Abstract
ANBL-6, a myeloma cell line, proliferates in response to interleukin 6 (IL-6) stimulation, coculture with bone marrow stromal cells, and when harboring a constitutively active mutant N-ras gene. Eighteen samples, including 4 IL-6-treated, 3 mutant N-ras-transfected, 3 normal stroma-stimulated, 2 multiple myeloma (MM) stroma-stimulated, and 6 untreated controls were profiled using microarrays interrogating 12 626 genes. Global hierarchical clustering analysis distinguished at least 6 unique expression signatures. Notably, the different stimuli altered distinct functional gene programs. Class comparison analysis (P = .001) revealed 138 genes (54% involved in cell cycle) that distinguished IL-6-stimulated versus nontreated samples. Eighty-seven genes distinguished stroma-stimulated versus IL-6-treated samples (22% encoded for extracellular matrix [ECM] proteins). A total of 130 genes distinguished N-ras transfectants versus IL-6-treated samples (26% involved in metabolism). A total of 157 genes, 20% of these involved in signaling, distinguished N-ras from stroma-interacting samples. All 3 stimuli shared 347 genes, mostly of metabolic function. Genes that distinguished MM1 from MM4 clinical groups were induced at least by one treatment. Notably, only 3 genes (ETV5, DUSP6, and KIAA0735) are uniquely induced in mutant ras-containing cells. We have demonstrated gene expression patterns in myeloma cells that distinguish an intrinsic genetic transformation event and patterns derived from both soluble factors and cell contacts in the bone marrow microenvironment. (Blood. 2003;102:2581-2592)
Introduction
Multiple myeloma (MM) is a B-cell neoplasia characterized by the clonal expansion of plasma cells in the bone marrow (BM). Interleukin 6 (IL-6) induces terminal differentiation of normal mature B cells.1 In contrast, IL-6 has been shown to be a potent mitogen and survival factor for myeloma cells and cell lines.2 Although no single common genetic mutation is associated with the disease, 40% to 50% of patients with myeloma display activating N- or K-ras mutations.3,4 In addition, our laboratory has demonstrated that an IL-6-dependent MM cell line, ANBL-6, stably transfected with a constitutively active N-ras does not require exogenous IL-6 addition for proliferation.5 Furthermore, cells containing the activated N-ras transgene become resistant to certain chemotherapeutic drugs such as dexamethasone and doxorubicin.6 Another hallmark of the disease is the requirement of the BM microenvironment for growth and survival of the malignant clone. Although normal plasma cells accumulate in the lamina propria of the gut, MM plasma cells rarely develop in this site, providing evidence that the BM provides a unique niche for the clone to proliferate.7 Indeed, several groups have demonstrated that the BM is a major source of IL-6, and myeloma cells can induce BM stroma (BMS) to produce this cytokine on cell interaction.8 We and others have shown that MM/BMS cell (BMSC) interactions drive ANBL-6 cells to proliferate and also alter their therapeutic response in vitro.9
A recent report by Zhan and coworkers has provided further evidence that myeloma plasma cells are significantly different from their normal counterparts in their gene expression profiles.10 Using microarray technology this group has demonstrated that MM can be subdivided into 4 genetically distinct gene expression profile groups that correlated with clinical prognosis. Our current study provides insight on how some of these genes might be induced and affect MM growth by comparing the effects of IL-6 response, N-ras-activating mutations, and BMSC stimulation. We examined these effects in a single cell line, thus allowing comparative gene profiling within a homogeneous cell population. All 3 treatments induce ANBL-6 cells to proliferate and were expected to share some overlap in gene expression. However, the genetic mechanism by which this proliferative phenotype is achieved may show distinct differences or gene expression signatures. Therefore, we used DNA microarray technology to globally investigate how similar and distinct these responses are and which genetic changes may uniquely characterize each group. Furthermore, we compared our results with the recently reported data from MM patient plasma cells and found that most genes that distinguished benign plasma cell dyscrasia, monoclonal gammopathies of undetermined significance (MGUS) from active end-stage MM were differentially induced by these activating signals.
Materials and methods
Cell culture
ANBL-6 cells, an IL-6-dependent MM cell line kindly provided by Diane Jelinek (Mayo Clinic, Rochester, MN), were grown in RPMI, 10% fetal calf serum (FCS), 50 U/mL each penicillin and streptomycin, and 2 mM l-glutamine (Gibco BRL, Gaithersburg, MD); the media were replaced every 4 days and supplemented with 0.5 ng/mL IL-6 (R&D Systems, Minneapolis, MN). Previous to the experiments, ANBL-6.plxsn (empty vector) and ANBL-6.N-ras 12 EE transfected cells, derived from mixed polyclonal cell populations to avoid artifacts of clonal selection and described previously,5,11 were incubated in the absence of IL-6 for 3 days to eliminate the IL-6 effect on the cells. On the third day, cells were centrifuged on Ficoll-Histopaque (Sigma Diagnostics, St Louis, MO) to remove any dead cells and allowed to recover for 24 hours. The IL-6-treated cells were stimulated with 10 ng/mL IL-6 at time 0 hour and once again at 24 hours. All control, N-ras- and IL-6-treated cells were cultured for 48 hours under respective treatment and subsequently harvested for RNA isolation. When cocultured with BMSCs, ANBL-6 cells were pretreated as described and seeded in contact with BMSCs in a 1:3 ratio for 48 hours. Normal BMSCs were obtained from femoral heads of healthy donors undergoing hip replacement surgery. Myeloma BMSC layers were derived from CD138- marrow aspirates after CD138 immunopurification, from patients with newly diagnosed MM (Mayo Clinic). All BMs were centrifuged on Ficoll-Histopaque to isolate mononuclear cells. The interface in the Histopaque density gradient was removed and cultured in α-minimum essential medium (α-MEM), 20% FCS, 50 U/mL each penicillin and streptomycin, and 2 mM l-glutamine, and the media were replaced every 7 days. Once cultures reached confluence, they were trypsinized and split 1:5. Only cell passages 2 to 5 were used for subsequent experiments. Twenty-four hours before coculturing with ANBL-6 cells, BMSC media were replaced with RPMI 1640, 10% FCS, 50 U/mL each penicillin and streptomycin, and 2 mM l-glutamine.
Cell sorting
After allowing 48-hour cell interaction, cocultures were trypsinized and immunostained with 20 μL/1 × 106 cells anti-CD38 conjugated to phycoerythrin (PE) or isotype control (PharMingen, San Diego, CA). Cocultures were sorted using the FACSVantage cytometer to obtain MM cell and BMSC separate populations more than 98% pure. Cells were collected in 1 mL FCS for subsequent total RNA extraction.
Total RNA extraction, preparation of c-RNA, and hybridization to microarray chips
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). Double-stranded cDNA was produced from total RNA using Superscript kit (Invitrogen, Carlsbad, CA) and biotinylated cRNA was synthesized using the Enzo array kit (Affymetrix, Santa Clara, CA). cRNA was hybridized to U95Av2 GeneChip microarrays (Affymetrix) according to the manufacturer's protocols. The arrays were scanned using the Agilant G2500A Gene Array Scanner (Agilant Technologies, Palo Alto, CA). Arrays were scaled to an average intensity of 1500 for interarray comparisons. The entire Affymetrix data set can be found on the Blood website; see the Supplemental Materials link at the top of the online article.
Data analysis
Statistical analysis of the average difference data (relative gene expression) derived from Affymetrix 5.0 software was performed using 3 software packages: BRB-Array Tools 2.0 (Dr. Simon, National Cancer Institute, Washington, DC), Expressionist, and Gene Cluster/TreeView (Eisen Laboratory, Stanford University, CA). Hierarchical clustering of average linkage with centered correlation metric was used on the average difference data of 12 626 genes. Multidimensional scaling analysis with the Euclidean distance metric (principal component analysis) of the average difference of the samples was performed. This analysis derived a 3-dimensional representation where samples whose expression profiles are very similar are shown close together.
To analyze genetic differences that distinguished and characterized the treatment groups uniquely, a class comparison tool was used, which performs univariate F tests that are differentially expressed between 2 or more phenotype classes. This tool also computes a global permutation test that excludes genes that differ significantly due to chance alone. Genes that were significant (P < .001) after 2000 permutations were considered to be significantly differentially expressed.
Nonvariable genes among phenotypic classes were determined using a variance test (10% level) in combination with the parametric t test and nonparametric Kruskal-Wallis test (P < .001).
To investigate which genes were newly expressed in the N-ras transfectants compared with nontreatment controls, average difference values of 200 or less in the noninterleukin (NIL) group were sorted for average difference values of 1000 or higher in the N-ras treatment group.
The complete data analysis set can be found on the Blood website; see the Supplemental Materials link.
Reverse transcription-polymerase chain reaction
Total RNA was isolated using the RNeasy Mini Kit (Qiagen); cDNA was made using the Superscript kit (Invitrogen) and amplified by polymerase chain reaction (PCR) using specific primers for MIP-1α, EZH2, and GAPDH as loading control. Primers: MIP-1 α: 5′-CAGAAGGACACGGGCAGCAG-3′,5′-GTGATGCAGAGAACTGGTTG-3′; EZH2:5′-GCCAGACTGGGAAGAAATCTG-3′, 5′-TGTC5GGAAAATCCAAGTCA-3′; GAPDH:5′-GGGCGCCTGGTCACCAGGGCTG-3′,5′-GGGGCCATCCACAGTCTTCTG -3′.
Cell cycle analysis
Myeloma cells (2 × 105) were collected before and after IL-6 treatment, coculture with BMSCs, and N-ras transfection. Pelleted cells were resuspended in 500 μL hypotonic solution containing propidium iodide (PI), which gives a measure of DNA content in the cell (50 μg/mL PI in 0.1% sodium citrate plus 0.1% Triton X-100) and incubated for 3 hours at 4°C in the dark. Analysis of nuclei was performed using FACSCalibur (Becton Dickinson, San Jose, CA).
Results
ANBL-6 MM cells proliferate in response to IL-6, N-ras-activating mutations, and interaction with BMSCs
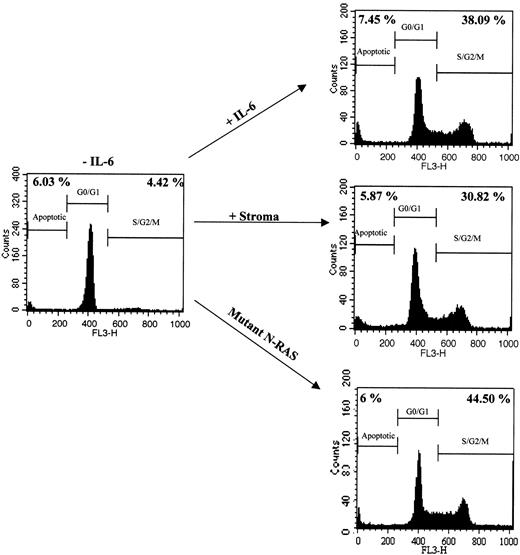
The proliferative response of ANBL-6 cells treated with IL-6, BMSCs, or those carrying an activating N-ras mutation, was investigated by PI staining and flow cytometry. Cell cycle analysis was performed by calculating the percentage of cells in the S/G2/M phase of the cell cycle. As shown in Figure 1, all 3 treatments increased the percentage of dividing cells when compared with growth factor-deprived control cells, up to 9-fold (38.09%, 30.82%, 44.50% in IL-6, stroma-treated, and N-ras cells, respectively, compared with 4.42% in control cells).
Proliferative response of MM cell line ANBL-6 to exogenous IL-6, stromal cell interactions, and activating N-ras mutations. Cell cycle analysis of ANBL-6.plxsn cells in the absence or presence of recombinant IL-6, after coculture with MM stromal cells, or stably transfected with a mutant N-ras gene. Numbers on the left indicate the percentage of subdiploid nuclei. Numbers on the right indicate percentage of nuclei in S/G2/M phases of the cell cycle.
Proliferative response of MM cell line ANBL-6 to exogenous IL-6, stromal cell interactions, and activating N-ras mutations. Cell cycle analysis of ANBL-6.plxsn cells in the absence or presence of recombinant IL-6, after coculture with MM stromal cells, or stably transfected with a mutant N-ras gene. Numbers on the left indicate the percentage of subdiploid nuclei. Numbers on the right indicate percentage of nuclei in S/G2/M phases of the cell cycle.
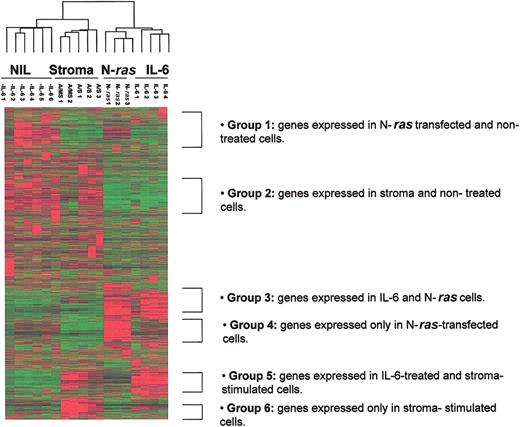
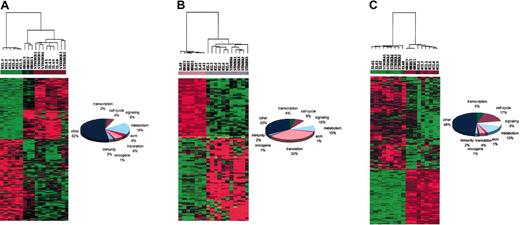
Hierarchical clustering of ANBL-6 gene expression reveals similarities and differences among IL-6 response, N-ras-activating mutation, and coculture with BMS
Global 2-dimensional hierarchical clustering was performed on 12 626 genes contained in the U95av2 Affymetrix gene chip. Eighteen samples, including 4 IL-6-treated, 3 N-ras-transfected, 3 normal stroma-stimulated, 2 MM stroma-stimulated, and 6 untreated control replicate samples were interrogated. This analysis revealed gene expression similarities and differences among cells that were proliferating under the 3 different stimuli (Figure 2). Although not reported here, we also profiled the stromal cells after coculture. Importantly, none of the highest expressed genes in the stroma expression profile were represented in the ANBL-6 cells after separation from the stromal coculture, indicating minimal stromal contamination of the myeloma cell expression pool.
Global 2-dimensional hierarchical clustering demonstrates distinct expression signatures for each activating signal. Global 2-dimensional hierarchical clustering was performed on 12 626 genes contained in the U95av2 Affymetrix gene chip using Cluster/Tree View (Eisen Laboratory). The dendrogram (top) shows clustering of ANBL-6. -IL-6 indicates ANBL-6.plxsn in the absence of IL-6; A/MS, ANBL-6.plxsn cells after contact with MM BMSCs; A/S, ANBL-6.plxsn cells after contact with normal BMSCs; N-Ras, ANBL-6 stably transfected with mutant N-ras; IL-6, ANBL-6.plxsn cells stimulated with 10 ng/mL recombinant IL-6.
Global 2-dimensional hierarchical clustering demonstrates distinct expression signatures for each activating signal. Global 2-dimensional hierarchical clustering was performed on 12 626 genes contained in the U95av2 Affymetrix gene chip using Cluster/Tree View (Eisen Laboratory). The dendrogram (top) shows clustering of ANBL-6. -IL-6 indicates ANBL-6.plxsn in the absence of IL-6; A/MS, ANBL-6.plxsn cells after contact with MM BMSCs; A/S, ANBL-6.plxsn cells after contact with normal BMSCs; N-Ras, ANBL-6 stably transfected with mutant N-ras; IL-6, ANBL-6.plxsn cells stimulated with 10 ng/mL recombinant IL-6.
Six groups of distinct gene expression patterns could be identified. Genes differentially expressed in a similar way in N-ras-transfected and empty vector samples when compared to all other response groups were designated group 1. Group 2 identified genetic similarities between stroma-treated samples and control samples. Commonly expressed genes in IL-6- and N-ras-treated samples are shown in group 3. Group 4 consists of genes that are expressed uniquely in the N-ras-transfected samples when compared to all other treatments. As expected, there is a group of genes (group 5) that is common to IL-6- and stroma-treated samples. This signature was to be expected because BMSCs are the main source of IL-6 for the MM cells; thus, these treatments probably activate many of the same genetic pathways. However, a sixth group of genes differentially expressed only in stroma-stimulated samples and not in the IL-6-treated or any other response group is noted, demonstrating that BMS provides signals to alter gene expression in MM cells other than through IL-6.
Principal component analysis and hierarchical clustering of MM cells under different treatments identifies phenotypic classes that correspond to each response group and the genetic changes that distinguish them
Principal component analysis and hierarchical clustering were performed (“Materials and methods”) to classify the data into phenotypic classes (data not shown and Figure 2). The 2-dimensional hierarchical clustering of all samples displayed as a dendrogram is shown in Figure 2 and demonstrates high reproducibility of the assay because all replicates for each treatment were found to cluster adjacent to one another, indicating that their gene expression profiles were more similar to each other than to other samples. Interestingly, samples carrying activated Ras were more similar to IL-6-treated samples than to stroma-stimulated or IL-6-starved samples, demonstrated by the 2 main branches of the dendrogram.
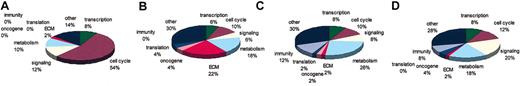
The class comparison tool (“Materials and methods”) was used to determine genes that distinguish the phenotypic classes or treatment groups at the level of significance of 0.001 for the F test and allowing 2000 data permutations to discard random values. A total of 138 genes were differentially expressed between the IL-6-starved and the IL-6-treated samples. Table 1 displays the top 50 genes that distinguish these 2 classes; among these, 37 genes were up-regulated and 13 genes were down-regulated in the IL-6-treated samples when compared with nontreated controls. Osteopontin (OPN) was the most up-regulated gene (65-fold), followed by several cell cycle-related genes such as CDC45, BUB1, WEE1, and CDC6. In fact, 54% of the genetic changes distinguishing IL-6-treated and nontreated samples were cell cycle genes (Figure 3A). Interferon-inducible gene (IFI56) was the most down-regulated gene in IL-6-treated samples as well as other growth suppressor genes such as IFI54 and KIP2. The fact that most up-regulated genes are involved in cell cycle progression is not surprising because IL-6 induces growth of MM cells, through Ras-mitogen-activated protein kinase (MAPK) signaling pathway targets.12 Thus, we demonstrate that IL-6 acts to stimulate proliferation through altering both growth-promoting and growth-suppressing genes.
Top 50 of 138 most differentially expressed genes in a comparison of IL-6 response versus IL-6 starvation effect
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| AF052124 | OPN/osteopontin | Bone/ECM | 65.61 | 8.72 × 10−5 |
| U73379 | E2C | Proteolysis | 42.32 | 9.35 × 10−5 |
| AJ223728 | CDC45 | Cell cycle | 37.12 | 1.33 × 10−8 |
| AF053305 | BUB1 | Cell cycle checkpoint | 30.43 | 1.95 × 10−5 |
| X62048 | WEE1 | Cell cycle | 29.07 | 7.55 × 10−7 |
| D14657 | KIAA0101 | Unknown | 26.32 | 2.73 × 10−5 |
| U77949 | CDC6 | Cell cycle | 25.81 | 1.05 × 10−5 |
| N58115 | EST | Unknown | 22.85 | .000162 |
| L25876 | CIP2 | Signaling | 19.5 | .000406 |
| AF067656 | Zw10 | Wint signaling | 18.17 | 1.38 × 10−6 |
| U74612 | FKHL16 | Transcription factor | 17.4 | 1.99 × 10−5 |
| AF025441 | OIP5 | Unknown | 17.34 | .000198 |
| U37426 | HKSP | Cell division | 17.28 | 1.22 × 10−5 |
| D80008 | KIAA0186 | Unknown | 15.97 | 1.56 × 10−5 |
| Y13115 | SAK | Signaling | 15.84 | .000811 |
| U63743 | KNSL6 | Cell division | 15.8 | .000119 |
| D14678 | MHCB | Cell division | 15.52 | .000154 |
| U14518 | CENP-A | Cell division | 15.45 | 1.04 × 10−7 |
| M25753 | Cyclin B | Cell cycle | 14.22 | 3.3 × 10−7 |
| U05340 | p55CDC | Cell cycle | 14.22 | 4.58 × 10−5 |
| Z15005 | CENP-E | Cell division | 13.71 | 6.88 × 10−7 |
| AF053306 | MAD3L | Checkpoint | 12.93 | 7.33 × 10−7 |
| AA203476 | PTTG1 | Sister chromatid separation/cell division | 12.84 | 1.33 × 10−6 |
| X13293 | B-myb | Transcription factor | 12.83 | 2.3 × 10−6 |
| AF016582 | Chk1 | Checkpoint | 12.67 | .00089 |
| AJ000186 | MAD2 | Mitotic checkpoint | 12.3 | 4.18 × 10−5 |
| M86699 | TTK | Signaling | 11.87 | .000384 |
| AB017430 | KNSL4 | Cell division/cytoskeleton | 11.84 | 3.35 × 10−7 |
| L33594 | ATX | Metabolism | 11.76 | .00019 |
| M36067 | LIG1 | Metabolism | 11.73 | 2.97 × 10−8 |
| AI375913 | TOP2A | DNA replication | 11.56 | .000182 |
| AI391564 | PTP4A3 | Signaling | 11.24 | 2.53 × 10−5 |
| J00140 | DHFR | Metabolism | 11.14 | .00078 |
| M80397 | POLD1 | DNA replication | 10.81 | 9.58 × 10−8 |
| M15205 | TK1 | Metabolism | 10.78 | 5.23 × 10−9 |
| AL080146 | CCNB2 | Cell cycle | 10.72 | 3.19 × 10−5 |
| X59618 | RRM2 | Metabolism | 10.44 | 6.47 × 10−5 |
| Down-regulated | ||||
| M24594 | IFI56 | Growth suppression | 21.22 | 8.85 × 10−5 |
| U81523 | EBAF | Signaling | 6.90 | 1.71 × 10−6 |
| AJ130718 | SCLA9 | Solute carrier | 6.83 | .000117 |
| AF093265 | HOMER3 | Metabolism | 6.18 | 1.11 × 10−5 |
| AB016811 | ARL7 | Metabolism | 5.59 | .000684 |
| M14660 | IFI54 | Growth suppression | 5.47 | 8.47 × 10−5 |
| D64137 | KIP2 | Growth suppression | 4.43 | .000208 |
| AB020689 | KIAA0882 | Unknown | 4.17 | .00036 |
| U12255 | FCGRT | Immunity | 3.85 | .000549 |
| M14949 | R-RAS | Oncogene/signaling | 3.77 | .000958 |
| M32011 | CGD | Metabolism | 3.67 | 3.21 × 10−6 |
| M80899 | AHNAK | Cell interaction/desmosome | 3.53 | .000138 |
| M15059 | CD23 | Immunity | 3.17 | .000399 |
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| AF052124 | OPN/osteopontin | Bone/ECM | 65.61 | 8.72 × 10−5 |
| U73379 | E2C | Proteolysis | 42.32 | 9.35 × 10−5 |
| AJ223728 | CDC45 | Cell cycle | 37.12 | 1.33 × 10−8 |
| AF053305 | BUB1 | Cell cycle checkpoint | 30.43 | 1.95 × 10−5 |
| X62048 | WEE1 | Cell cycle | 29.07 | 7.55 × 10−7 |
| D14657 | KIAA0101 | Unknown | 26.32 | 2.73 × 10−5 |
| U77949 | CDC6 | Cell cycle | 25.81 | 1.05 × 10−5 |
| N58115 | EST | Unknown | 22.85 | .000162 |
| L25876 | CIP2 | Signaling | 19.5 | .000406 |
| AF067656 | Zw10 | Wint signaling | 18.17 | 1.38 × 10−6 |
| U74612 | FKHL16 | Transcription factor | 17.4 | 1.99 × 10−5 |
| AF025441 | OIP5 | Unknown | 17.34 | .000198 |
| U37426 | HKSP | Cell division | 17.28 | 1.22 × 10−5 |
| D80008 | KIAA0186 | Unknown | 15.97 | 1.56 × 10−5 |
| Y13115 | SAK | Signaling | 15.84 | .000811 |
| U63743 | KNSL6 | Cell division | 15.8 | .000119 |
| D14678 | MHCB | Cell division | 15.52 | .000154 |
| U14518 | CENP-A | Cell division | 15.45 | 1.04 × 10−7 |
| M25753 | Cyclin B | Cell cycle | 14.22 | 3.3 × 10−7 |
| U05340 | p55CDC | Cell cycle | 14.22 | 4.58 × 10−5 |
| Z15005 | CENP-E | Cell division | 13.71 | 6.88 × 10−7 |
| AF053306 | MAD3L | Checkpoint | 12.93 | 7.33 × 10−7 |
| AA203476 | PTTG1 | Sister chromatid separation/cell division | 12.84 | 1.33 × 10−6 |
| X13293 | B-myb | Transcription factor | 12.83 | 2.3 × 10−6 |
| AF016582 | Chk1 | Checkpoint | 12.67 | .00089 |
| AJ000186 | MAD2 | Mitotic checkpoint | 12.3 | 4.18 × 10−5 |
| M86699 | TTK | Signaling | 11.87 | .000384 |
| AB017430 | KNSL4 | Cell division/cytoskeleton | 11.84 | 3.35 × 10−7 |
| L33594 | ATX | Metabolism | 11.76 | .00019 |
| M36067 | LIG1 | Metabolism | 11.73 | 2.97 × 10−8 |
| AI375913 | TOP2A | DNA replication | 11.56 | .000182 |
| AI391564 | PTP4A3 | Signaling | 11.24 | 2.53 × 10−5 |
| J00140 | DHFR | Metabolism | 11.14 | .00078 |
| M80397 | POLD1 | DNA replication | 10.81 | 9.58 × 10−8 |
| M15205 | TK1 | Metabolism | 10.78 | 5.23 × 10−9 |
| AL080146 | CCNB2 | Cell cycle | 10.72 | 3.19 × 10−5 |
| X59618 | RRM2 | Metabolism | 10.44 | 6.47 × 10−5 |
| Down-regulated | ||||
| M24594 | IFI56 | Growth suppression | 21.22 | 8.85 × 10−5 |
| U81523 | EBAF | Signaling | 6.90 | 1.71 × 10−6 |
| AJ130718 | SCLA9 | Solute carrier | 6.83 | .000117 |
| AF093265 | HOMER3 | Metabolism | 6.18 | 1.11 × 10−5 |
| AB016811 | ARL7 | Metabolism | 5.59 | .000684 |
| M14660 | IFI54 | Growth suppression | 5.47 | 8.47 × 10−5 |
| D64137 | KIP2 | Growth suppression | 4.43 | .000208 |
| AB020689 | KIAA0882 | Unknown | 4.17 | .00036 |
| U12255 | FCGRT | Immunity | 3.85 | .000549 |
| M14949 | R-RAS | Oncogene/signaling | 3.77 | .000958 |
| M32011 | CGD | Metabolism | 3.67 | 3.21 × 10−6 |
| M80899 | AHNAK | Cell interaction/desmosome | 3.53 | .000138 |
| M15059 | CD23 | Immunity | 3.17 | .000399 |
Functional characterization of genes that distinguish each phenotypic class. Functional annotations of the genes that distinguished each treatment were obtained by comparing 4 public databases, GenBank, ONIM, Unigene, and AMIGO. (A) IL-6 versus nontreatment. (B) Stroma versus IL-6. (C) N-ras versus IL-6. (D) N-rasversus stroma.
Functional characterization of genes that distinguish each phenotypic class. Functional annotations of the genes that distinguished each treatment were obtained by comparing 4 public databases, GenBank, ONIM, Unigene, and AMIGO. (A) IL-6 versus nontreatment. (B) Stroma versus IL-6. (C) N-ras versus IL-6. (D) N-rasversus stroma.
Eighty-seven genes were differentially expressed in those treated with IL-6 versus those cocultured with stromal cell samples (Table 2). Among them, insulin growth factor-binding protein 8 (IGFBP8) was the most up-regulated gene in the stroma-stimulated samples when compared to IL-6-treated samples. Other significant genes encoding chemokines were human macrophage inflammatory protein 1 alpha (LD78/hMIP-1α) and Gro-beta (CXCL2). Figure 3B demonstrates that most genes distinguishing these groups are extracellular matrix (ECM) genes (22%) such as osteonectin (SPARC), collagen (COLA2, COL6A3) and versican (CSPG2). From this comparison we demonstrate that stromal cells contribute to alteration of gene expression in myeloma cells beyond growth-promoting effects.
Top 50 of 87 most differentially expressed genes in a comparison of stroma stimulation and IL-6 response
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X78947 | IGFBP8 | Growth factor | 123.88 | 5.13 × 10−5 |
| M77349 | TGFBIG | Adhesion/ECM | 121.08 | 5.25 × 10−5 |
| V01512 | cFos | Oncogene | 91.72 | 8.02 × 10−8 |
| X04430 | IFN-β2A | Growth factor | 76.19 | .000492 |
| J03040 | SPARC/osteonectin | ECM | 56.96 | 7.97 × 10−5 |
| M12125 | tropomyosin/TPM2 | Cytoskeleton | 56.47 | 3.91 × 10−5 |
| J03464 | CO1A2 | ECM | 45.27 | .000677 |
| X15998 | versican | ECM | 42.57 | 1.78 × 10−5 |
| K00650 | cFos | Oncogene | 41.98 | 1.86 × 10−5 |
| V00503 | CO1A2 | ECM | 38.02 | 4.84 × 10−5 |
| X68277 | CL100 | Signaling | 34.36 | 3.22 × 10−6 |
| U24389 | LOXL1 | Metabolism | 31.69 | 9.37 × 10−6 |
| M17733 | TMSB4X | Cytoskeleton | 30.18 | .000114 |
| X52022 | COL6A3 | ECM | 28.31 | 2.56 × 10−5 |
| X13839 | ACTSA | Cytoskeleton | 27.78 | 9.71 × 10−5 |
| S77154 | NR4A2 | Transcription factor | 22.62 | 1.44 × 10−6 |
| M35878 | IGFBP3 | Angiogenesis | 20.92 | .000238 |
| L12350 | THBS2 | ECM | 16.81 | .000253 |
| M19267 | TPM1 | Cytoskeleton | 16.64 | .000244 |
| U03057 | HSN | Cytoskeleton | 15.98 | .000723 |
| U08021 | NNMT | Metabolism | 15.79 | .000352 |
| M55153 | TGM2 | Metabolism | 14.42 | .000228 |
| S81914 | IER3 | Transcription factor | 14.08 | .000231 |
| J04130 | Act-2/(MIP-1β) | Chemokine | 13.67 | 6.8 × 10−7 |
| L19182 | IGFBP4 | IGF regulation/growth | 13.59 | .000327 |
| M36820 | GRO-β | Chemokine | 11.09 | .000229 |
| AB029000 | KIAA1077 | Unknown | 10.96 | 9.56 × 10−5 |
| S78569 | LAMA 4 | ECM | 10.58 | .000138 |
| M14083 | PAI 1 | ECM | 10.52 | .000262 |
| U97669 | NOTCH 3 | Signaling | 10.09 | 5.05 × 10−6 |
| M27830 | 28S ribosomal RNA | Translation | 9.9 | .000126 |
| L49169 | RGS2 | Cell cycle | 9.74 | .000186 |
| L13463 | GOS3 | Cell cycle | 9.59 | .000949 |
| AB018293 | KIAA0750 | Unknown | 9.16 | .000933 |
| M58459 | RPS4Y | Translation | 9.16 | .000423 |
| S59049 | RGS1 | B-cell activation | 8.99 | .000579 |
| D32039 | CSPG2 | Proteoglycan/ECM | 8.89 | .000748 |
| D90144 | LD78/Mip-1α | Osteoclastogenic factor/chemokine | 8.86 | 1.87 × 10−5 |
| AF052119 | EST | Unknown | 8.13 | .000464 |
| AL096713 | EST | Unknown | 8.10 | .000728 |
| AI246894 | EST | Unknown | 7.87 | .000453 |
| AF056085 | GPR 51 | GABA receptor | 7.71 | .000891 |
| X15880 | COL6A1 | ECM | 7.50 | .000988 |
| M69043 | NFKBIA | Signaling | 7.11 | 4.57 × 10−6 |
| L16895 | LOX | Metabolism | 6.87 | .000644 |
| Down-regulated | ||||
| AJ001612 | PSPHL | Signaling | 3.50 | 4.69 × 10−5 |
| S68805 | GATM | Metabolism | 2.66 | 1.24 × 10−5 |
| M55067 | CGD | Metabolism | 2.32 | .00083 |
| U77949 | CDC6 | Cell cycle | 2.19 | 6.47 × 10−5 |
| AB014597 | KIAA0697 | Unknown | 2.12 | .000558 |
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X78947 | IGFBP8 | Growth factor | 123.88 | 5.13 × 10−5 |
| M77349 | TGFBIG | Adhesion/ECM | 121.08 | 5.25 × 10−5 |
| V01512 | cFos | Oncogene | 91.72 | 8.02 × 10−8 |
| X04430 | IFN-β2A | Growth factor | 76.19 | .000492 |
| J03040 | SPARC/osteonectin | ECM | 56.96 | 7.97 × 10−5 |
| M12125 | tropomyosin/TPM2 | Cytoskeleton | 56.47 | 3.91 × 10−5 |
| J03464 | CO1A2 | ECM | 45.27 | .000677 |
| X15998 | versican | ECM | 42.57 | 1.78 × 10−5 |
| K00650 | cFos | Oncogene | 41.98 | 1.86 × 10−5 |
| V00503 | CO1A2 | ECM | 38.02 | 4.84 × 10−5 |
| X68277 | CL100 | Signaling | 34.36 | 3.22 × 10−6 |
| U24389 | LOXL1 | Metabolism | 31.69 | 9.37 × 10−6 |
| M17733 | TMSB4X | Cytoskeleton | 30.18 | .000114 |
| X52022 | COL6A3 | ECM | 28.31 | 2.56 × 10−5 |
| X13839 | ACTSA | Cytoskeleton | 27.78 | 9.71 × 10−5 |
| S77154 | NR4A2 | Transcription factor | 22.62 | 1.44 × 10−6 |
| M35878 | IGFBP3 | Angiogenesis | 20.92 | .000238 |
| L12350 | THBS2 | ECM | 16.81 | .000253 |
| M19267 | TPM1 | Cytoskeleton | 16.64 | .000244 |
| U03057 | HSN | Cytoskeleton | 15.98 | .000723 |
| U08021 | NNMT | Metabolism | 15.79 | .000352 |
| M55153 | TGM2 | Metabolism | 14.42 | .000228 |
| S81914 | IER3 | Transcription factor | 14.08 | .000231 |
| J04130 | Act-2/(MIP-1β) | Chemokine | 13.67 | 6.8 × 10−7 |
| L19182 | IGFBP4 | IGF regulation/growth | 13.59 | .000327 |
| M36820 | GRO-β | Chemokine | 11.09 | .000229 |
| AB029000 | KIAA1077 | Unknown | 10.96 | 9.56 × 10−5 |
| S78569 | LAMA 4 | ECM | 10.58 | .000138 |
| M14083 | PAI 1 | ECM | 10.52 | .000262 |
| U97669 | NOTCH 3 | Signaling | 10.09 | 5.05 × 10−6 |
| M27830 | 28S ribosomal RNA | Translation | 9.9 | .000126 |
| L49169 | RGS2 | Cell cycle | 9.74 | .000186 |
| L13463 | GOS3 | Cell cycle | 9.59 | .000949 |
| AB018293 | KIAA0750 | Unknown | 9.16 | .000933 |
| M58459 | RPS4Y | Translation | 9.16 | .000423 |
| S59049 | RGS1 | B-cell activation | 8.99 | .000579 |
| D32039 | CSPG2 | Proteoglycan/ECM | 8.89 | .000748 |
| D90144 | LD78/Mip-1α | Osteoclastogenic factor/chemokine | 8.86 | 1.87 × 10−5 |
| AF052119 | EST | Unknown | 8.13 | .000464 |
| AL096713 | EST | Unknown | 8.10 | .000728 |
| AI246894 | EST | Unknown | 7.87 | .000453 |
| AF056085 | GPR 51 | GABA receptor | 7.71 | .000891 |
| X15880 | COL6A1 | ECM | 7.50 | .000988 |
| M69043 | NFKBIA | Signaling | 7.11 | 4.57 × 10−6 |
| L16895 | LOX | Metabolism | 6.87 | .000644 |
| Down-regulated | ||||
| AJ001612 | PSPHL | Signaling | 3.50 | 4.69 × 10−5 |
| S68805 | GATM | Metabolism | 2.66 | 1.24 × 10−5 |
| M55067 | CGD | Metabolism | 2.32 | .00083 |
| U77949 | CDC6 | Cell cycle | 2.19 | 6.47 × 10−5 |
| AB014597 | KIAA0697 | Unknown | 2.12 | .000558 |
IGF indicates insulin-like growth factor; GABA, γ-aminobutyric acid.
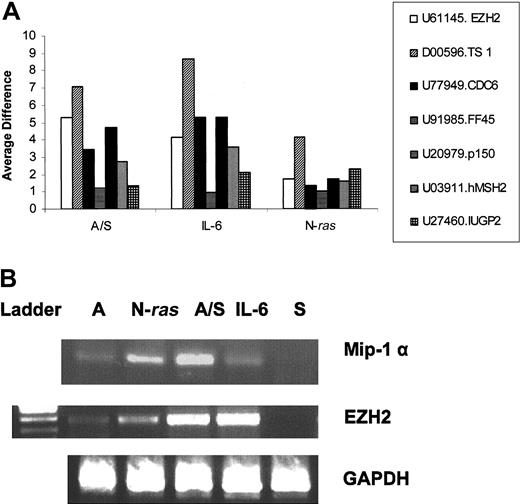
A total of 130 genes distinguished the IL-6-treated and N-ras samples, with N-ras (HSNRASR) being the most up-regulated gene as expected in the N-ras samples and an unknown gene (KIAA0963) the most down-regulated in the N-ras samples compared with IL-6-treated samples (Table 3). Most genes in this group are of metabolic function (26%) followed by genes with immunologic activity (12%) such as the costimulatory molecule CD86 (Figure 3C). Interestingly, hmip-1α is also differentially expressed between these classes. Reverse transcriptase-polymerase chain reaction (RT-PCR) confirmed that hmip-1α is overexpressed on interaction with BMSCs, as well as, constitutively active in N-ras-transfected cells, compared with low basal levels of expression in control, untreated cells (Figure 5B).
Top 50 of 130 most differentially expressed genes in a comparison of N-ras-activating mutation and IL-6 response
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X02751 | HSNRASR | Oncogene | 52.41 | 3.41 × 10−7 |
| AI142565 | RNASE6 | Transcription | 34.33 | 4.4 × 10−7 |
| Y14737 | IGHM | Immunity | 27.78 | .000209 |
| J04130 | Act-2/(MIP-1β) | Chemokine | 16.43 | 1.28 × 10−5 |
| M37766 | MEM-102 | B-cell activation | 12.08 | 2.24 × 10−5 |
| AB013382 | DUSP6 | Sinaling | 11.37 | .000243 |
| U20350 | V28 | G protein-coupled receptor | 11.03 | .000866 |
| U33147 | MGB1 | ECM | 9.21 | .000391 |
| D90144 | LD78 | Chemokine/osteoclastogenic factor | 6.79 | 2.16 × 10−5 |
| AL040446 | ORP1 | Drug metabalism | 6.38 | .000642 |
| D13265 | MSR1 | Macrophage scavenger receptor | 6.31 | 4.15 × 10−5 |
| AI660656 | IGJ | Immunity | 6.17 | 7.47 × 10−5 |
| AB018278 | SV2B | Synaptic vesicle transporter | 5.77 | .000259 |
| X16354 | BGPa/CEACAM1 | Adhesion/angiogenesis | 5.95 | .000253 |
| AL023653 | XPNPEP2 | ECM | 5.57 | .000697 |
| M15059 | IgE receptor | Immunity | 5.5 | 8.44 × 10−6 |
| AB020499 | MD-1 | B-cell activation | 5.21 | .000214 |
| U04343 | CD86 | Costimulatory signal/immunity | 5.2 | 2.16 × 10−5 |
| M24594 | IFIT1 | Unknown | 5.15 | .000881 |
| AI535946 | LGALS1 | Cell cycle inhibition | 4.99 | 1.92 × 10−5 |
| AF093265 | homer-3 | Calcium release | 4.34 | .000621 |
| M32011 | NSF2 | Metabolism | 4.23 | .000254 |
| X04011 | CGD | Metabolism | 3.96 | 2 × 10−4 |
| NP001060 | Tubulin beta | Cytoskeleton | 3.87 | .000952 |
| M21121 | RANTES | Chemokine/immunity | 3.8 | .000289 |
| Down-regulated | ||||
| AB023180 | KIAA0963 | Unknown | 9.08 | .000675 |
| AB017430 | KNSL4 | Mitosis | 8.31 | .000264 |
| L41668 | Gale | Metabolism | 7.75 | 4.33 × 10−5 |
| AI391564 | PTP4A3 | Signaling | 7.38 | .000179 |
| M80397 | POLD1 | DNA replication | 7.36 | .000337 |
| AF091754 | EXO1 | DNA repair | 5.24 | 6.93 × 10−5 |
| M25753 | cyclin B1 | Cell cycle | 4.85 | .000496 |
| U73379 | E2C | Ubiquination | 4.63 | 2.78 × 10−5 |
| U77949 | CDC6 | Cell cycle control | 4.55 | 1.89 × 10−5 |
| X62048 | WEE1 | Cell cycle control | 4.31 | .000138 |
| L25876 | CDKN3 | Cell cycle control | 4.27 | 2.54 × 10−5 |
| U77845 | hTRIP | Signaling | 4.15 | .000402 |
| U05340 | CDC20 | Cell cycle control | 4.13 | .00014 |
| M34065 | CDC25C | Cell cycle | 4.09 | .0000294 |
| X13293 | B-myb | Oncogene | 4.06 | 3.81 × 10−6 |
| AF025441 | OIP5 | Adhesion/invasion | 4.02 | .000414 |
| U14518 | CENPA | Cell division | 4.00 | .000269 |
| U74612 | FKHL16 | Transcription factor | 3.91 | 6.59 × 10−5 |
| U63743 | KNSL6 | Mitosis | 3.89 | .000552 |
| AL080146 | cyclin B2 | Cell cycle | 3.88 | .000243 |
| D38073 | MCM3 | DNA replication | 3.77 | .000107 |
| AF053306 | BUB3 | Cell cycle control | 3.77 | 3.52 × 10−6 |
| X65550 | MK167 | Cell cycle | 3.76 | .000159 |
| X05360 | CDC2 | Cell cycle | 3.75 | .00099 |
| N58115 | AD024 | Unknown | 3.75 | .000115 |
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X02751 | HSNRASR | Oncogene | 52.41 | 3.41 × 10−7 |
| AI142565 | RNASE6 | Transcription | 34.33 | 4.4 × 10−7 |
| Y14737 | IGHM | Immunity | 27.78 | .000209 |
| J04130 | Act-2/(MIP-1β) | Chemokine | 16.43 | 1.28 × 10−5 |
| M37766 | MEM-102 | B-cell activation | 12.08 | 2.24 × 10−5 |
| AB013382 | DUSP6 | Sinaling | 11.37 | .000243 |
| U20350 | V28 | G protein-coupled receptor | 11.03 | .000866 |
| U33147 | MGB1 | ECM | 9.21 | .000391 |
| D90144 | LD78 | Chemokine/osteoclastogenic factor | 6.79 | 2.16 × 10−5 |
| AL040446 | ORP1 | Drug metabalism | 6.38 | .000642 |
| D13265 | MSR1 | Macrophage scavenger receptor | 6.31 | 4.15 × 10−5 |
| AI660656 | IGJ | Immunity | 6.17 | 7.47 × 10−5 |
| AB018278 | SV2B | Synaptic vesicle transporter | 5.77 | .000259 |
| X16354 | BGPa/CEACAM1 | Adhesion/angiogenesis | 5.95 | .000253 |
| AL023653 | XPNPEP2 | ECM | 5.57 | .000697 |
| M15059 | IgE receptor | Immunity | 5.5 | 8.44 × 10−6 |
| AB020499 | MD-1 | B-cell activation | 5.21 | .000214 |
| U04343 | CD86 | Costimulatory signal/immunity | 5.2 | 2.16 × 10−5 |
| M24594 | IFIT1 | Unknown | 5.15 | .000881 |
| AI535946 | LGALS1 | Cell cycle inhibition | 4.99 | 1.92 × 10−5 |
| AF093265 | homer-3 | Calcium release | 4.34 | .000621 |
| M32011 | NSF2 | Metabolism | 4.23 | .000254 |
| X04011 | CGD | Metabolism | 3.96 | 2 × 10−4 |
| NP001060 | Tubulin beta | Cytoskeleton | 3.87 | .000952 |
| M21121 | RANTES | Chemokine/immunity | 3.8 | .000289 |
| Down-regulated | ||||
| AB023180 | KIAA0963 | Unknown | 9.08 | .000675 |
| AB017430 | KNSL4 | Mitosis | 8.31 | .000264 |
| L41668 | Gale | Metabolism | 7.75 | 4.33 × 10−5 |
| AI391564 | PTP4A3 | Signaling | 7.38 | .000179 |
| M80397 | POLD1 | DNA replication | 7.36 | .000337 |
| AF091754 | EXO1 | DNA repair | 5.24 | 6.93 × 10−5 |
| M25753 | cyclin B1 | Cell cycle | 4.85 | .000496 |
| U73379 | E2C | Ubiquination | 4.63 | 2.78 × 10−5 |
| U77949 | CDC6 | Cell cycle control | 4.55 | 1.89 × 10−5 |
| X62048 | WEE1 | Cell cycle control | 4.31 | .000138 |
| L25876 | CDKN3 | Cell cycle control | 4.27 | 2.54 × 10−5 |
| U77845 | hTRIP | Signaling | 4.15 | .000402 |
| U05340 | CDC20 | Cell cycle control | 4.13 | .00014 |
| M34065 | CDC25C | Cell cycle | 4.09 | .0000294 |
| X13293 | B-myb | Oncogene | 4.06 | 3.81 × 10−6 |
| AF025441 | OIP5 | Adhesion/invasion | 4.02 | .000414 |
| U14518 | CENPA | Cell division | 4.00 | .000269 |
| U74612 | FKHL16 | Transcription factor | 3.91 | 6.59 × 10−5 |
| U63743 | KNSL6 | Mitosis | 3.89 | .000552 |
| AL080146 | cyclin B2 | Cell cycle | 3.88 | .000243 |
| D38073 | MCM3 | DNA replication | 3.77 | .000107 |
| AF053306 | BUB3 | Cell cycle control | 3.77 | 3.52 × 10−6 |
| X65550 | MK167 | Cell cycle | 3.76 | .000159 |
| X05360 | CDC2 | Cell cycle | 3.75 | .00099 |
| N58115 | AD024 | Unknown | 3.75 | .000115 |
Expression pattern of genes that distinguish MM1 from MM4 under different treatments. (A) Twenty-four of 30 genes that distinguished MM1 from MM4 classes reported by Zhan et al10 were found to be present in at least one response group. Seven genes are depicted as a ratio of their average difference over the average difference of NIL controls. A/S indicates stroma-stimulated samples; IL-6, IL-6-treated samples, N-ras, mutant N-ras transfectants. (B) RT-PCR confirms the microarray data. Total RNA was isolated using the RNA Easy kit (Qiagen); cDNA was made and amplified by PCR using specific primers for mip-1α, EZH2, and GAPDH as loading control. A indicates ANBL6 IL-6-starved; N-Ras, ANBL-6 stably transfected with activated N-ras; A/S, ANBL-6 stimulated with stroma; IL-6, ANBL-6 stimulated with 10 ng recombinant IL-6; S, stroma.
Expression pattern of genes that distinguish MM1 from MM4 under different treatments. (A) Twenty-four of 30 genes that distinguished MM1 from MM4 classes reported by Zhan et al10 were found to be present in at least one response group. Seven genes are depicted as a ratio of their average difference over the average difference of NIL controls. A/S indicates stroma-stimulated samples; IL-6, IL-6-treated samples, N-ras, mutant N-ras transfectants. (B) RT-PCR confirms the microarray data. Total RNA was isolated using the RNA Easy kit (Qiagen); cDNA was made and amplified by PCR using specific primers for mip-1α, EZH2, and GAPDH as loading control. A indicates ANBL6 IL-6-starved; N-Ras, ANBL-6 stably transfected with activated N-ras; A/S, ANBL-6 stimulated with stroma; IL-6, ANBL-6 stimulated with 10 ng recombinant IL-6; S, stroma.
A total of 157 genes distinguished stroma-stimulated and N-ras samples (Table 4). Once again N-ras (HSNRASR) is the most up-regulated gene followed by other ras-related signaling molecules such as the guanine exchange factor RAP2. In fact, 20% of genetic changes comprise signaling molecules (Figure 3D). Thus, although signaling at the protein level occurs, transcriptional regulation of signaling factor genes occurs as well.
Top 50 of 157 most differentially expressed genes in a comparison of N-ras-activating mutation and stroma response
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X02751 | HSNRASR | Oncogene | 304.88 | .000274 |
| AI142565 | RNASE 6 | Transcription | 78.79 | 2.26 × 10−5 |
| U20350 | V28 | G protein-coupled receptor | 29.09 | .000275 |
| AI660656 | EST | Unknown | 23.69 | 5.34 × 10−6 |
| U04343 | CD86 | Costimulatory signal | 12.62 | .000805 |
| AF070641 | IGJ | Immunity | 12.42 | .000308 |
| AF043722 | Rap2 | GEF/signaling | 12.03 | 9.71 × 10−5 |
| D21205 | ERFP | Ubiquitin ligase/cell cycle | 11.65 | 6 × 10−5 |
| S71326 | BGPc/CEACAM1 | Adhesion | 11.56 | 3.86 × 10−5 |
| X81438 | amphiphysin | Synaptic vesicle protein | 11.38 | .000535 |
| X04011 | CGD | Metabolism | 10.50 | .000728 |
| Y14737 | IGHM | Immunity | 9.68 | .000561 |
| AB013382 | DUSP6 | Signaling | 9.52 | 6.95 × 10−5 |
| U48807 | DUSP4 | Signaling | 9.28 | 9.37 × 10−5 |
| AL512715 | GEF2 | Signaling | 8.98 | 3.14 × 10−6 |
| Z54367 | PLECTIN | Cytoskeleton | 8.84 | .000168 |
| AB020689 | KIAA0882 | Unknown | 8.45 | .000161 |
| M55067 | NCF1 | Metabolism | 7.69 | 9.12 × 10−6 |
| J04430 | ACP5 | Metabolism | 7.42 | .000149 |
| AF055004 | EST | Unknown | 7.00 | .000496 |
| AB018278 | SVB2 | Synaptic vesicle transporter | 6.96 | .000573 |
| AB002351 | DMN | Cytoskeleton | 6.85 | .000287 |
| M80335 | p47-phox | Metabolism | 6.78 | .000483 |
| M62397 | MCC | Tumor supressor | 6.62 | .00057 |
| X99802 | ZYG | Unknown | 6.34 | 3.46 × 10−5 |
| Down-regulated | ||||
| V01512 | cFos | Oncogene | 304.43 | 3.38 × 10−6 |
| M77349 | TGFBI | Adhesion | 140.46 | .000748 |
| M12125 | TPM2 | Cytoskeleton | 77.33 | .000394 |
| X78947 | IGFBP8 | Signaling | 67.11 | 3.58 × 10−5 |
| X04430 | IL-6 | Cytokine | 64.43 | .000822 |
| J03464 | COL1A2 | ECM | 56.21 | .000724 |
| K00650 | c-Fos | Oncogene | 54.48 | 1.84 × 10−5 |
| D13666 | OSF-2 | ECM | 52.91 | 3.28 × 10−5 |
| J03040 | Sparc | ECM | 49.84 | 7.16 × 10−5 |
| X52022 | COL6A3 | ECM | 45.36 | .000489 |
| S77154 | NR4A2 | Signaling | 38.75 | 3.19 × 10−7 |
| X13839 | ACTSA | Cytoskeleton | 33.11 | .00033 |
| U03057 | FSCN1 | Cytoskeleton | 30.31 | .000256 |
| M19267 | TPM1 | Cytoskeleton | 29.27 | 3.27 × 10−5 |
| X52947 | GJA1 | Gap junction | 29.03 | .000323 |
| X68277 | DUSP1 | Signaling | 27.11 | 4.62 × 10−5 |
| M17733 | TMSB4 | Cytoskeleton | 23.31 | .000299 |
| L19182 | IGFBP7 | Signaling | 21.15 | .00024 |
| M35878 | IGFBP3 | Signaling | 19.38 | .000308 |
| X75918 | NR4A2 | Signaling | 19.09 | .000212 |
| AB029000 | SULF1 | Metabolism | 18.51 | 9.69 × 10−5 |
| AI557295 | EST | Unknown | 15.75 | .000395 |
| M58459 | RPS4X | Translation | 12.56 | .000231 |
| M33680 | CD81 | Membrane protein | 11.87 | .000117 |
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X02751 | HSNRASR | Oncogene | 304.88 | .000274 |
| AI142565 | RNASE 6 | Transcription | 78.79 | 2.26 × 10−5 |
| U20350 | V28 | G protein-coupled receptor | 29.09 | .000275 |
| AI660656 | EST | Unknown | 23.69 | 5.34 × 10−6 |
| U04343 | CD86 | Costimulatory signal | 12.62 | .000805 |
| AF070641 | IGJ | Immunity | 12.42 | .000308 |
| AF043722 | Rap2 | GEF/signaling | 12.03 | 9.71 × 10−5 |
| D21205 | ERFP | Ubiquitin ligase/cell cycle | 11.65 | 6 × 10−5 |
| S71326 | BGPc/CEACAM1 | Adhesion | 11.56 | 3.86 × 10−5 |
| X81438 | amphiphysin | Synaptic vesicle protein | 11.38 | .000535 |
| X04011 | CGD | Metabolism | 10.50 | .000728 |
| Y14737 | IGHM | Immunity | 9.68 | .000561 |
| AB013382 | DUSP6 | Signaling | 9.52 | 6.95 × 10−5 |
| U48807 | DUSP4 | Signaling | 9.28 | 9.37 × 10−5 |
| AL512715 | GEF2 | Signaling | 8.98 | 3.14 × 10−6 |
| Z54367 | PLECTIN | Cytoskeleton | 8.84 | .000168 |
| AB020689 | KIAA0882 | Unknown | 8.45 | .000161 |
| M55067 | NCF1 | Metabolism | 7.69 | 9.12 × 10−6 |
| J04430 | ACP5 | Metabolism | 7.42 | .000149 |
| AF055004 | EST | Unknown | 7.00 | .000496 |
| AB018278 | SVB2 | Synaptic vesicle transporter | 6.96 | .000573 |
| AB002351 | DMN | Cytoskeleton | 6.85 | .000287 |
| M80335 | p47-phox | Metabolism | 6.78 | .000483 |
| M62397 | MCC | Tumor supressor | 6.62 | .00057 |
| X99802 | ZYG | Unknown | 6.34 | 3.46 × 10−5 |
| Down-regulated | ||||
| V01512 | cFos | Oncogene | 304.43 | 3.38 × 10−6 |
| M77349 | TGFBI | Adhesion | 140.46 | .000748 |
| M12125 | TPM2 | Cytoskeleton | 77.33 | .000394 |
| X78947 | IGFBP8 | Signaling | 67.11 | 3.58 × 10−5 |
| X04430 | IL-6 | Cytokine | 64.43 | .000822 |
| J03464 | COL1A2 | ECM | 56.21 | .000724 |
| K00650 | c-Fos | Oncogene | 54.48 | 1.84 × 10−5 |
| D13666 | OSF-2 | ECM | 52.91 | 3.28 × 10−5 |
| J03040 | Sparc | ECM | 49.84 | 7.16 × 10−5 |
| X52022 | COL6A3 | ECM | 45.36 | .000489 |
| S77154 | NR4A2 | Signaling | 38.75 | 3.19 × 10−7 |
| X13839 | ACTSA | Cytoskeleton | 33.11 | .00033 |
| U03057 | FSCN1 | Cytoskeleton | 30.31 | .000256 |
| M19267 | TPM1 | Cytoskeleton | 29.27 | 3.27 × 10−5 |
| X52947 | GJA1 | Gap junction | 29.03 | .000323 |
| X68277 | DUSP1 | Signaling | 27.11 | 4.62 × 10−5 |
| M17733 | TMSB4 | Cytoskeleton | 23.31 | .000299 |
| L19182 | IGFBP7 | Signaling | 21.15 | .00024 |
| M35878 | IGFBP3 | Signaling | 19.38 | .000308 |
| X75918 | NR4A2 | Signaling | 19.09 | .000212 |
| AB029000 | SULF1 | Metabolism | 18.51 | 9.69 × 10−5 |
| AI557295 | EST | Unknown | 15.75 | .000395 |
| M58459 | RPS4X | Translation | 12.56 | .000231 |
| M33680 | CD81 | Membrane protein | 11.87 | .000117 |
GEF indicates guanine exchange factor.
Similar gene expression profiles displayed by different activating signals
We also determined which genes were commonly expressed by all 3 activating signals but different from the nontreated control samples. We found 347 genes that differed from each other by less than 10% among all treatment groups, but were differentially expressed from the nontreated samples at a significance level of P < .001. Of these genes, 149 genes were up-regulated in a similar fashion in IL-6-treated and N-ras- and stroma-stimulated samples when compared with nontreated controls. In contrast, 198 genes were down-regulated in these groups compared with nontreated samples. The results of this analysis were visualized and confirmed by Cluster and TreeView (Eisen Laboratory) and are depicted in Figure 4A. The most invariant gene among all 3 treatment groups was F-box and leucine-rich repeat protein 11 (FBXL11) with a variance score of 0.006073, and significantly down-regulated from nontreated controls with a t test P of 1.60 × 10-7 and a Kruskal-Wallis P of 1.54 × 10-5. The most invariant gene among all response groups was an unknown protein (KIAA0062) with a variance of 0.006522, and significantly up-regulated from nontreated controls, with a t test P of 3.52 × 10-8 and Kruskal-Wallis P value of 4.10 × 10-5. Among genes up-regulated commonly in all treatment groups when compared to nontreated samples are mitogen activated kinase 6 (MAPK-6), signal transducer and activator of transcription 3 (STAT 3), and integrin B1 (CD29). Some of the genes down-regulated by all 3 signals when compared with control samples comprised interferon regulatory factor 4 (IRF4), B-cell chronic lymphocytic leukemia (CLL)/lymphoma 2 (BCL2), and β2-microglobulin (B2M). Functional characterization of these common genes among response groups showed that a high percentage of them (19%) are metabolic genes. Again, we note that growth promotion alters not only cytoplasmic signaling events, but levels of expression of a variety of signaling encoding genes.
Similar gene expression profiles displayed by different activating signals. Nonvariable common genes among phenotypic classes were determined using a variance test (10% level) in combination with the parametric t test and nonparametric Kruskal-Wallis test (P < .001). Data were visualized using Cluster/Tree View. Stroma 1-3, normal stroma-treated samples; stroma 4-5, MM stroma-treated samples. (A) Genes shared by all 3 treatments when compared to NIL controls; 347 genes were found to be commonly expressed by IL-6, stroma, and N-ras samples and not by NIL controls (left). Functional characterization of these common genes indicates that metabolic genes are the most represented functional group (right). (B) Genes expressed similarly by IL-6 and N-ras samples but different from stroma and NIL samples; 114 genes were shared by IL-6 and N-ras samples but different from stroma and NIL samples (left). Most of these genes encode translation proteins (right). (C) Genes shared by IL-6- and stroma-treated samples but different from NIL and N-ras samples; 169 genes were shared by IL-6- and stroma-treated samples but not by NIL and N-ras samples (left). Most of these were cell cycle genes (right).
Similar gene expression profiles displayed by different activating signals. Nonvariable common genes among phenotypic classes were determined using a variance test (10% level) in combination with the parametric t test and nonparametric Kruskal-Wallis test (P < .001). Data were visualized using Cluster/Tree View. Stroma 1-3, normal stroma-treated samples; stroma 4-5, MM stroma-treated samples. (A) Genes shared by all 3 treatments when compared to NIL controls; 347 genes were found to be commonly expressed by IL-6, stroma, and N-ras samples and not by NIL controls (left). Functional characterization of these common genes indicates that metabolic genes are the most represented functional group (right). (B) Genes expressed similarly by IL-6 and N-ras samples but different from stroma and NIL samples; 114 genes were shared by IL-6 and N-ras samples but different from stroma and NIL samples (left). Most of these genes encode translation proteins (right). (C) Genes shared by IL-6- and stroma-treated samples but different from NIL and N-ras samples; 169 genes were shared by IL-6- and stroma-treated samples but not by NIL and N-ras samples (left). Most of these were cell cycle genes (right).
We next examined nonvariant genes between IL-6-treated and N-ras-transfected samples that also significantly differed from all other treatment groups (stroma-stimulated and controls) at the level of P < .001. A total of 114 genes fit these criteria; 48 genes were commonly up-regulated and 66 genes down-regulated in the IL-6-treated and N-ras-transfected samples when compared with the other response groups (Figure 4B). The most invariant gene between the IL-6-treated and N-ras-transfected samples with a variance of 0.003183 was mitogen-activated protein kinase kinase 1 (MAP2K1), being significantly up-regulated in these response groups when compared with stroma and nontreated samples. The most invariant and commonly down-regulated gene in the IL-6-treated and N-ras-transfected samples was poly(rC)-binding protein 1 (PCBP1) with a variance of 0.003255. Most genes shared by IL-6 stimulation and N-ras activating mutation were involved in translation (33%; Figure 4B).
IL-6- and stroma-stimulated samples shared 169 genes that were also differentially expressed (P < .001) with respect to N-ras and control samples. A total of 107 genes were commonly up-regulated and 62 genes were down-regulated by IL-6 and stroma stimulation when compared with the other groups. The most invariant gene among IL-6- and stroma-stimulated samples was small nuclear ribonucleoprotein polypeptides B and B1 (SNRPB), which was up-regulated with respect to the rest of the samples and had a variance of 0.006137 (Figure 4C). The most invariant and commonly down-regulated gene was dimethylarginine dimethylaminohydrolase 2 (DDAH2) with a variance of 0.00705. Seventeen percent of the genes shared by IL-6- and stroma-treated samples are involved in cell-cycle control (Figure 4C).
Finally, N-ras- and stroma-treated samples were analyzed for genetic similarities that also fit the criterion of being statistically different from IL-6-treated and control samples. Only 3 genes were found that satisfied these criteria, suggesting that at this level of significance P < .001 and variance of 10%, these 2 groups are the most dissimilar classes (data not shown).
Genes that distinguish MM1 and MM4 subgroups have distinct expression patterns under different conditions
We next compared our data to the patient sample data that distinguished the MM1 and MM4 subgroups reported by Zhan and colleagues.10 Twenty-four of 30 most differentially expressed genes between these subgroups were found to be present in at least one response group. Seven genes from this group were randomly chosen and their expression pattern, normalized to NIL controls, under different treatments is depicted in Figure 5A. Interestingly, some of these genes are differentially expressed under different treatments. One such gene, enhancer of Zeste homologue 2 (EZH2), is expressed at very low levels in control IL-6-starved samples. In contrast, it becomes activated after treatment with stromal cells or IL-6, whereas it is constitutively active in N-ras-transfected cells (Figure 5A-B). Conversely, the DNA fragmentation gene DFFA/FF45, although present in all samples, did not vary significantly among treatments, suggesting common targets of induction.
Mutant N-ras induces expression of known Ras/MAPK targets
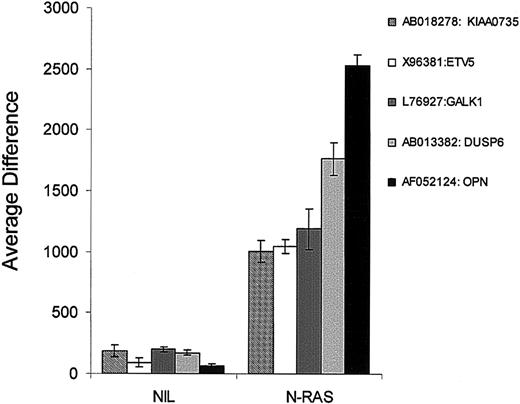
We next examined which genes were induced de novo in the N-ras transfectants when compared with empty vector controls to determine if we could identify a unique mutant ras signature expression pattern. The criterion chosen for gene selection was an average difference value of less than 200 in the controls and consistently more than 1000 in the N-ras samples. Notably, only 5 genes met these criteria (Figure 6). The ets transcription factor (ETV5/ERM) was virtually absent in the NIL group and was induced significantly on mutant N-ras transfection (P = 6.7 × 10-7). The dual-specificity phosphatase 6 (DUSP6/MKP-3) expression was also induced in the N-ras samples while absent in NIL controls (P = 6.5 × 10-8). An unknown protein (KIAA0735) was also induced from absent levels in NIL controls, to high levels in N-ras transfectants (P = 2.2 × 10-5). Interestingly, none of these 3 genes were induced by any other treatments and are most likely part of the N-ras unique signature observed in group 4 of Figure 2. Finally, galactokinase 1 (GALK1) and osteopontin (OSP) were also newly expressed in N-ras transfectants when compared with controls (P = 1.11 × 10-5 and P = 1.25 × 10-5, respectively; Figure 6). In contrast to ETV5, DUSP6 and KIAA0735, both GALK and OSP were also induced significantly by interaction with stromal cells and IL-6 stimulation (data not shown).
Mutant N-Ras induces expression of known Ras/MAPK targets. Five genes were found to be virtually absent in NIL controls (average difference of < 200) and significantly induced in the mutant N-ras-transfected samples (average difference of > 1000). Three of these genes were solely induced by N-ras activating mutation. Among these ETV5 has been shown to be a target of Ras/MAPK signaling.
Mutant N-Ras induces expression of known Ras/MAPK targets. Five genes were found to be virtually absent in NIL controls (average difference of < 200) and significantly induced in the mutant N-ras-transfected samples (average difference of > 1000). Three of these genes were solely induced by N-ras activating mutation. Among these ETV5 has been shown to be a target of Ras/MAPK signaling.
Discussion
We have demonstrated in this report that, although distinct activating signals share some genetic targets and pathways, they also affect MM cells distinctively through up-regulation and deactivation of sets of genes that have different functional effects. We have shown this to be the case for the IL-6-dependent MM cell line ANBL-6, treated with IL-6, cocultured with BMSCs and carrying an N-ras activating mutation. For example, it may have been expected that because IL-6 activates the Ras pathway, most genes differentially expressed in N-ras-transfected cells would constitute a subset of the genes activated by IL-6 treatment. However, this was not the case. Although IL-6 treatment and mutant N-ras samples clustered closer together than stroma-stimulated samples or untreated controls, N-ras-transfected samples displayed a distinct group of genes that characterized this phenotypic class. This can be explained by our previous report showing that although IL-6 activates Ras signaling, this activation is transient and incomplete, whereas constitutively active mutant Ras has a significant and constant effect on downstream genes, which confers a distinct genetic profile in cells that carry this mutant oncogene.13 Indeed, relevant functional consequences have been noted in which IL-6 enhances apoptotic response to doxorubicin, whereas constitutively active Ras mutations confer doxorubicin resistance.
It has been proposed that IL-6, which is produced by the BMSCs, has a major role in MM growth and protection from chemotherapeutic agent-induced apoptosis.9 Thus, it was expected that most differentially expressed genes in these 2 groups would overlap. However, hierarchical clustering as well as class comparison analysis demonstrated that although some genes are shared by these 2 treatment groups, there is a distinct group of genes activated in MM cells only by BMS interaction. This is in agreement with evidence provided by several groups demonstrating that the BM microenvironment provides important factors other than IL-6 that contribute to MM resistance to apoptosis,14 bone disease,15 and neovascularization.16,17 In contrast, the ANBL-6 gene expression profile after interaction with normal stroma versus MM stroma was not significantly different (only 9 genes distinguished these treatments at P = .001, data not shown). One report has suggested that the MM microenvironment is significantly different from the normal marrow in the cytokines that it produces.18 Our data suggest that ANBL-6 cells “reprogram” BMSCs from MM patients and healthy donors in a similar manner. Nevertheless, our study has not focused on the analysis of gene expression profiles of MM and normal stromal cells themselves; therefore we cannot conclude that these microenvironments are not different. On the other hand, MM cells do significantly alter their genetic profile in response to BMS because 176 genes distinguished ANBL-6 cells before and after interaction with BMSCs at P = .001.
MM cell line expression patterns closely resemble active MM patient samples, which validates their use as models for MM.10 MM cell lines become especially useful when studying the effects of a single mutation or treatment on an otherwise homogeneous genetic background. For example, several groups have implicated hMIP-1α (LD78) as an osteoclastogenic factor during MM progression. It has been demonstrated that MM cells produce this factor, which, in turn, induces osteoclast differentiation in vitro and in an in vivo model of bone disease.15 Antisense directed against hMIP-1α reduced bone destruction in vivo.19 Interestingly, hMIP-1α was significantly up-regulated in ANBL-6 cells on interaction with stromal cells as well as constitutively expressed in N-ras-transfected cells, but not in cells treated with IL-6 or control samples. In fact, hMIP-1α was one of the genes that significantly distinguished the stroma-treated from IL-6-treated samples and the N-ras-transfected versus the IL-6-treated samples by class comparison analysis. In addition, hMIP-1α was one of the top 10 most up-regulated genes in ANBL-6 cells after interaction with stroma when compared with control cells (data not shown). Furthermore, RT-PCR results confirmed this expression profile under all 3 signals (Figure 5B). This report provides insight into the molecular mechanism by which hMIP-1α may be induced, through constitutive Ras signaling, and on interaction with the BMS, thus identifying potential new targets for treatment of bone destruction in MM.
We have also compared our data to the primary patient data recently reported by Zhan et al.10 In particular, we compared those genes that were differentially expressed between benign plasma cell dyscrasia (MM1) and aggressive MM (MM4). Twenty-four of the 30 reported signature genes were found to be present in at least one response group. Seven genes were randomly chosen to demonstrate their pattern of expression under the 3 activating signals. TS1 or TYMS (thymidylate synthase) displayed the highest levels of expression as in the clinical data presented by Zhan et al.10 Interestingly, this gene's highest expression was induced on IL-6 stimulation and stromal interaction, followed by lower levels in the N-ras-transfected cells but still above control levels. Other genes followed a similar pattern such as p150 or CHAF1A involved in DNA replication and MSH 2 (mutS homologue 2), a DNA repair gene. In contrast, EZH 2 (enhancer of Zeste homologue 2), a polycomb group gene involved in transcriptional regulation, was induced mostly by stromal interaction, followed by IL-6 stimulation and constitutive but lower expression in the N-ras samples. Other genes such as CDC 6 involved in cell-cycle control were activated only on IL-6 and stroma stimulation. Interestingly, IUGP2 was most highly induced in N-ras samples, followed by IL-6 stimulation and stroma interaction. Finally, FF45 or DFFA, a DNA fragmentation factor, was present and invariable in all groups including control samples. Thus, we have demonstrated that expression profiles of myeloma cells are influenced by multiple interactions and intrinsic genetic factors.
An interesting candidate for the study of MM progression is EZH2. This gene is a chromatin-remodeling factor that belongs to the polycomb group (PcG) family controlling homeotic expression. Recently, it has been shown that metastatic prostate cancer patient tissues overexpress this gene.20 This group has also shown that this transcriptional repressor induces proliferation of prostate cancer cells when transiently overexpressed and its absence results in growth inhibition as assayed by small interfering RNA duplexes targeting this gene.20 Furthermore, EZH2 is not expressed by normal BM plasma cells. Nevertheless, it becomes progressively up-regulated as MM progresses with highest expression in aggressive disease, which recapitulates levels found in tonsil B cells during normal development.21 The fact that EZH2 confers a proliferative phenotype in other cancer cells, is overexpressed in active aggressive MM, and can be induced on all 3 of our activating signals as assayed by microarray and RT-PCR in this report, suggests its involvement in MM progression.
We also investigated mutant N-Ras-induced genes in 2 ways. First, we searched for genes newly induced in N-ras transfectants that were virtually absent in nontreatment controls. We found that only 5 genes met our criteria and of these 5, 3 were completely unique to N-ras mutants when compared with other treatments. ETV5 was one such gene, which has been previously shown to be a Ras/MAPK target.22 In addition, ETV5 expression appears to correlate with other MM cell lines harboring an endogenous mutant N-ras when compared with cell lines that carry the wild-type copy of the gene (J. Shaughnessy, written personal communication, March 2003) making it a potential predictive marker for patients who may carry N-ras-activating mutations. The other unique gene to N-ras samples is DUSP6, a dual-specificity phosphatase whose substrate is ERK2.23 The fact that only mutant N-Ras induces this phosphatase and not IL-6 or stromal interaction suggests once again that mutant N-Ras may activate different downstream targets or alternatively prolongs activation of the signaling pathways, which results in a different gene expression outcome. Secondly, we directly interrogated which genes were induced to a higher level of expression uniquely by N-ras samples and not induced by any other treatment or control samples (group 4 of Figure 2). We found that 39 genes distinguished N-ras transfectants from all other samples at the level of P = .001 (Table 5). The most up-regulated gene was N-ras (HSNRAS; P = 5.13e-05) and the most down-regulated gene was an EST (AL021786). Other genes found in our analysis were DUSP6 and the costimulatory molecule CD86 that we report here also distinguished N-ras from IL-6 stimulation. CD86 is normally induced in activated B cells when presenting antigen to T cells. This molecule interacts with CD28 expressed on the T-cell surface and is absolutely required for T-cell activation. It is not expressed on normal B cells. However, most MM plasma cells and cells lines are CD28+. The fact that mutant N-ras MM plasma cells constitutively express CD86 suggests a potential autocrine signaling loop and perhaps a deficiency in T-cell activation that would occur in the subset of patients harboring mutant ras genes.
39 genes distinguish mutant N-Ras transfectants from all other treatments
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X02751 | HSNRASR | Oncogene | 33.99 | 5.13 × 10−5 |
| U20350 | CXCR1 | Chemokine receptor | 13.7 | 1.52 × 10−5 |
| Y14737 | IGHG3 | Immunity | 13.54 | 1.5 × 10−5 |
| D50640 | PDE3B | Metabolism | 12.32 | .00015 |
| AI142565 | RNASE6 | Metabolism | 11.74 | 1.07 × 10−5 |
| AB013382 | DUSP6 | Signaling | 10.38 | 1.56 × 10−9 |
| S71326 | BGPc | Adhesion | 8.63 | 9.96 × 10−6 |
| AF070641 | LOC221810 | Unknown | 7.91 | .000606 |
| AI660656 | IGJ | Immunity | 6.82 | .000303 |
| X81438 | AMPH | Synaptic vesicle protein | 6.5 | .000643 |
| AB018278 | KIAA0735 | Unknown | 6.46 | 1.65 × 10−5 |
| AB020499 | MD1 | Immunity | 5.71 | .000147 |
| AF022797 | KCNN4 | KCa channel | 5.66 | .000768 |
| AF016004 | GPM6B | Neural development | 5.63 | .000355 |
| U04343 | CD86 | Immunity | 5.0 | .000496 |
| D50918 | KIAA0128 | Unknown | 5.34 | .000156 |
| AL040446 | OSBPLIA | Metabolism | 4.89 | .000869 |
| X16354 | BGP | Adhesion | 4.66 | .000353 |
| AL023653 | XPNPEP2 | Metabolism | 4.44 | .000166 |
| S68805 | GATM | Metabolism | 4.37 | .00067 |
| M31315 | Coagulation factor XII | Coagulation | 4.37 | .000814 |
| M55067 | CGD | Metabolism | 4.34 | .00046 |
| Down-regulated | ||||
| AL021786 | ITM2A | Osteogenesis | 11.15 | 2.52 × 10−6 |
| AB018343 | KIAA0800 | Unknown | 7.09 | .000323 |
| D88308 | SLC27A2 | Metabolism | 6.67 | 8.65 × 10−5 |
| U59209 | UGT2B17 | Metabolism | 6.24 | .000432 |
| L01664 | CLC | Metabolism | 6.00 | 6.58 × 10−5 |
| AB000897 | PCDHGA12 | Adhesion | 5.59 | .000701 |
| AF043938 | M-RAS | Signaling | 5.57 | .000129 |
| AW024467 | HSPF2 | Signaling | 5.56 | .000669 |
| U45975 | PIB5PA | Signaling | 5.49 | 5.25 × 10−6 |
| AF029900 | ADAM21 | ECM | 5.01 | .000675 |
| L19314 | HES1 | Transcription | 4.93 | .00033 |
| AB007869 | KIAA0409 | Unknown | 4.85 | .000948 |
| AA584202 | c-ERB-3 | Signaling | 4.66 | .00051 |
| X70340 | TGFA | Signaling | 4.60 | .000802 |
| U94346 | CAPN5 | Signaling | 4.38 | .000276 |
| L13436 | NPR2 | Metabolism | 4.31 | .000944 |
| AF052497 | EST | Unknown | 4.31 | .000658 |
Accession no. . | Gene symbol . | Function . | Fold difference in geometric means . | F test P . |
|---|---|---|---|---|
| Up-regulated | ||||
| X02751 | HSNRASR | Oncogene | 33.99 | 5.13 × 10−5 |
| U20350 | CXCR1 | Chemokine receptor | 13.7 | 1.52 × 10−5 |
| Y14737 | IGHG3 | Immunity | 13.54 | 1.5 × 10−5 |
| D50640 | PDE3B | Metabolism | 12.32 | .00015 |
| AI142565 | RNASE6 | Metabolism | 11.74 | 1.07 × 10−5 |
| AB013382 | DUSP6 | Signaling | 10.38 | 1.56 × 10−9 |
| S71326 | BGPc | Adhesion | 8.63 | 9.96 × 10−6 |
| AF070641 | LOC221810 | Unknown | 7.91 | .000606 |
| AI660656 | IGJ | Immunity | 6.82 | .000303 |
| X81438 | AMPH | Synaptic vesicle protein | 6.5 | .000643 |
| AB018278 | KIAA0735 | Unknown | 6.46 | 1.65 × 10−5 |
| AB020499 | MD1 | Immunity | 5.71 | .000147 |
| AF022797 | KCNN4 | KCa channel | 5.66 | .000768 |
| AF016004 | GPM6B | Neural development | 5.63 | .000355 |
| U04343 | CD86 | Immunity | 5.0 | .000496 |
| D50918 | KIAA0128 | Unknown | 5.34 | .000156 |
| AL040446 | OSBPLIA | Metabolism | 4.89 | .000869 |
| X16354 | BGP | Adhesion | 4.66 | .000353 |
| AL023653 | XPNPEP2 | Metabolism | 4.44 | .000166 |
| S68805 | GATM | Metabolism | 4.37 | .00067 |
| M31315 | Coagulation factor XII | Coagulation | 4.37 | .000814 |
| M55067 | CGD | Metabolism | 4.34 | .00046 |
| Down-regulated | ||||
| AL021786 | ITM2A | Osteogenesis | 11.15 | 2.52 × 10−6 |
| AB018343 | KIAA0800 | Unknown | 7.09 | .000323 |
| D88308 | SLC27A2 | Metabolism | 6.67 | 8.65 × 10−5 |
| U59209 | UGT2B17 | Metabolism | 6.24 | .000432 |
| L01664 | CLC | Metabolism | 6.00 | 6.58 × 10−5 |
| AB000897 | PCDHGA12 | Adhesion | 5.59 | .000701 |
| AF043938 | M-RAS | Signaling | 5.57 | .000129 |
| AW024467 | HSPF2 | Signaling | 5.56 | .000669 |
| U45975 | PIB5PA | Signaling | 5.49 | 5.25 × 10−6 |
| AF029900 | ADAM21 | ECM | 5.01 | .000675 |
| L19314 | HES1 | Transcription | 4.93 | .00033 |
| AB007869 | KIAA0409 | Unknown | 4.85 | .000948 |
| AA584202 | c-ERB-3 | Signaling | 4.66 | .00051 |
| X70340 | TGFA | Signaling | 4.60 | .000802 |
| U94346 | CAPN5 | Signaling | 4.38 | .000276 |
| L13436 | NPR2 | Metabolism | 4.31 | .000944 |
| AF052497 | EST | Unknown | 4.31 | .000658 |
In summary, we have demonstrated that different proliferating signals share some genetic overlap but can also be distinguished by defined sets of genes, suggesting that malignant plasma cells have available to them distinct stimuli to achieve different functions. Notably, the different stimuli tended to alter distinct functional gene programs in signaling, metabolic functions, extracellular matrix, or translational genes. Thus, as we have demonstrated, gene expression patterns in myeloma cells are not only derived from the intrinsic transformation events (such as ras mutations), but also significantly from both soluble factors and cell contacts in the BM microenvironment.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2003-04-1227.
Supported by grants from the National Institutes of Health (PO1 CA62242), from the Multiple Myeloma Research Foundation (B.G.V.N.), and National Institutes of Health Cancer Biology Training Grant CA09138 (P.A.C. and M.A.L.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to acknowledge the assistance of the Flow Cytometry Core Facility of the University of Minnesota Cancer Center (UMCC), a comprehensive cancer center designated by the National Cancer Institute, supported in part by P30 CA77598. We would also like to thank the Biomedical Genomics Center Microarray Facility of the University of Minnesota for assistance with gene chip processing; Suzanne Grindle from UMCC Informatics Core for providing invaluable help with data analysis; Greg J. Ahmann from the Department of Immunology at the Mayo Clinic for kindly providing the myeloma bone marrow stromal cells; and John Shaughnessy, PhD, from the Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, for helpful discussion and technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal