIG-transduced) cells (lane 1); 3 clones of BCR-ABL– transduced +/+ cells (lanes 2-4, clones 1-3); and 3 clones of BCR-ABL–transduced SHIP–/– cells (lanes 5-7, clones 1-3). Lysates of equal numbers of cells were separated on 8% polyacrylamide gradient gels. The filter was first probed with antiphosphotyrosine (4G10) (top panel), then stripped, and reprobed with anti-SHIP antibodies against the SH2 domain and c-terminal region of SHIP and anti-STAT5 (STAT5A) (middle and bottom panels). The positions of prestained molecular weight markers are indicated on the left side of each blot.
Evidence of BCR-ABL–mediated deregulation of SHIP expression in expanded clones of Sca-1+lin– cells
Because the biologic properties displayed by primitive BCR-ABL–transduced SHIP–/– cell findings were contrary to what had been anticipated from previous studies, it was of interest to investigate whether the primary cells studied here would also show differences in the effect of BCR-ABL on their expression of SHIP. Figure 7B shows a representative Western blot of extracts from one example of an expanded clone of MIG-transduced +/+ cells (n = 4), 3 examples of BCR-ABL–transduced +/+ cells (n = 6), and 3 examples of BCR-ABL–transduced SHIP–/– cells (n = 6). All 4 isoforms of SHIP protein are readily evident in the +/+ cells, and there is no evidence of their down-regulation in the BCR-ABL–transduced +/+ cells. In fact, the clones exhibiting evidence of the most highly tyrosine phosphorylated p210BCR-ABL showed higher expression of full-length (145 kDa) SHIP than control (MIG)–transduced cells (Figure 7B, lanes 2 and 4 versus lanes 1 and 3). As expected, SHIP expression could not be detected from expanded clones of BCR-ABL–transduced SHIP–/– BM cells (Figure 7B, lanes 5-7). These results are consistent with a positive (enhancing) effect of SHIP on the autonomous growth potential of primitive BCR-ABL–transduced mouse BM cells.
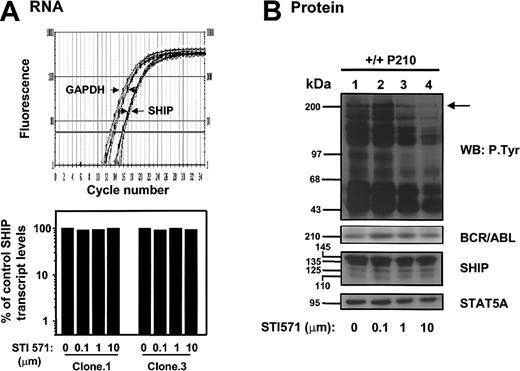
To determine whether the effects on SHIP expression could be directly related to the kinase activity of p210BCR-ABL in the BCR-ABL–transduced +/+ cells, SHIP expression was examined in cells treated with the ABL tyrosine kinase inhibitor, STI571. The results of this experiment are shown in Figure 8. Quantitative real-time RT-PCR measurements revealed that SHIP mRNA levels were not affected by in vitro treatment of the cells with STI571 (0-10 μMol) for 16 to 24 hours (n = 5, Figure 8A), and Western analysis of SHIP protein levels showed these were also not changed, even though this treatment was sufficient to reduce the extent of tyrosine phosphorylation of the p210BCR-ABL being expressed (Figure 8B).
STI571 does not reduce expression of SHIP RNA or protein in expanded clones of BCR-ABL–transduced +/+ cells. (A) Top panel shows real-time RT-PCR SYBR Green1 fluorescence versus cycle number of murine SHIP and GAPDH cDNAs prepared from RNAisolated from BCR-ABL–transduced +/+ cells cultured with 0 to 10 μm STI571 for 16 hours. Bottom panel shows the calculated levels of SHIP transcripts (after normalization to GAPDH transcript levels in the same extracts) in each treated clone expressed as a percentage of the SHIP transcript levels in the untreated control. The levels of SHIP transcripts relative to GAPDH in each sample were calculated using the formula described.26 (B) Western analysis of cells from a BCR-ABL–transduced +/+ clone (no. 1) cultured with varying concentrations of STI571 as described in panelA. Antibodies were the same as in panel B of Figure 7.
STI571 does not reduce expression of SHIP RNA or protein in expanded clones of BCR-ABL–transduced +/+ cells. (A) Top panel shows real-time RT-PCR SYBR Green1 fluorescence versus cycle number of murine SHIP and GAPDH cDNAs prepared from RNAisolated from BCR-ABL–transduced +/+ cells cultured with 0 to 10 μm STI571 for 16 hours. Bottom panel shows the calculated levels of SHIP transcripts (after normalization to GAPDH transcript levels in the same extracts) in each treated clone expressed as a percentage of the SHIP transcript levels in the untreated control. The levels of SHIP transcripts relative to GAPDH in each sample were calculated using the formula described.26 (B) Western analysis of cells from a BCR-ABL–transduced +/+ clone (no. 1) cultured with varying concentrations of STI571 as described in panelA. Antibodies were the same as in panel B of Figure 7.
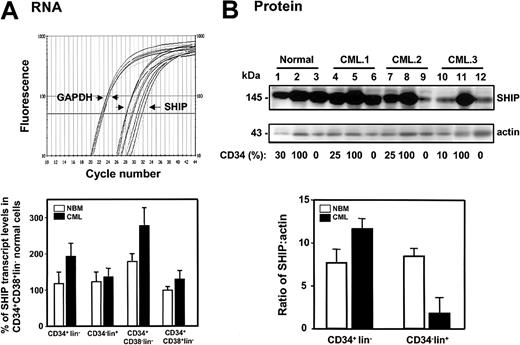
Evidence of up-regulated SHIP expression in primitive CML cells
We then compared SHIP expression in various FACS-purified subpopulations of cells obtained from 8 patients with chronic phase CML and primary normal human BM samples. As shown in Figure 9, real-time RT-PCR and Western analysis indicated both SHIP transcript (Figure 9A) and protein levels (Figure 9B) to be similar in subpopulations of normal BM cells (n = 4) at different stages of differentiation from the most primitive CD34+CD38–lin– subset to the most mature CD34–lin+ cells. In the corresponding leukemic populations isolated from 8 CML samples in which BCR-ABL+ cells were predominant, there were consistently higher levels of SHIP transcripts in all subpopulations of CML cells (n = 8). In the more primitive (CD34+lin–) CML cells this was also associated with elevated levels of SHIP protein in these cells. These results reinforce the findings obtained in the BCR-ABL–transduced murine BM cells in which up-regulated expression of full-length SHIP protein in the expanded progeny of primitive BCR-ABL–transduced cells was also observed. Nevertheless, as previously reported,16 expression of the full-length 145-kDa SHIP protein was significantly lower in the more mature CML cells that exhibit a CD34–lin+ phenotype (7 ± 3-fold lower compared with normal BM CD34–lin+ cells P < .05, Figure 9B). Thus, in these leukemic cells, transcript levels were not predictive of protein expression. Reprobing of the Western blots with an antibody to SHIP217 showed that expression of SHIP2 protein was also down-regulated in these cells (data not shown).
SHIP protein expression is up-regulated in CD34+ CML cells. (A) The top panel shows an example of real-time RT-PCR SYBR Green1 fluorescence versus cycle number of human SHIP and GAPDH cDNAs prepared from the RNA isolated from FACS-sorted CD34+lin–, CD34–lin+, CD34+CD38–lin–, and CD34+CD38+lin– normal BM (n = 4) and CML (n = 8) cells. Bottom panel shows the levels of human SHIP transcripts relative to GAPDH in each population expressed as a percentage of the levels of SHIP transcripts detected in the CD34+CD38+lin– subset of normal BM cells. The level of SHIP transcripts in each individual sample was calculated using the formula described.26 (B) The top panel shows a Western blot analysis of cell lysates prepared from one normal BM sample (lanes 1-3) and 3 CML samples (lanes 4-12). The lysates from equal numbers of cells were separated on 8% polyacrylamide gradient gels. The filter was first probed with a human anti-SHIP antibody (P1C1) (upper blot), then stripped and reprobed with an antiactin antibody (lower blot). The percentage of CD34+ cells in each population is indicated below each lane. In the lower part of panel B, the measured levels of human SHIP protein relative to actin are shown.
SHIP protein expression is up-regulated in CD34+ CML cells. (A) The top panel shows an example of real-time RT-PCR SYBR Green1 fluorescence versus cycle number of human SHIP and GAPDH cDNAs prepared from the RNA isolated from FACS-sorted CD34+lin–, CD34–lin+, CD34+CD38–lin–, and CD34+CD38+lin– normal BM (n = 4) and CML (n = 8) cells. Bottom panel shows the levels of human SHIP transcripts relative to GAPDH in each population expressed as a percentage of the levels of SHIP transcripts detected in the CD34+CD38+lin– subset of normal BM cells. The level of SHIP transcripts in each individual sample was calculated using the formula described.26 (B) The top panel shows a Western blot analysis of cell lysates prepared from one normal BM sample (lanes 1-3) and 3 CML samples (lanes 4-12). The lysates from equal numbers of cells were separated on 8% polyacrylamide gradient gels. The filter was first probed with a human anti-SHIP antibody (P1C1) (upper blot), then stripped and reprobed with an antiactin antibody (lower blot). The percentage of CD34+ cells in each population is indicated below each lane. In the lower part of panel B, the measured levels of human SHIP protein relative to actin are shown.
Discussion
Mouse models of human leukemogenesis offer powerful systems to investigate the mechanisms involved in disease latency and the role of specific gene products in disease progression. Transplantation of HSC-enriched fractions of mouse BM cells transduced with the BCR-ABL oncogene encoded in a retroviral vector is now widely used for this purpose.37 However, when large numbers of transduced cells are transplanted, the recipients develop rapidly lethal polyclonal leukemias that preclude investigation of factors that affect the relatively slow amplification in vivo of isolated leukemic clones characteristic of the early stages of development of human CML. Recently, we7 and others38 have shown that prior culture of the BCR-ABL–transduced mouse BM cells offers one approach to increasing by several weeks the latency of disease obtained. However, it is difficult to control the composition of the population ultimately transplanted using this type of strategy, and the culture conditions used can also variably alter mechanisms initially operative in the transduced cells.
In the present study, we have used an alternative approach. This approach involved isolating highly HSC-enriched populations of BCR-ABL–transduced mouse BM cells (the GFP+Sca-1+lin– subset) immediately after transduction and then assessing their leukemogenic activity in lethally irradiated Ly5-congenic recipients of transplants with limiting numbers of transduced cells and a radioprotective dose of normal BM cells. In this way, clonally derived, exclusively multilineage leukemias could be reproducibly generated in vivo. However, we also noted that this occurred at a frequency that was 7-fold lower than predicted by the concentration of BCR-ABL–transduced HSCs that should have been present on the basis of the HSC content measured in GFP+Sca-1+lin– cells transduced with a control vector. Thus, less than 15% of HSCs transduced with BCR-ABL appeared to have acquired an ability to generate transplantable progressively growing leukemic clones and thereby meet the definition of L-ICs.
Of additional interest was the observation that none of the recipients of BCR-ABL+ cells that survived long term had GFP+ WBCs in their circulation beyond the first 3 weeks. The present experiments do not discriminate whether these short-lived nonleukemogenic GFP+ clones were derived from cells that already had only short-term repopulating activity prior to transduction or whether they were originally HSCs whose long-term repopulating activity was then compromised by the expression of BCR-ABL. However, in either case, it can be seen that there was a significant loss of HSC function that cannot be accounted for by transformation to L-ICs. Previous studies have shown that CML stem cells have an increased probability of differentiation, possibly because of their exposure to increased levels of autocrine IL-3,6,39 and we have also demonstrated that IL-3 expression is rapidly up-regulated in primitive BCR-ABL–transduced mouse BM cells.7 The present findings are thus consistent with a model of BCR-ABL–mediated HSC transformation in which the leukemic stem cells accumulate in vivo relatively slowly because of their continuous loss by differentiation despite an increased proliferative activity.
The introduction of a method for quantitating BCR-ABL–transduced L-ICs in highly enriched HSC populations also provides a sensitive approach to assessing the contribution of other genes to the leukemogenic process. Here, we used this method to examine the role of SHIP because of reported evidence that SHIP might have a tumor suppressor function in BCR-ABL–induced leukemia.16 However, we found that primitive SHIP–/– BM cells are not more readily transformed than +/+ cells, and the latency of the disease they generate and their pattern of differentiation in vivo are also the same. In vitro, evidence of reduced growth factor independence was found in directly isolated primitive BCR-ABL–transduced SHIP–/– cells and in their in vitro expanded mast cell progeny. These results suggest that SHIP is required for optimal activation of factor-independent proliferation and/or survival by BCR-ABL when cells are maintained in the complete absence of exogenous growth factors. In fact, analysis of expanded lines of +/+ cells showed the full-length 145-kDa SHIP protein to be expressed at a higher level and to be more highly phosphorylated in the BCR-ABL–transduced cells than in similarly expanded lines of cells transduced with a control vector. However, SHIP is unlikely to be a direct target of the kinase activity of p210BCR-ABL because the changes in SHIP levels were unaffected by the presence of STI571. Other candidates that could be involved in the observed activation of SHIP in BCR-ABL–transduced cells include PKB/AKT (a serine/threonine kinase) and BAD (a proapoptotic protein). It has been reported that a dominant-negative mutant of PKB/AKT inhibits BCR-ABL–mediated transformation,40 and AKT is highly phosphorylated in BCR-ABL–transduced BM cells but not in cells transduced with an SH3 deletion mutant of c-abl.41 BAD has been reported to be an key substrate of AKT.42 Interestingly, SHIP has also been found to be involved in the regulation of AKT in normal hematopoietic cells.43 Future investigations will be required to elucidate how altered levels of SHIP contribute to perturbed PI3 kinase signaling in BCR-ABL–expressing cells.
The regulation of expression of the SHIP gene in normal hematopoietic cells is known to be complex with different isoforms being differentially expressed in various stages of differentiation and in different cell lineages.11,44-47 In BCR-ABL–transformed BaF3 cells, SHIP protein levels were found to be reduced, and this effect was regulated by the level of expression of the introduced BCR-ABL.16 The same group noted that SHIP protein expression was also reduced in several BCR-ABL+ human cell lines as well as unseparated preparations of primary CML cells.16 Here, we confirmed the finding of reduced levels of SHIP protein in terminally differentiating CML cells and also found that this late stage down-regulation of SHIP expression is not a result of decreased levels of SHIP transcripts. In addition, we obtained evidence that SHIP expression is elevated in the most primitive (CD34+CD38–lin–) CML cells relative to their normal counterparts. Together, these observations support our experimental findings that leukemogenesis at the stem cell level is not enhanced in the absence of SHIP and, indeed, may be adversely affected, as shown by the reduced factor-independence exhibited by BCR-ABL–transduced SHIP–/– cells in vitro. However, it is important to note that the role of SHIP in human CML may still not be fully accessible to study in the experimental mouse model used because of the higher level of virus-induced BCR-ABL expression as compared with that seen in primary human CML cells. Using quantitative real-time RT-PCR to measure absolute transcript levels in both types of cells, we have recently found these to differ by an order of magnitude (X.J., Y. Zhao, W.Y.C., et al, manuscript in preparation, February 5, 2003). In addition, our findings do not exclude the possibility that down-regulation of SHIP in terminally differentiating myeloid cells may contribute to the final overproduction of granulocytes characteristic of CML.
In summary, we describe here an experimental protocol for monitoring the function of single transplanted BCR-ABL–transduced L-ICs from adult mouse BM cells and show that, under these conditions, fatal transplantable multilineage leukemias are generated after a prolonged latency. This model should facilitate future analyses of the early stages of BCR-ABL–mediated leukemogenesis and the identification of other genes that cooperate to accelerate or inhibit specific steps that are important to disease progression. An example of this is provided by the evidence presented here of a positive role of SHIP in contributing to the BCR-ABL–stimulated acquisition by primitive hematopoietic cells of an autonomous growth potential.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2003-05-1550.
Supported by grants from the Leukemia Research Fund of Canada and the National Cancer Institute of Canada (NCIC), with funds from the Canadian Cancer Society and from the Terry Fox Run. M.S. and A.L. were both recipients of BC Cancer Foundation Summer Studentships. Y.C. held a Terwindt Foundation Fellowship. W.E. held a grant from the Fonds zur Forderung der Wissenschaftlichen Forschung (J17777-MED).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank G. Thornbury and R. Zapf for FACS operation; D. Reid, M. Hale, K. Lambie, and K. Saw for technical assistance; K. Humphries and P. Lansdorp (Terry Fox Laboratory, Vancouver, BC, Canada) for the MSCV retroviral vector and antibodies; Novartis (Basel, Switzerland) for the ABL tyrosine kinase inhibitor (STI571); and Rozmina Premji for assistance in preparing the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal