Abstract
We present 3 cases of large B-cell lymphoma (LBCL) with a granular cytoplasmic staining for anaplastic lymphoma kinase (ALK). All of the cases showed striking similarities in morphology and immunohistochemical profile characterized by a massive monomorphic proliferation of CD20-/CD138+ plasmablast-like cells. In one of the cases, initially diagnosed as a null-type anaplastic large cell lymphoma (ALCL), the B-cell phenotype became evident only at recurrence. Fluorescent in situ hybridization (FISH) and molecular studies led to the detection of a CLTC-ALK rearrangement in all 3 cases, without any evidence of full-length ALK receptor expression. The associated t(2;17)(p23;q23) was demonstrated in the karyotype of 2 cases. Although a similar CLTC-ALK aberration was previously identified in ALK-positive T-/null cell ALCL and inflammatory myofibroblastic tumor, its association with ALK-positive LBCL seems to be specific and intriguing. (Blood. 2003;102:2638-2641)
Introduction
Anaplastic lymphoma kinase (ALK)-positive lymphomas represent a distinct entity within the group of CD30+ T-/null cell anaplastic large cell lymphoma (ALCL).1 The aberrant expression of ALK in these tumors results from a rearrangement of the ALK gene with various partner genes including NPM/5q35, TPM3/1q25, ATIC/2q35, TFG/3q21, CLTC/17q23, MSN/Xq12,2-4 and ALO17/17q21.5 The common molecular feature of all 2p23/ALK-related aberrations is the fusion of the ALK tyrosine kinase domain to the 5′ region of partners, which provides a promotor and most likely an oligomerization motif involved in the constitutive activation of the ALK kinase.
The possible involvement of ALK in B-cell lymphomagenesis also has been hypothesized by Delsol et al6 based upon a series of ALK-positive large B-cell lymphomas (LBCLs) assumed to express full-length ALK receptor by an unknown molecular mechanism.
Here, we report 3 LBCL cases characterized by a granular ALK cytoplasmic immunostaining found to result from an underlying t(2;17)(p23;q23)/CLTC-ALK rearrangement.
Study design
Patients
Cases 1, 2, and 3 are from Ghent, Nijmegen, and Leuven, respectively.
Polymerase chain reaction (PCR) and Southern blot
Cytogenetics and fluorescent in situ hybridization (FISH)
G-banding analysis and FISH were performed according to previously described protocols.9 Applied probes included locus-specific identifier (LSI) ALK, whole chromosome paint (WCP) 2, and WCP17 (Vysis, Bergish-Gladbach, Germany), the P1 clone 1111H1 (ALK),10 and bacterial artificial chromosome (BAC) 758H9 (CLTC).5
Combined immunophenotyping and interphase cytogenetic analysis (FICTION)11 using LSI ALK with either an ALK1 monoclonal antibody (MoAb)12 or anti-immunoglobulin A (IgA), or anti-IgG (DAKO, Glostrub, Denmark) antibodies were performed on tissue imprints of frozen material and frozen tissue sections (10 μm).
Reverse transcription (RT)-PCR
RNA isolation, cDNA synthesis, nested PCR, and sequencing were carried out to identify ALK fusion transcripts as described elsewhere.5 To analyze the possible presence of full-length ALK transcripts, 2 RT-PCR experiments were designed amplifying the 5′ end of ALK, primers ALK-ex28f (5′-GCAACATCAGCCTGAAGACA-3′) and ALK-ex29r (5′-GCCTGTTGAGAGACCAGGAG-3′), or the 3′ end of ALK, ALK-ex1f (5′-CTCAGCGAGCTGTTCAGTTG) and ALK-ex2r (5′-GGAGAAGGCATGTTTGTTGG-3′). As a positive control for both 5′- and 3′-ALK, cDNA derived from human fetal poly(A)+ RNA (Clontech, Palo Alto, CA) was used. mRNA (1 μg) was reverse transcribed with random hexamer primers in 20 μL of buffer provided by manufacturer; 3 μL cDNA was used for each RT-PCR. With an annealing temperature at 59°C, 35 cycles were performed. The identity of the PCR products was confirmed by sequencing.
Results and discussion
Clinical, pathologic, (cyto)genetic, and molecular features of the cases are summarized in Table 1. Of interest is that the B-cell phenotype of case 2, originally diagnosed as null-type ALCL and included in our previous study,5 became evident only at recurrence. Since monoclonal IGH/K/L rearrangements were found at relapse (Table 1), diagnostic and relapse biopsies were reanalyzed with a wider spectrum of B-cell markers resulting in a final diagnosis of ALK-positive LBCL. Case 3 was initially diagnosed as diffuse LBCL (DLBCL) with plasmablastic differentiation. ALK expression in this case was found after revision.
Characteristics of 3 patients with ALK-positive LBCL
Characteristics . | Case 1 . | . | Case 2 . | Case 3 . |
|---|---|---|---|---|
| Age, y | 10 | 13 | 26 | |
| Sex | M | F | M | |
| Date of examination mo/y | 2/2002 | 5/2000 (at diagnosis) | 9/2001 (at relapse) | 2/1998 |
| Clinical presentation | Cervical mass | Cervical mass | Cervical lymph nodes, mediastinal mass, spleen, liver | Cervical adenopathy, tumor of base of tongue |
| Stage | 2 | 2 | 3 | 2 |
| Phenotype | CD3−, CD4−, CD5−, CD45−, CD15−, CD30−, EMA−, CD68−, ALK1+, CD20−, CD79a+ (w), CD138+ (s), lgκ−, lgλ+, lgA+, lgG− | Initial: CD2−, CD3−, CD4−, CD5−, CD45−, CD15−, CD30+, EMA+, ALK1+, CD20−, CD79a− | CD2−, CD3−, CD4−, CD5−, CD45−, CD15−, CD30+, EMA+, ALK1+, CD20−, CD22−, CD23−, CD79a+ (w), CD138+, lgκ−, lgλ+, lgA−, lgG+ | Initial: CD2−, CD3−, CD5−, CD20−, lgκ+, lgλ−, lgA+ |
| Reviewed: CD138+, lgκ−, lgλ+, lgA−, lgG+ | Reviewed: CD138+, ALK1+, CD30− | |||
| EBV* | Negative | Negative | Negative | nd |
| Genotype | PCR: IGH(FR3)-R; TCRβ-R, TCRγ-R | PCR: IGH(FR1)-R; TCRγ-R | PCR: IGH-R (FR1/2/3), Kde-R, IGL-R | SB: IGH-R, Kde-R, TCRγ-G |
| SB: IGH-R, Kde-R, TCRγ-G | ||||
| Diagnosis | ALK-positive LBCL | Initial: ALK-positive null-type ALCL | ALK-positive LBCL | Initial: DLBCL with plasma cell differentiation |
| Revised: ALK-positive LBCL | Revised: ALK-positive LBCL | |||
| Therapy | SFOP-LMB 96, group B | ALCL-99 HR | NHL-BFM ALCL99 with ALCL relapse, ABMT | CHOP 4×, VIM 1×, DHAP 1×: NR, RT 40 Gy (Waldeyer ring and neck), 30 Gy upper mediastinum: PD with new skeletal lesions in 1/1999: Hyper-C-VAD 3×: CR, 6/1999 ABMT after BEAM |
| Outcome | CR | CR | PR | CR |
| Follow-up, mo | 6 | 12 | 3‡ | 44 |
| Cytogenetics | Not successful | Not successful | 46-47,XX,i(1)(q10),der(2) add(2)(p13)t(2;3)(q37;q21), der(5)t(3;5)(q25;q34), add(6)(q10),+der(6) t(6;14)(p21;q11),+8,del(10) (p12),?inv(12)(q15q24),−14, add(17)(p11),add(17)(q15), +19,inc[cp7]§ | 44-46,XY,-Y,idic(1)(p11),del(2)(p23), del(5)(q23),+14,add(17)(?q23)[cp5]§ |
| RT-PCR | ALK-EC mRNA− | ALK-EC mRNA− | nd | ALK-EC mRNA− |
| ALK-C mRNA+ | ALK-C mRNA+ | nd | ALK-C mRNA+ | |
| FISH† (LSI ALK) | 1GO/2O/1G→ALK rearrangement | 1GO/1O/1G→ALK rearrangement | 1GO/1O/1G→ALK rearrangement→ der(2)add(2)(p13)t(2;3) (q37;q21).ish der(2)t(2;17) (p23;q23)t(2;3)(q37;q21); add(17)(q15).ish der(17)t(2;17) (p23;q23)§ | 1GO/1O/1G→ALK rearrangement→ del(2)(p23).ish der(2)t(2;17)(p23;q23); add(17)(?q23).ish der(17)t(2;17)(p23;q23)§ |
Characteristics . | Case 1 . | . | Case 2 . | Case 3 . |
|---|---|---|---|---|
| Age, y | 10 | 13 | 26 | |
| Sex | M | F | M | |
| Date of examination mo/y | 2/2002 | 5/2000 (at diagnosis) | 9/2001 (at relapse) | 2/1998 |
| Clinical presentation | Cervical mass | Cervical mass | Cervical lymph nodes, mediastinal mass, spleen, liver | Cervical adenopathy, tumor of base of tongue |
| Stage | 2 | 2 | 3 | 2 |
| Phenotype | CD3−, CD4−, CD5−, CD45−, CD15−, CD30−, EMA−, CD68−, ALK1+, CD20−, CD79a+ (w), CD138+ (s), lgκ−, lgλ+, lgA+, lgG− | Initial: CD2−, CD3−, CD4−, CD5−, CD45−, CD15−, CD30+, EMA+, ALK1+, CD20−, CD79a− | CD2−, CD3−, CD4−, CD5−, CD45−, CD15−, CD30+, EMA+, ALK1+, CD20−, CD22−, CD23−, CD79a+ (w), CD138+, lgκ−, lgλ+, lgA−, lgG+ | Initial: CD2−, CD3−, CD5−, CD20−, lgκ+, lgλ−, lgA+ |
| Reviewed: CD138+, lgκ−, lgλ+, lgA−, lgG+ | Reviewed: CD138+, ALK1+, CD30− | |||
| EBV* | Negative | Negative | Negative | nd |
| Genotype | PCR: IGH(FR3)-R; TCRβ-R, TCRγ-R | PCR: IGH(FR1)-R; TCRγ-R | PCR: IGH-R (FR1/2/3), Kde-R, IGL-R | SB: IGH-R, Kde-R, TCRγ-G |
| SB: IGH-R, Kde-R, TCRγ-G | ||||
| Diagnosis | ALK-positive LBCL | Initial: ALK-positive null-type ALCL | ALK-positive LBCL | Initial: DLBCL with plasma cell differentiation |
| Revised: ALK-positive LBCL | Revised: ALK-positive LBCL | |||
| Therapy | SFOP-LMB 96, group B | ALCL-99 HR | NHL-BFM ALCL99 with ALCL relapse, ABMT | CHOP 4×, VIM 1×, DHAP 1×: NR, RT 40 Gy (Waldeyer ring and neck), 30 Gy upper mediastinum: PD with new skeletal lesions in 1/1999: Hyper-C-VAD 3×: CR, 6/1999 ABMT after BEAM |
| Outcome | CR | CR | PR | CR |
| Follow-up, mo | 6 | 12 | 3‡ | 44 |
| Cytogenetics | Not successful | Not successful | 46-47,XX,i(1)(q10),der(2) add(2)(p13)t(2;3)(q37;q21), der(5)t(3;5)(q25;q34), add(6)(q10),+der(6) t(6;14)(p21;q11),+8,del(10) (p12),?inv(12)(q15q24),−14, add(17)(p11),add(17)(q15), +19,inc[cp7]§ | 44-46,XY,-Y,idic(1)(p11),del(2)(p23), del(5)(q23),+14,add(17)(?q23)[cp5]§ |
| RT-PCR | ALK-EC mRNA− | ALK-EC mRNA− | nd | ALK-EC mRNA− |
| ALK-C mRNA+ | ALK-C mRNA+ | nd | ALK-C mRNA+ | |
| FISH† (LSI ALK) | 1GO/2O/1G→ALK rearrangement | 1GO/1O/1G→ALK rearrangement | 1GO/1O/1G→ALK rearrangement→ der(2)add(2)(p13)t(2;3) (q37;q21).ish der(2)t(2;17) (p23;q23)t(2;3)(q37;q21); add(17)(q15).ish der(17)t(2;17) (p23;q23)§ | 1GO/1O/1G→ALK rearrangement→ del(2)(p23).ish der(2)t(2;17)(p23;q23); add(17)(?q23).ish der(17)t(2;17)(p23;q23)§ |
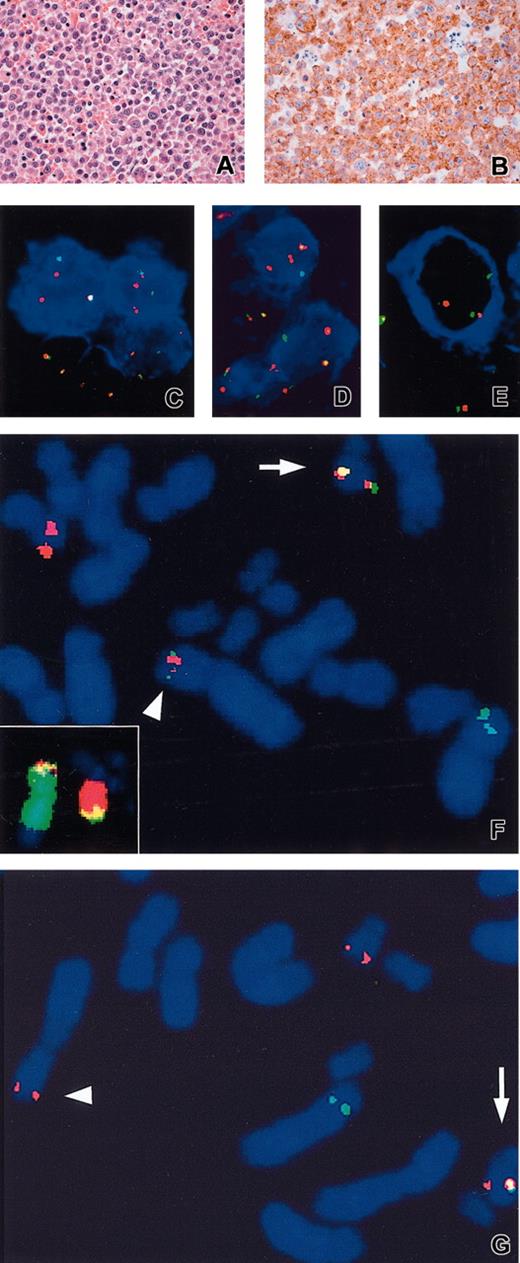
All cases showed monomorphic proliferation of large cells and a granular cytoplasmic ALK1 immunostaining pattern.
EMA indicates epithelial membrane antigen; EBV, Epstein-Barr virus; PCR, polymerase chain reaction; M, male; F, female; s, strong; w, weak; SB, Southern blot; SFOP-LMB, Société Française d'Oncologie Pédiatrique, étude lymphomes B; HR, high risk; NHL-BFM, non-Hodgkin lymphoma-Berlin-Frankfurt-Munster; ABMT, autologous bone marrow transplantation; CHOP, cyclophosphamide, doxorubicin, vincristine, and prednisone; VIM, etoposide, ifosfamide, and mitoxantrone; DHAP, dexamethasone, cytosine, arabinoside, and cisplatin; NR, no remission; RT, radiation therapy; PD, progressive disease; BEAM, BCNU (1,3-bis(2-chloroethyl)-1-nitrosourea), etoposide, cytosine, arabinoside, and melphalan; CR, complete remission; PR, partial remission; ALK-EC, extracellular portion of ALK; ALK-C, cytoplasmic portion of ALK; nd, not done; G, green signal; and O, orange signal.
Analyzed using anti-latent membrane protein 1 antibody and EBV-encoded RNA in situ hybridization.
Interphase and metaphase FISH experiments were performed on tissue imprints (case 1), cytogenetic specimen (case 2 [at diagnosis and relapse], case 3), and frozen section (case 2 [at relapse]).
Death.
Chromosomal aberrations are presented in accordance with the International System for Human Cytogenetic Nomenclature.13
All 3 cases showed a granular cytoplasmic ALK expression using the ALK1 MoAb12 (Figure 1B), and the molecular events underlying this expression pattern were investigated. Dual-color FISH with LSI ALK revealed a prominent population of cells with split signals indicative of ALK rearrangement in all analyzed samples (Table 1). The direct association of ALK rearrangements with the aberrant ALK expression and B-cell immunophenotype of the affected cells was demonstrated by FICTION (fluorescence immunophenotyping and interphase cytogenetics as a tool for the investigation of neoplasm) where LSI ALK was combined with antibodies for ALK or IgA (case 1) (Figure 1C-D), or IgG (case 2) (Figure 1E).
Histologic, immunophenotypic, and molecular cytogenetic findings. An example of hematoxylin and eosin (A) and ALK1 (B) staining performed in case 3 showing, respectively, a massive proliferation of plasmablast-like cells and a granular cytoplasmic localization of ALK. FICTION with LSI ALK and ALK1 (blue) (C), or anti-IgA (blue) (case 1) (D), and anti-IgG (case 2) (E). Metaphase FISH with ALK P1 (1111H1) (green) and BAC 758H9 (CLTC) (red) in case 1 (F) and case 2 (G). Arrowhead and arrow indicate the der(2) and der(17), respectively. Inset in panel F: WCP2 (green) and WCP17 (red). Note 1F2O1G FISH signals in case 1 (C-D) pointing the ALK gene rearrangement in ALK1+/IgA+ cells. Extra red signal possibly reflects an additional copy of the der(17) chromosome. Original magnifications: × 200 (A-B); × 630 (C-G).
Histologic, immunophenotypic, and molecular cytogenetic findings. An example of hematoxylin and eosin (A) and ALK1 (B) staining performed in case 3 showing, respectively, a massive proliferation of plasmablast-like cells and a granular cytoplasmic localization of ALK. FICTION with LSI ALK and ALK1 (blue) (C), or anti-IgA (blue) (case 1) (D), and anti-IgG (case 2) (E). Metaphase FISH with ALK P1 (1111H1) (green) and BAC 758H9 (CLTC) (red) in case 1 (F) and case 2 (G). Arrowhead and arrow indicate the der(2) and der(17), respectively. Inset in panel F: WCP2 (green) and WCP17 (red). Note 1F2O1G FISH signals in case 1 (C-D) pointing the ALK gene rearrangement in ALK1+/IgA+ cells. Extra red signal possibly reflects an additional copy of the der(17) chromosome. Original magnifications: × 200 (A-B); × 630 (C-G).
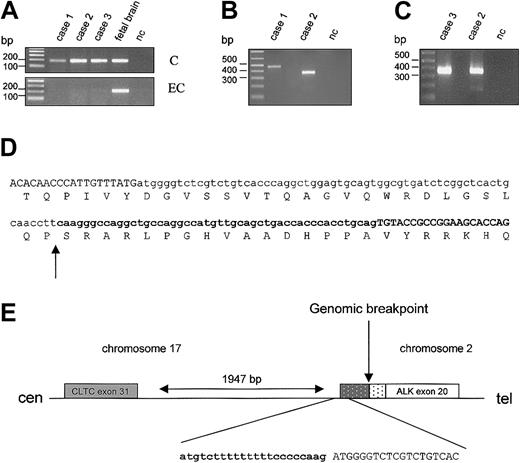
The diagnostic sample of case 2 was already included in our previous study of ALK-positive tumors and shown to contain the CLTC-ALK rearrangement.5 The same CLTC-ALK fusion product (confirmed by direct sequencing) was detected in the sample at relapse (Figure 2B/case 2). To identify the fusion product of case 1, RT-PCR analysis for some of the known ALK fusions (TPM3-ALK, ATIC-ALK, CLTC-ALK, ALO17-ALK) was carried out. In this case also a CLTC-ALK fusion transcript was present. However, the RT-PCR product was about 100 nucleotides (nt) longer than the product obtained in case 2.5 Sequencing of the RT-PCR product revealed a fusion of exon 31 of CTLC to exon 20 of ALK, separated by an additional 111 nt (Figure 2D). Sequence analysis suggests that this is the result of the presence of the genomic fusion site 48 nt upstream of ALK exon 20 and the use of a cryptic splice signal in the CLTC intron following exon 31, 63 nt upstream of this genomic breakpoint (Figure 2E). The AG acceptor sequence of this cryptic site is preceded by a 17 nt polypyrimidine stretch, probably generating a stronger splice signal than the one preceding ALK exon 20. The open reading frame is conserved by this rearrangement. For case 3, again a CLTC-ALK fusion transcript was detected by RT-PCR (Figure 2C) and sequence analysis indicated the same fusion as reported for case 2.
Identification and schematic representation of theCLTC-ALKfusion transcripts. (A) RT-PCR specific for the cytoplasmic (“C”) or the extracellular (“EC”) portion of ALK in cases 1 to 3 and in fetal brain cDNA (positive control); nc indicates negative control. (B) RT-PCR performed on cases 1 and 2 (relapsed) with primers for the CLTC-ALK fusion. (C) RT-PCR for case 3 with primers for CLTC-ALK; case 2 served as a positive control. (D) Sequence of the fusion transcript/fusion protein of case 1. Exon sequences are in uppercase, intron sequences are in lowercase, and ALK sequences are in bold. Arrow indicates the genomic junction. (E) Schematic representation of the breakpoint region on the der(17) chromosome. ( ) CLTC intron sequence present in the fusion transcript. (▦) ALK intron sequence present in the transcript. Arrow indicates the chromosomal breakpoint.
) CLTC intron sequence present in the fusion transcript. (▦) ALK intron sequence present in the transcript. Arrow indicates the chromosomal breakpoint.
Identification and schematic representation of theCLTC-ALKfusion transcripts. (A) RT-PCR specific for the cytoplasmic (“C”) or the extracellular (“EC”) portion of ALK in cases 1 to 3 and in fetal brain cDNA (positive control); nc indicates negative control. (B) RT-PCR performed on cases 1 and 2 (relapsed) with primers for the CLTC-ALK fusion. (C) RT-PCR for case 3 with primers for CLTC-ALK; case 2 served as a positive control. (D) Sequence of the fusion transcript/fusion protein of case 1. Exon sequences are in uppercase, intron sequences are in lowercase, and ALK sequences are in bold. Arrow indicates the genomic junction. (E) Schematic representation of the breakpoint region on the der(17) chromosome. ( ) CLTC intron sequence present in the fusion transcript. (▦) ALK intron sequence present in the transcript. Arrow indicates the chromosomal breakpoint.
) CLTC intron sequence present in the fusion transcript. (▦) ALK intron sequence present in the transcript. Arrow indicates the chromosomal breakpoint.
The results of the molecular analysis were further confirmed by metaphase FISH on cases 2 and 3 using LSI ALK, clones covering ALK (1111H1) and CLTC (758H9), and painting probes (Figure 1F-G). The FISH patterns were in line with the t(2;17)(p23;q23)/CLTC-ALK.
So far, only very few data on ALK-positive B-cell lymphomas are available in literature.14-17 Of interest is the report of Gascoyne et al18 who found among 70 adult ALCLs, 5 B-cell ALCL cases with ALK expression in both the nucleus and cytoplasm. Only one of these cases displayed a phenotype similar to our cases; in this particular case a t(2;5) was documented by cytogenetics. These data are in contrast with the findings of Delsol et al6 who reported 7 cases of ALK-positive LBCL showing morphologic and phenotypic features of a plasmablastic differentiation. All of these cases were characterized by a granular cytoplasmic staining for ALK1 and a membranous staining for ALK-EC (serum reactive with the extracellular ALK region), assumed to represent expression of full-length ALK receptor through an unknown molecular mechanism. Interestingly, by morphology and phenotypic analysis our 3 cases are very similar to those reported by Delsol et al.6 However, RT-PCR analysis of the present cases with primer sets designed to amplify the 5′ or the 3′ end of the ALK cDNA, respectively, detected the presence of 3′-ALK but not of 5′-ALK sequences showing that in these cases the ALK reactivity is due to the exclusive expression of CLTC-ALK fusion (Figure 2A).
Occurrence of the same CLTC-ALK rearrangement in all 3 LBCL cases analyzed is intriguing. Although this variant aberration was already found in T-/null cell ALCL19 and in the inflammatory myofibroblastic tumor,20 it could be a specific event in ALK-positive LBCL. Considering the peculiar morphology and phenotype of these lymphoma cells (CD20-, CD138+, ALK1+, CD30+/-) one might wonder how many similar cases are hidden in the null-type ALCL and DLBCL groups.
Of interest, the finding of ALK expression/rearrangement in T-/null cell ALCL as well as in LBCL is in line with results published by Kuefer et al,21 Lange et al,22 and Chiarle et al23 who showed development of either B- or T- and B-cell lymphoma in mice with experimentally overexpressed NPM-ALK.
In conclusion, presented data indicate that ALK activation by CLTC-ALK also plays a role in the pathogenesis of large B-cell lymphomas. The clear correlation of CLTC rearrangement with LBCL is of particular interest and warrants further investigations.
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2003-04-1050.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Ursula Pluys, Gerard Merkx, and Femmy Stellink for skillful technical assistance in FISH and cytogenetic analysis; Elisabeth Moreau and Philip Kluin (chairman of the pathology panel of the SNWLK) for pathologic data; and Rita Logist for editorial help.
P.V. is a senior clinical investigator of Fonds voor Wetenschappelijk Onderzoek Vlaanderen.
This text presents research results of the Belgian Programme of Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming. The scientific responsibility is assumed by the authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal