Abstract
Transgenic mice expressing PML-RARα in early myeloid cells under control of human cathepsin G regulatory sequences all develop a myeloproliferative syndrome, but only 15% to 20% develop acute promyelocytic leukemia (APL) after a latent period of 6 to 14 months. However, this transgene is expressed at very low levels in the bone marrow cells of transgenic mice. Because the transgene includes only 6 kb of regulatory sequences from the human cathepsin G locus, we hypothesized that sequences required for high-level expression of the transgene might be located elsewhere in the cathepsin G locus and that a knock-in model might yield much higher expression levels and higher penetrance of disease. We, therefore, targeted a human PML-RARα cDNA to the 5′ untranslated region of the murine cathepsin G gene, using homologous recombination in embryonic stem cells. This model produced a high-penetrance APL phenotype, with more than 90% of knock-in mice developing APL between 6 and 16 months of age. The latent period and phenotype of APL (including a low frequency of an interstitial deletion of chromosome 2) was similar to that of the previous transgenic model. Remarkably, however, the expression level of PML-RARα in bone marrow cells or APL cells was less than 3% of that measured in the low-penetrance transgenic model. Although the explanation for this result is not yet clear, one hypothesis suggests that very low levels of PML-RARα expression in early myeloid cells may be optimal for the development of APL in mice.
Introduction
Several groups, including our own, have modeled acute promyelocytic leukemia (APL) in the mouse.1-5 When a PML-RARα cDNA (PR) derived from a t(15;17) translocation is placed under control of human cathepsin G regulatory sequences, it is expressed in early myeloid cells at low levels.1 Virtually all mice expressing PML-RARα in early myeloid cells develop a myeloproliferative syndrome, and 15% to 20% go on to develop a disease that closely resembles acute promyelocytic leukemia after a latent period of 6 to 14 months.1,2,5 The penetrance of that APL-like disease can be increased nearly 4-fold by coexpressing the reciprocal RARα-PML cDNA(RP) in the same early myeloid compartment, but the long latency persists in these doubly transgenic mice.6 These results have suggested that PML-RARα is the primary determinant of the phenotype of this disease and that it is a bona fide leukemia-initiating protein.7
However, even though this molecule is necessary for the development of APL in the mouse, the previous models suggest that it is not sufficient. The long latency and low penetrance suggest that additional genetic events are required for leukemia progression. Several events associated with progression have been identified cytogenetically, including an interstitial deletion of chromosome 2, gain of chromosome 15, and loss of a sex chromosome.8,9 In addition, overexpression of bcl-2 in early myeloid cells,10 or the coexpression of an activated FLT3 allele,11 also increases the penetrance of the APL phenotype in transgenic mice expressing PML-RARα.
The precise mechanism by which PML-RARα expression facilitates the development of APL is not yet known. This molecule has been proposed to act in a dominant-negative fashion to suppress the normal function of both RARα and PML.12-16 When overexpressed, it interferes with the assembly of RARα-RXR heterodimers, and its ability to be displaced from target sequences by its physiologic ligand is dramatically reduced.17 Furthermore, when human cathepsin G (hCG)–PML-RARα transgenic mice are intercrossed with mice that are deficient for PML, the penetrance of APL increases, suggesting that enforcement of a dominant-negative activity against PML may increase the susceptibility of mice to leukemia.18
Because hCG–PML-RARα mice express the transgene at very low levels compared with the endogenous cathepsin G allele,1 we hypothesized that the penetrance of APL might be greatly increased in these mice if we could increase levels of PML-RARα expression in early myeloid cells. To accomplish this end, we targeted the same bcr-1–derived PML-RARα cDNA used previously in the hCG–PML-RARα transgenic mice to the 5′ untranslated region of the endogenous murine cathepsin G locus, using homologous recombination techniques in embryonic stem cells (we decided not to target PML-RARα into the endogenous mouse PML locus because PML expression is ubiquitous; widespread expression of PML-RARα in transgenic mice may be toxic2 ). We found that the retained PGK-neo cassette in the mutant cathepsin G allele caused transcriptional shutdown of the gene. We, therefore, removed the PGK-neo cassette from the locus in targeted embryonic stem (ES) cells using Lox P-Cre–mediated recombination. These ΔPGK-neo mice did express PML-RARα, and more than 90% developed APL with a latency similar to that of the original transgenic model. Surprisingly, however, the expression of PML-RARα in the bone marrow and inAPL cells was not higher than that of the transgenic mice; in fact, it was less than 3% that of the transgenic model. These results suggest that this high-penetrance model does not arise because of a simple dominant-negative effect, but rather an optimal, low level of PML-RARα expression that facilitates its gain-of-function effects.
Materials and methods
Generation of knock-in mice
A targeting vector for inserting a bcr-1 PML-RARα cDNA1 into the 5′ untranslated region of the murine cathepsin G locus (mCG) was generated using the polymerase chain reaction (PCR) to generate 5′ and 3′ targeting arms flanking the insertion site. Oligonucleotide primers were used to generate a 1.9-kb 5′ targeting arm flanked by SacI and BamHI sites at its 5′ and 3′ ends, respectively, and extending to within 1 bp of the translation initiation site within exon 1. Oligonucleotide primers were used to generate a 2.0-kb 3′ targeting arm flanked by SalI and HindIII sites at its 5′ and 3′ ends, respectively, and extending from the translational initiation site in exon 1 to within exon 4 (nucleotides 461-2441).19 The targeting vector was assembled within a pUC19 backbone, together with a 1.6-kb PGK-neo selectable marker cassette flanked by LoxP1 sequences. The RW-4 embryonic stem (ES) cell line20 was transfected with the targeting vector by electroporation, and G418-resistant clones were isolated. To screen for homologous recombination at the mCG locus, DNA samples isolated from resistant clones were digested with HindIII, Southern blotted, and probed using a PCR-generated 512-bp random primer-labeled DNA probe that spanned exons 3-4 and intron 3 of the mCG locus (nucleotide 2415-292619 ), located downstream of the 3′ targeting arm (Figure 1A). To remove the PGK-neo selectable marker cassette, a correctly targeted ES cell clone (no. 100) was transfected with the pTurbo-Cre expression cassette and grown in the absence of G418 as described.20 Loss of PGK-neo was determined by Southern blotting of HindIII-digested ES cell DNA, which was hybridized with the 3′ CG probe described earlier.
Targeting PML-RARα to the murine cathepsin G locus by homologous recombination and generation of transgenic mice. (A) A targeting vector consisting of a PML-RARα cDNA and PGK-neo selectable marker cassette, flanked by targeting arms from the murine cathepsin G (mCG) locus were transfected into embryonic stem cells. Coding exons of the mCG gene are represented as black boxes (not to scale). The PML-RARα cDNA and PGK-neo selectable marker cassette are represented as open boxes. Transcriptional start sites for the mCG gene and PGK-neo selectable marker cassette are indicated by arrows. The mCG 5′ untranslated region is represented by a gray box. LoxP sites used for CRE recombinase-mediated cassette excision are represented as shaded ovals. The position of an external 3′-mCG DNA probe is indicated as a hatched box under mCG exons 3 and 4. The PGK-neo cassette was subsequently removed from a homologously recombinant ES cell clone by transient transfection of targeted ES cells with a Cre recombinase expression vector. (B) The targeted mCG locus was identified in ES cells by a size shift from 3.2 kb (wild type [WT]) to 7.9 kb (targeted, +PGK-neo) following HindIII digestion of genomic DNA. Removal of the PGK-neo cassette by Cre recombinase resulted in a shift in the size of the recombinant band to 6.4 kb (a loading artifact caused the slight apparent differences in the sizes of the WT mCG bands at 3.2 kb). (C) Tail DNA from the offspring a heterozygous CGPR/+ intercross was screened by Southern blotting to identify wild-type (CG+/+), heterozygous (CG+/PR), and homozygous (CGPR/PR) animals. (D) Tail DNA from the offspring of CGPR/PR × CG+/– animals was screened by Southern blotting to identify animals that carried one PML-RARα allele together with either an intact CG gene (CG+/PR) or a CG null mutation (CG–/PR) at the other allele.
Targeting PML-RARα to the murine cathepsin G locus by homologous recombination and generation of transgenic mice. (A) A targeting vector consisting of a PML-RARα cDNA and PGK-neo selectable marker cassette, flanked by targeting arms from the murine cathepsin G (mCG) locus were transfected into embryonic stem cells. Coding exons of the mCG gene are represented as black boxes (not to scale). The PML-RARα cDNA and PGK-neo selectable marker cassette are represented as open boxes. Transcriptional start sites for the mCG gene and PGK-neo selectable marker cassette are indicated by arrows. The mCG 5′ untranslated region is represented by a gray box. LoxP sites used for CRE recombinase-mediated cassette excision are represented as shaded ovals. The position of an external 3′-mCG DNA probe is indicated as a hatched box under mCG exons 3 and 4. The PGK-neo cassette was subsequently removed from a homologously recombinant ES cell clone by transient transfection of targeted ES cells with a Cre recombinase expression vector. (B) The targeted mCG locus was identified in ES cells by a size shift from 3.2 kb (wild type [WT]) to 7.9 kb (targeted, +PGK-neo) following HindIII digestion of genomic DNA. Removal of the PGK-neo cassette by Cre recombinase resulted in a shift in the size of the recombinant band to 6.4 kb (a loading artifact caused the slight apparent differences in the sizes of the WT mCG bands at 3.2 kb). (C) Tail DNA from the offspring a heterozygous CGPR/+ intercross was screened by Southern blotting to identify wild-type (CG+/+), heterozygous (CG+/PR), and homozygous (CGPR/PR) animals. (D) Tail DNA from the offspring of CGPR/PR × CG+/– animals was screened by Southern blotting to identify animals that carried one PML-RARα allele together with either an intact CG gene (CG+/PR) or a CG null mutation (CG–/PR) at the other allele.
Mutant mice were generated by injection of C57Bl/6 blastocysts with correctly targeted ES cells that were implanted into pseudopregnant Swiss Webster females. Chimeric male offspring were identified on the basis of coat color and bred to C57Bl/6 female mice. Germ line transmission of the mutant CG locus was assessed by Southern blotting of HindIII-digested tail DNA as described earlier. To generate animals homozygous for the targeted mutation, heterozygous animals were intercrossed, and homozygous male offspring were subsequently bred with heterozygous females to generate a colony of PML-RARα heterozygous and homozygous littermates. To generate knock-in heterozygotes with or without a functional CG gene on the residual allele, mCGPR/PR homozygotes were bred with mCG+/– mice.21
Leukemia development
To determine the incidence of leukemia among PML-RARα animals, cohorts of each genotype were generated and followed over time. To screen for leukemia development, peripheral blood was obtained for automated complete blood count analysis by retro-orbital plexus bleeding at 1- to 2-month intervals. Animals that became moribund were killed, and blood and spleen samples were analyzed for evidence of acute leukemia, using Coulter analysis, morphology, flow cytometry, and histopathologic analysis.
Cryopreservation of tumor cells
Splenocytes from killed leukemic animals were harvested under sterile conditions and cryopreserved in 10% dimethyl sulfoxide (DMSO) media in liquid nitrogen as previously described.6
Flow cytometry
Cryopreserved tumor samples were thawed and washed with phosphate-buffered saline, and samples were prepared for flow cytometric analysis by red cell lysis and incubation with antibodies to Gr-1, Mac-1, Sca-1, CD34, Ter119, B220, CD3, CD34, or isotype controls (Becton Dickinson, San Jose, CA). Flow cytometry was performed using a BD FACscan, and data were analyzed using CellQuest software (Becton Dickinson).
Real time quantitative RT-PCR
Bone marrow was collected from the tibias and femurs of 3- to 6-month old C57Bl/6 wild-type, mCG+/PR (+PGK-neo), mCG+/PR (ΔPGK-neo), and transgenic hCG–PML-RARα nonleukemic animals. Whole marrow was washed in phosphate-buffered saline (PBS), and red cells were removed by incubation in red cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA (ethylenediaminetetraacetic acid)). Cryopreserved splenic tumor cells obtained from overtly leukemic mCG+/PR (ΔPGK-neo) and transgenic hCG–PML-RARα animals were thawed, and RNA was purified from 5 × 106 cells using an RNEasy kit (Qiagen, Valencia, CA) with on-column DNAseI treatment, per the manufacturer's protocols.
RNA was subjected to real-time quantitative one-step reverse transpcription (RT)–PCR. Briefly, PCR assays were performed using 500 ng total RNA per reaction in TaqMan One-Step RT-PCR Master Mix (Applied Biosystems, Foster City, CA), 400 nM each oligonucleotide primer, and 250 nM TaqMan probe (Applied Biosystems), with or without 1.25 U/μL MultiScribe reverse transcriptase. Primers for human PML-RARα were forward (5′-CCCAGGAGCCCCGTCATAGG-3′) and reverse (5′-CTTGTAGATGCGGGGTAGAGG-3′). Primers for mouse neutrophil elastase were forward (5′-CCTTCTCTGTCAGCGGATCTTC-3′) and reverse (5′-ACATGGAGTTCTGTCACCCAC-3′). Primers for mouse MMP9 were forward (5′-CAGGGAGATGCCCATTTCG-3′) and reverse (5′-GGGCACCATTTGGAGTTTCCA-3′). Fluorescent probes were synthesized by Applied Biosystems for human PML-RARα (5′-VIC-TCCTGCCCAACAGCAACCACGT-TAMRA-3′), for mouse neutrophil elastase (5′-VICCCAACGTGCAGGTGGCCCAG-TAMRA-3′), and for mouse MMP9 (5′-VIC-TCGCTGGGCAAAGGCGTCG-TAMRA-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification was performed on the same cDNA using the ABI protocol and ABI reagents. RT-PCR was performed on a GeneAmp 5700 (Applied Biosystems) as follows: 25°C for 10 minutes, 48°C for 30 minutes, 95°C for 5 minutes, then 40 cycles of 95°C for 15 seconds and 62°C for 1 minute. Fluorescence ΔCT values were used to calculate mRNA levels of PML-RARα relative to neutrophil elastase, or of MMP9 relative to GAPDH. Data represent samples obtained from 3 animals or 1 tumor; each was assayed in duplicate in 3 independent experiments.
Western blotting
Bone marrow and splenic tumor cells were washed with PBS and subjected to red cell lysis. Cells (2 × 106) were dissolved in 100 μL RIPA (radioimmunoprecipitation assay) buffer, protein was quantitated by a BCA (bicinchoninic acid) assay (Pierce, Rockford, IL), and 50 μg total protein was electrophoresed on 8% SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) and transferred to PVDF (polyvinylidene difluoride) according to standard protocols. Blotting was performed using polyclonal rabbit antisera against human PML, 2 monoclonal antimouse PML antibodies (a generous gift from Scott Lowe Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), a rabbit polyclonal antibody against RARα (C-20; Santa Cruz Biotechnology, Santa Cruz, CA), or a goat anti-actin polyclonal antibody (C-11; Santa Cruz Biotechnology). After staining with the appropriate secondary horseradish peroxidase (HRP)–conjugated antibodies (Amersham, Arlington Heights, IL) diluted 1:10 000 in TBST (Tris (tris(hydroxymethyl)aminomethane)–buffered saline Tween-20 (polyoxyethylene sorbitane monolaureate)), protein was visualized using Maximum Sensitivity Fempto electrogenerated chemiluminescence (ECL; Pierce).
In vitro ATRA differentiation
Spleen cells from banked APL tumors were incubated in vitro (1 × 106 cells per mL) with 1 μM ATRA (all-trans retinoic acid) dissolved in ethanol (EtOH) or with EtOH alone (total 0.1% EtOH by volume) for 72 hours at 37°C in RPM1-1640 with 10% fetal calf serum (FCS). Total RNA was then prepared and subjected to real time quantitative RT-PCR analysis as described in “Real time quantitative RT-PCR.”
Molecular cytogenetics
Chromosomes were prepared from APL spleen cells as previously described.9 Chromosome preparations prepared from cells cultured for 4 to 5 days were stained with Giemsa for chromosome counting or were used for fluorescence in situ hybridization (FISH) with a painting probe for mouse chromosome 2. The conditions of hybridization, the detection of hybridization signals, and digital-image acquisition and processing were performed as previously described.22
Results
Generation of mCG+/PR knock-in mice by homologous recombination
To generate a transgenic mouse line in which a single copy of PML-RARα cDNA was inserted into the murine cathepsin G locus, ES cells were transfected with the targeting vector shown in Figure 1. The PML-RARα cDNA (the same bcr-1 fusion cDNA used in our previous studies),1,6 in tandem with a PGK-neo selection cassette, was inserted by homologous recombination into the 5′ untranslated region of the murine cathepsin G locus. On the basis of evidence that a transcriptionally active selectable marker cassette might alter the expression of nearby genes,20 we designed the targeting vector with loxP sites flanking the PGK-neo cassette to facilitate its subsequent removal, which was achieved via transient expression of CRE recombinase in the targeted ES clone (Figure 1A). After CRE transfection, a subclone was identified that retained the PML-RARα cDNA in the 5′ untranslated region of the mCG locus, but from which the PGK-neo cassette was excised, leaving only a single loxP site in its place (Figure 1B). Blastocyst injection of both unmanipulated (+PGK-neo) and pTurboCRE-transfected (ΔPGK-neo) clones was performed to generate chimeric male founder animals, and germ line transmission of the targeted loci was observed with predicted Mendelian frequency in both +PGK-neo and ΔPGK-neo lines.
Southern blot analysis of genomic DNA from mCG+/PR animals revealed that the pTurboCRE plasmid had stably integrated into the genome of the ES cell at the time of transfection. The integrated CRE gene was inherited independently from the mCG+/PR locus, and its presence was not associated with development of APL (data not shown).
High-penetrance acute myeloid leukemia in mCG+/PR (ΔPGK-neo) mice
Transgenic mCG+/PR (+PGK-neo) animals were phenotypically normal, with peripheral blood and bone marrow counts that were indistinguishable from wild-type littermates (data not shown). None of these animals developed acute leukemia during observation for up to 2 years (Figure 2A). In contrast, all tested mCG+/PR (ΔPGK-neo) animals showed evidence of a myeloid expansion in the bone marrow similar to that described in our previously characterized hCG–PML-RARα model, with progressive splenomegaly and extramedullary splenic hematopoiesis. Peripheral blood counts were not significantly different from wild-type littermates during the “preleukemic” phase (Figure 3B; Table 1). Between 6 and 19 months of age, however, these animals exhibited a high probability of developing an abrupt onset, rapidly fatal acute leukemia, characterized by pronounced leukocytosis with increased myeloblasts and promyelocytes, anemia, thrombocytopenia, and massive hepatosplenomegaly with leukemic cell infiltration (Figure 2A; Table 1). Leukemic cells from the spleens of moribund animals displayed an APL phenotype that was similar to that of PR transgenic mice, with markedly increased numbers of immature myeloid cells, including promyelocytes (Figure 3A). The median age of overt leukemia development was 10 months. Leukemia was readily transferable to genetically compatible wild-type secondary recipients (C57Bl/6 × 129Sv/J F1), which developed similar, rapidly fatal leukemias within 6 to 8 weeks of tumor inoculation (data not shown).
Development of acute myeloid leukemia in transgenic and knock-in animals. (A) Kaplan-Meier probability of leukemia-free survival of wild-type (CG+/+; ▪), mCG+/PR (+PGK-neo; g▾), and mCG+/PR (ΔPGK-neo; ▴) animals is plotted against time and is compared with leukemia-free survival of a simultaneous cohort of hCG–PML-RARα animals from line no. 134 (the same data were used in Pollock et al6 ; ♦). The number of mice in each cohort was as follows: CG+/+ = 25; mCG+/PR (+PGK-neo) = 26; mCG+/PR (ΔPGK-neo) = 118; hCG-PR = 27. (B) Kaplan-Meier probability of leukemia-free survival of heterozygous mCG+/PR (ΔPGK-neo; ▪) versus homozygous (mCGPR/PR; ▴) animals is plotted against time. The difference between the curves is statistically significant (P = .002). mCG+/PR (ΔPGK-neo) = 118 mice; mCGPR/PR (ΔPGK-neo) = 99 mice. (C) Kaplan-Meier probability of leukemia-free survival of animals carrying a single PML-RARα gene copy together with either an intact CG gene (CG+/PR; ▴) or a null mutation (CG–/PR; ▪) at the other allele is plotted against time. The differences between the curves are not statistically significant (P = .48). mCG+/PR (ΔPGK-neo) = 60 mice; mCG–/PR (ΔPGK-neo) = 71 mice.
Development of acute myeloid leukemia in transgenic and knock-in animals. (A) Kaplan-Meier probability of leukemia-free survival of wild-type (CG+/+; ▪), mCG+/PR (+PGK-neo; g▾), and mCG+/PR (ΔPGK-neo; ▴) animals is plotted against time and is compared with leukemia-free survival of a simultaneous cohort of hCG–PML-RARα animals from line no. 134 (the same data were used in Pollock et al6 ; ♦). The number of mice in each cohort was as follows: CG+/+ = 25; mCG+/PR (+PGK-neo) = 26; mCG+/PR (ΔPGK-neo) = 118; hCG-PR = 27. (B) Kaplan-Meier probability of leukemia-free survival of heterozygous mCG+/PR (ΔPGK-neo; ▪) versus homozygous (mCGPR/PR; ▴) animals is plotted against time. The difference between the curves is statistically significant (P = .002). mCG+/PR (ΔPGK-neo) = 118 mice; mCGPR/PR (ΔPGK-neo) = 99 mice. (C) Kaplan-Meier probability of leukemia-free survival of animals carrying a single PML-RARα gene copy together with either an intact CG gene (CG+/PR; ▴) or a null mutation (CG–/PR; ▪) at the other allele is plotted against time. The differences between the curves are not statistically significant (P = .48). mCG+/PR (ΔPGK-neo) = 60 mice; mCG–/PR (ΔPGK-neo) = 71 mice.
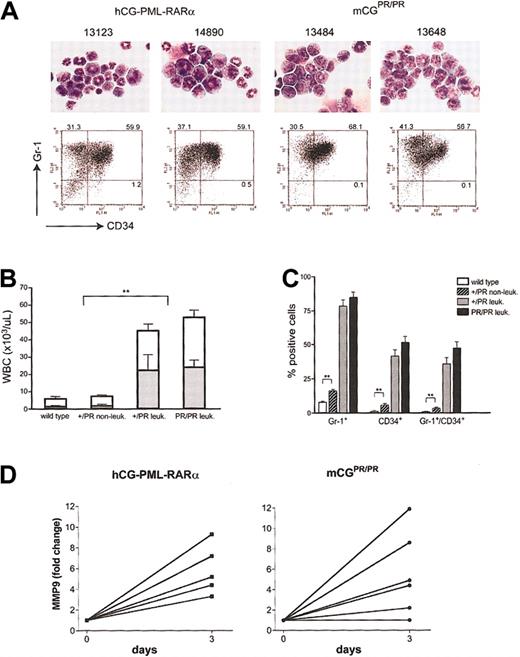
Characterization of APL cells. (A) Morphology by May-Grünwald-Giemsa staining and flow cytometric analysis for Gr-1 and CD34 is shown for 2 independent splenic tumors arising in hCG–-PML-RARα animals and in 2 mCGPR/PR animals. Numbers represent the percentage of cells in each quadrant. Original magnification, ×400. (B) Peripheral white blood cell (WBC; □) counts and absolute neutrophil counts (ANCs; ▦) determined by automated counting are shown for wild-type littermates, nonleukemic mCG+/PR, and leukemic mCG+/PR and mCGPR/PR animals. For panels B and C, wild type = 7 mice; nonleukemic mCG+/PR = 9 mice; leukemic mCG+/PR = 9 mice; leukemic mCGPR/PR = 10 mice. (C) The fraction of Gr-1+, CD34+, and Gr-1+/CD34+ cells in the spleens of wild-type, nonleukemic, and leukemia cells from mCG+/PR (+/–) and mCGPR/PR (+/+) knock-in animals are shown. Error bars indicate ± SEM. (D) The relative increase in abundance of MMP9 mRNA in individual APL samples after 3 days of treatment with 1 μM ATRA relative to diluent alone is shown. Left panel: 5 independent hCG–PML-RARα tumors; right panel: 6 independent mCGPR/PR tumors. MMP9 is normally expressed at its highest levels in terminally differentiated myeloid cells. Expression levels from each sample were normalized by measuring the abundance of GAPDH mRNA, which is expressed at all stages of myeloid development. Pairwise comparisons that yielded statistical significance of P < .01 are indicated by asterisks (**).
Characterization of APL cells. (A) Morphology by May-Grünwald-Giemsa staining and flow cytometric analysis for Gr-1 and CD34 is shown for 2 independent splenic tumors arising in hCG–-PML-RARα animals and in 2 mCGPR/PR animals. Numbers represent the percentage of cells in each quadrant. Original magnification, ×400. (B) Peripheral white blood cell (WBC; □) counts and absolute neutrophil counts (ANCs; ▦) determined by automated counting are shown for wild-type littermates, nonleukemic mCG+/PR, and leukemic mCG+/PR and mCGPR/PR animals. For panels B and C, wild type = 7 mice; nonleukemic mCG+/PR = 9 mice; leukemic mCG+/PR = 9 mice; leukemic mCGPR/PR = 10 mice. (C) The fraction of Gr-1+, CD34+, and Gr-1+/CD34+ cells in the spleens of wild-type, nonleukemic, and leukemia cells from mCG+/PR (+/–) and mCGPR/PR (+/+) knock-in animals are shown. Error bars indicate ± SEM. (D) The relative increase in abundance of MMP9 mRNA in individual APL samples after 3 days of treatment with 1 μM ATRA relative to diluent alone is shown. Left panel: 5 independent hCG–PML-RARα tumors; right panel: 6 independent mCGPR/PR tumors. MMP9 is normally expressed at its highest levels in terminally differentiated myeloid cells. Expression levels from each sample were normalized by measuring the abundance of GAPDH mRNA, which is expressed at all stages of myeloid development. Pairwise comparisons that yielded statistical significance of P < .01 are indicated by asterisks (**).
Peripheral blood counts and splenic flow cytometry on preleukemic and leukemic animals
. | . | Age, mo . | WBC count, × 106/mL . | . | PLT count, × 106/mL . | . | Spleen . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse . | Genotype . | . | . | %PMNs . | . | HCT . | % Gr-1+ . | % CD34+ . | % Gr-1+/%CD34+ . | ||
| Wild type | C57B1/6 | 1 | 4.24 | 16.78 | 306 | 23.1 | 8.68 | 0.89 | 0.48 | ||
| Wild type | C57B1/6 | 1 | 2.6 | 18.68 | 511 | 50.2 | 8.59 | 0.87 | 0.41 | ||
| Wild type | C57B1/6 | 2 | 4.38 | 10.62 | 608 | 48.4 | 8.18 | 1.99 | 0.94 | ||
| Wild type | C57B1/6 | 6 | 7.88 | 20.98 | 756 | 55.1 | 4.85 | 1.02 | 0.46 | ||
| Wild type | 129/SvJ | 3 | 4.22 | 23.33 | 452 | 69.1 | 9.42 | 1.52 | 0.71 | ||
| Wild type | 129/SvJ | 4 | 14.44 | 29.8 | 457 | 29.8 | 9.32 | 1.58 | 0.99 | ||
| Wild type | 129/SvJ | 6 | 7.74 | 20.7 | 284 | 36 | 6.41 | 1.06 | 0.47 | ||
| Average | 6.5 | 20.04 | 482 | 44.53 | 7.92 | 1.28 | 0.64 | ||||
| Nonleukemic | |||||||||||
| 17776 | +/PR | 1 | 6.42 | 21.46 | 647 | 41 | 18.47 | 8.33 | 4.39 | ||
| 17283 | +/PR | 3 | 4.86 | 21.38 | 953 | 44.3 | 20.01 | 6.04 | 4.22 | ||
| 16098 | +/PR | 6 | 5.62 | 11.79 | 354 | 21.2 | 19.23 | 3.92 | 2.7 | ||
| 15458 | +/PR | 10 | 7.72 | 12.6 | 481 | 27.4 | 22.25 | 5.62 | 3.77 | ||
| 14111 | +/PR | 10 | 9.4 | 37 | 2179 | 43.8 | 14.3 | 2.9 | 1.7 | ||
| 14142 | +/PR | 10 | 9.0 | 56 | 1230 | 46 | 11.2 | 6.3 | 2.6 | ||
| 14143 | +/PR | 10 | 13.2 | 34 | 1163 | 42.5 | 13.5 | 10.8 | 7.48 | ||
| 14586 | +/PR | 9 | 6.7 | 25 | 1124 | 45.2 | 11.6 | 3.42 | 1.9 | ||
| 14584 | +/PR | 9 | 6.7 | 12 | 954 | 46.7 | 12.5 | 5.8 | 2.5 | ||
| Average | 7.74 | 25.69 | 1009 | 39.79 | 15.9 | 5.9 | 3.47 | ||||
| Leukemic | |||||||||||
| 13646 | +/PR | 5 | 47 | 48 | 254 | 32.7 | 24.7 | 26 | 25.9 | ||
| 13843 | +/PR | 5 | 45.2 | 64 | 362 | 27.9 | 73.4 | 26.6 | 26.4 | ||
| 13487 | +/PR | 6 | 25.8 | 44 | 135 | 24.1 | 88 | 61.1 | 61 | ||
| 13498 | +/PR | 6 | 126 | 77 | 184 | 26 | 54.4 | 55.4 | 30.5 | ||
| 13499 | +/PR | 6 | 69 | 33 | 246 | 21.7 | 60.2 | 53 | 32.6 | ||
| 13659 | +/PR | 7 | 29.8 | 35 | 368 | 30.3 | 87 | 33 | 32.9 | ||
| 13346 | +/PR | 8 | 17 | 34.5 | 328 | 34.5 | 76.5 | 23.3 | 24.7 | ||
| 13437 | +/PR | 8 | 23.7 | 23 | 414 | 30.9 | 85.6 | 52.4 | 51.2 | ||
| 13355 | +/PR | 9 | 18.8 | 40.3 | 322 | 32.8 | 86.1 | 42.2 | 40.3 | ||
| Average | 44.7 | 43.53 | 290 | 29.0 | 78.4 | 41.1 | 36.2 | ||||
| Leukemic | |||||||||||
| 13834 | PR/PR | 6 | 50 | 81 | 413 | 31 | 65.8 | 38.6 | 31 | ||
| 13445 | PR/PR | 7 | 61.4 | 15 | 358 | 26.1 | 77.9 | 59.6 | 48.1 | ||
| 13484 | PR/PR | 7 | 36.6 | 25 | 141 | 31.6 | 92.5 | 46.9 | 42.2 | ||
| 13647 | PR/PR | 7 | 48.4 | 50 | 361 | 31.7 | 89.9 | 64.4 | 64.2 | ||
| 13648 | PR/PR | 7 | 49.9 | 55.6 | 331 | 35.3 | 97.9 | 26 | 26 | ||
| 13663 | PR/PR | 7 | 84.2 | 51 | 98 | 32.4 | 91.9 | 45.4 | 45.2 | ||
| 13178 | PR/PR | 8 | 56 | 65 | 213 | 35.7 | 97 | 50.6 | 44.7 | ||
| 13451 | PR/PR | 8 | 34.6 | 39 | 282 | 35.8 | 84.9 | 83.6 | 75.6 | ||
| 13442 | PR/PR | 10 | 66 | 39 | 697 | 19.7 | 61 | 43.4 | 42.9 | ||
| 13349 | PR/PR | 11 | 38.2 | 30.5 | 529 | 36.5 | 85.2 | 54.7 | 53.9 | ||
| Average | 52.5 | 45.2 | 342 | 31.6 | 84.4 | 51.3 | 47.4 | ||||
. | . | Age, mo . | WBC count, × 106/mL . | . | PLT count, × 106/mL . | . | Spleen . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse . | Genotype . | . | . | %PMNs . | . | HCT . | % Gr-1+ . | % CD34+ . | % Gr-1+/%CD34+ . | ||
| Wild type | C57B1/6 | 1 | 4.24 | 16.78 | 306 | 23.1 | 8.68 | 0.89 | 0.48 | ||
| Wild type | C57B1/6 | 1 | 2.6 | 18.68 | 511 | 50.2 | 8.59 | 0.87 | 0.41 | ||
| Wild type | C57B1/6 | 2 | 4.38 | 10.62 | 608 | 48.4 | 8.18 | 1.99 | 0.94 | ||
| Wild type | C57B1/6 | 6 | 7.88 | 20.98 | 756 | 55.1 | 4.85 | 1.02 | 0.46 | ||
| Wild type | 129/SvJ | 3 | 4.22 | 23.33 | 452 | 69.1 | 9.42 | 1.52 | 0.71 | ||
| Wild type | 129/SvJ | 4 | 14.44 | 29.8 | 457 | 29.8 | 9.32 | 1.58 | 0.99 | ||
| Wild type | 129/SvJ | 6 | 7.74 | 20.7 | 284 | 36 | 6.41 | 1.06 | 0.47 | ||
| Average | 6.5 | 20.04 | 482 | 44.53 | 7.92 | 1.28 | 0.64 | ||||
| Nonleukemic | |||||||||||
| 17776 | +/PR | 1 | 6.42 | 21.46 | 647 | 41 | 18.47 | 8.33 | 4.39 | ||
| 17283 | +/PR | 3 | 4.86 | 21.38 | 953 | 44.3 | 20.01 | 6.04 | 4.22 | ||
| 16098 | +/PR | 6 | 5.62 | 11.79 | 354 | 21.2 | 19.23 | 3.92 | 2.7 | ||
| 15458 | +/PR | 10 | 7.72 | 12.6 | 481 | 27.4 | 22.25 | 5.62 | 3.77 | ||
| 14111 | +/PR | 10 | 9.4 | 37 | 2179 | 43.8 | 14.3 | 2.9 | 1.7 | ||
| 14142 | +/PR | 10 | 9.0 | 56 | 1230 | 46 | 11.2 | 6.3 | 2.6 | ||
| 14143 | +/PR | 10 | 13.2 | 34 | 1163 | 42.5 | 13.5 | 10.8 | 7.48 | ||
| 14586 | +/PR | 9 | 6.7 | 25 | 1124 | 45.2 | 11.6 | 3.42 | 1.9 | ||
| 14584 | +/PR | 9 | 6.7 | 12 | 954 | 46.7 | 12.5 | 5.8 | 2.5 | ||
| Average | 7.74 | 25.69 | 1009 | 39.79 | 15.9 | 5.9 | 3.47 | ||||
| Leukemic | |||||||||||
| 13646 | +/PR | 5 | 47 | 48 | 254 | 32.7 | 24.7 | 26 | 25.9 | ||
| 13843 | +/PR | 5 | 45.2 | 64 | 362 | 27.9 | 73.4 | 26.6 | 26.4 | ||
| 13487 | +/PR | 6 | 25.8 | 44 | 135 | 24.1 | 88 | 61.1 | 61 | ||
| 13498 | +/PR | 6 | 126 | 77 | 184 | 26 | 54.4 | 55.4 | 30.5 | ||
| 13499 | +/PR | 6 | 69 | 33 | 246 | 21.7 | 60.2 | 53 | 32.6 | ||
| 13659 | +/PR | 7 | 29.8 | 35 | 368 | 30.3 | 87 | 33 | 32.9 | ||
| 13346 | +/PR | 8 | 17 | 34.5 | 328 | 34.5 | 76.5 | 23.3 | 24.7 | ||
| 13437 | +/PR | 8 | 23.7 | 23 | 414 | 30.9 | 85.6 | 52.4 | 51.2 | ||
| 13355 | +/PR | 9 | 18.8 | 40.3 | 322 | 32.8 | 86.1 | 42.2 | 40.3 | ||
| Average | 44.7 | 43.53 | 290 | 29.0 | 78.4 | 41.1 | 36.2 | ||||
| Leukemic | |||||||||||
| 13834 | PR/PR | 6 | 50 | 81 | 413 | 31 | 65.8 | 38.6 | 31 | ||
| 13445 | PR/PR | 7 | 61.4 | 15 | 358 | 26.1 | 77.9 | 59.6 | 48.1 | ||
| 13484 | PR/PR | 7 | 36.6 | 25 | 141 | 31.6 | 92.5 | 46.9 | 42.2 | ||
| 13647 | PR/PR | 7 | 48.4 | 50 | 361 | 31.7 | 89.9 | 64.4 | 64.2 | ||
| 13648 | PR/PR | 7 | 49.9 | 55.6 | 331 | 35.3 | 97.9 | 26 | 26 | ||
| 13663 | PR/PR | 7 | 84.2 | 51 | 98 | 32.4 | 91.9 | 45.4 | 45.2 | ||
| 13178 | PR/PR | 8 | 56 | 65 | 213 | 35.7 | 97 | 50.6 | 44.7 | ||
| 13451 | PR/PR | 8 | 34.6 | 39 | 282 | 35.8 | 84.9 | 83.6 | 75.6 | ||
| 13442 | PR/PR | 10 | 66 | 39 | 697 | 19.7 | 61 | 43.4 | 42.9 | ||
| 13349 | PR/PR | 11 | 38.2 | 30.5 | 529 | 36.5 | 85.2 | 54.7 | 53.9 | ||
| Average | 52.5 | 45.2 | 342 | 31.6 | 84.4 | 51.3 | 47.4 | ||||
Peripheral blood white cell counts (WBC count), percent neutrophils (% PMNs), platelet count (PLT count), and hematocrit (HCT) were determined by automated counting for wild-type, nonleukemic mCG+/PR, leukemic mCG+/PR, and mCGPR/PR animals at the indicated ages. Spleens of the same animals were analyzed by flow cytometry for expression of the surface markers Gr-1 and CD34.
Aberrant coexpression of CD34 and myeloid differentiation markers in knock-in PML-RARα tumors
To characterize the leukemias that developed in the mCG+/PR (ΔPGK-neo) knock-in animals, flow cytometric analyses were performed on the spleen cells of affected animals and compared with tumor spleens from hCG–PML-RARα mice. Analysis of multiple independent tumors demonstrated coexpression of myeloid differentiation markers Gr-1 and Mac-1 in leukemic cells and the absence of B- or T-lymphoid marker expression or the erythroid lineage marker Ter119 (Table 1; Figure 3C; and data not shown). The abnormal coexpression of the primitive hematopoietic marker CD34 with Gr-1 was detected in a large population of cells in leukemic spleens, similar to that previously described in hCG–PML-RARα mice6,9 (Figure 3A; Table 1). Furthermore, the analysis of spleens prior to the development of leukemia demonstrated increased numbers of Gr-1–positive cells compared with wild-type mice. A modest but statistically significant increase in Gr-1/CD34 coexpressing cells was also detected in the nonleukemic spleens (Figure 3C).
Induction of myeloid differentiation following ATRA treatment
To determine whether leukemia cells derived from the knock-in mice respond to ATRA treatment, we devised an assay to measure the induction of MMP9, a gene that is expressed at its highest levels during the late stages of myeloid development.23 RNA isolated from hCG–PML-RARα or mCGPR/PR tumor spleens was reverse transcribed into DNA and subjected to quantitative real time PCR amplification. We observed 1.5- to 12-fold increases in MMP9 mRNA abundance following treatment of tumor cells from either model with 1 μM ATRA for 72 hours over treatment with vehicle alone, after normalization against GAPDH expression levels (Figure 3D). Increases in MMP9 mRNA levels correlated with an increase in the proportion of mature myeloid cells in the cultures over time (data not shown). ATRA sensitivity was similar in the transgenic and knock-in tumor cells. Spleen cells from one knock-in tumor did not exhibit an increase in MMP9 expression level or in myeloid differentiation over time.
A small decrease in leukemia latency with a 2-fold increase in PML-RARα gene dosage
To determine the effect of gene dosage on leukemia development, heterozygous mCG+/PR animals (one PML-RARα copy) were bred together to generate offspring homozygous at the CG locus for the PML-RARα knock-in mutation (2 PML-RARα copies). Although the high lifetime probability of leukemia development was the same for heterozygous and homozygous animals, the median age at leukemia development was decreased by approximately 6 weeks among homozygous animals (Figure 2B). Flow cytometric analysis demonstrated no differences in expression of differentiation markers between PML-RARα heterozygous and homozygous tumors (Figure 3; Table 1). Moreover, tumor inoculation into secondary recipients showed no difference in the ability of heterozygous versus homozygous tumors to transfer leukemia to wild-type recipients (data not shown).
Endogenous cathepsin G loss-of-function does not contribute to leukemia development
Because the PML-RARα knock-in mutation results in disruption of one or both mCG loci, it was formally possible that the difference in leukemia incidence between the transgenic and knock-in models resulted from loss of cathepsin G function. We considered this possibility to be unlikely, because both CG+/– and CG–/– mice lack a detectable hematopoietic phenotype,21 and because acute myelogenous leukemia (AML) has never been observed in any CG+/– or CG–/– animal (C. Pham and T.J.L., unpublished observation, 2000). To formally test the effect of mCG loss-of-function in the setting of PML-RARα expression, we bred mCGPR/PR homozygous animals with CG+/– animals. The offspring of these animals all carried a single PML-RARα cDNA inserted into one mCG allele and either a wild-type mCG gene or a null mutation at the other allele. These mice were, therefore, either haploinsufficient or null for cathepsin G. The probability of leukemia development among these animals was identical, indicating that homozygous loss-of-function for cathepsin G played no detectable role in the development of leukemia in this system (Figure 2C).
Expression of PML-RARα mRNA in transgenic versus knock-in mice
We designed a quantitative RT-PCR assay to determine the relative levels of PML-RARα mRNA expression in the knock-in model and the hCG–PML-RARα transgenic model previously described. Total RNA was harvested from the bone marrow of 3-month-old C57Bl/6 wild-type mice and from the nonleukemic bone marrow of 3-month-old knock-in and transgenic animals. Additionally, RNA was harvested from cryopreserved splenic tumors arising from both APL models. Primers and probes specific for the PML-RARα cDNA, mouse neutrophil elastase, and mouse GAPDH were designed to quantitate PML-RARα abundance with respect to whole cell mRNA abundance (GAPDH) and promyelocyte-specific mRNA abundance (neutrophil elastase). Neutrophil elastase (NE) is coordinately regulated with cathepsin G, both of which demonstrate maximal expression in the promyelocyte compartment.24,25 Normalizing PML-RARα mRNA abundance to NE mRNA abundance controls for variation in the number of early myeloid cells in each sample (ie, expression of PML-RARα will not be overestimated in samples with increased numbers of early myeloid cells). Levels of PML-RARα mRNA normalized to GAPDH mRNA yielded similar relative expression values in all samples tested (data not shown).
Because the transgenic model contains approximately 50 concatamerized copies of the PML-RARα cDNA1 (versus only one copy in the knock-in model), the following steps were taken to avoid DNA contamination in the PCR reaction: (1) RNA samples were treated with DNAseI on a purification column; (2) samples were then digested with HaeIII, which cleaves the PML-RARα cDNA within the 200–base pair PCR amplicon; and (3) lack of contamination was verified by requiring at least a 5-cycle difference in ΔCT between reactions with and without reverse transcriptase.
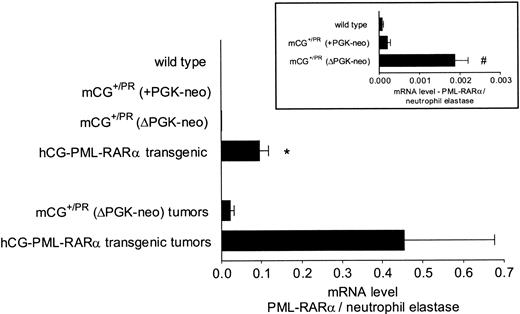
Bone marrow cells derived from mice with the retained PGK-neo cassette lacked PML-RARα expression that was detectable above the background levels observed in wild-type littermates (Figure 4, inset). Removal of the PGK-neo cassette, however, led to consistently detectable PML-RARα expression (Figure 4). Surprisingly, PML-RARα mRNA levels from preleukemic knock-in bone marrow cells were significantly lower than those of similar hCG–PML-RARα transgenic mice (48 ± 5.8-fold, P < .005). These data were highly reproducible and represent the average expression for 3 mice of each genotype assayed in duplicate in 3 separate experiments. This expression relationship was also true for independent tumors that arose in both models (Figure 4). In these samples, in which the cells were nearly all leukemic, the levels of PML-RARα mRNA were again much higher in the tumors derived from the transgenic mice. These data represent average data from 3 tumors in each model, assayed in duplicate in 2 separate experiments.
Expression of PML-RARα mRNA in nonleukemic and leukemic spleen cells. Real time quantitative PCR was performed on the bone marrow RNA derived from 3 nonleukemic animals of each genotype at 3 to 6 months of age and on 3 independent splenic tumors each from leukemic mCG+/PR (ΔPGK-neo) or transgenic hCG–PML-RARα animals. Each sample was analyzed in duplicate at least twice. Levels of human PML-RARα mRNA were normalized to levels of endogenous mouse neutrophil elastase mRNA to control for the contribution of early myeloid cells, which express abundant neutrophil elastase mRNA, in each sample. All of the normalized data were pooled and averaged; means and standard errors of the mean are shown. Statistical significance was determined by a 2-tailed Student t test. Inset: enlarged view of PML-RARα expression in the nonleukemic bone marrow RNAs of C57Bl/6 wild-type, mCG+/PR (+PGK-neo), and mCG+/PR (ΔPGK-neo) animals. *P < .005 versus mCG+/PR (ΔPGK-neo). #P < .005 versus mCG+/PR (+PKG-neo).
Expression of PML-RARα mRNA in nonleukemic and leukemic spleen cells. Real time quantitative PCR was performed on the bone marrow RNA derived from 3 nonleukemic animals of each genotype at 3 to 6 months of age and on 3 independent splenic tumors each from leukemic mCG+/PR (ΔPGK-neo) or transgenic hCG–PML-RARα animals. Each sample was analyzed in duplicate at least twice. Levels of human PML-RARα mRNA were normalized to levels of endogenous mouse neutrophil elastase mRNA to control for the contribution of early myeloid cells, which express abundant neutrophil elastase mRNA, in each sample. All of the normalized data were pooled and averaged; means and standard errors of the mean are shown. Statistical significance was determined by a 2-tailed Student t test. Inset: enlarged view of PML-RARα expression in the nonleukemic bone marrow RNAs of C57Bl/6 wild-type, mCG+/PR (+PGK-neo), and mCG+/PR (ΔPGK-neo) animals. *P < .005 versus mCG+/PR (ΔPGK-neo). #P < .005 versus mCG+/PR (+PKG-neo).
Undetectable expression of PML-RARα protein in transgenic and knock-in APL cells
Total cellular protein was extracted from wild-type C57Bl/6 bone marrow cells, 3-month-old nonleukemic transgenic, and knock-in bone marrow cells and from several independent tumors arising in each mouse model. Western blotting was performed using several different antibodies directed against RARα or PML. In Figure 5, lane 3 represents U937 cells transiently expressing the human bcr-1 PML-RARα cDNA that was used to create the knock-in and transgenic mouse models. Full-length PML-RARα protein is detected at approximately 120 kDa. Despite using highly sensitive chemiluminescence reagents and long exposure times, full-length PML-RARα protein was not detected in nonleukemic bone marrow cells from either model (data not shown). Furthermore, in the tumor cells, which represent a more homogeneous cell population, endogenous mouse RARα protein was detectable at approximately 55 kDa, but, again, full-length PML-RARα was undetectable (Figure 5, lanes 5-9). Similarly, if anti-PML blots were highly overexposed, endogenous mouse PML was visible at approximately 70 kDa, but full-length PML-RARα remained below the limit of detection (data not shown). These sensitive Western blots were repeated with a second rabbit polyclonal antibody to PML and with 2 different monoclonal antibodies against mouse PML. In no case was full-length PML-RARα detectable nor was it detectable in several other tumors from each mouse model (data not shown). Further, lysis by boiling in loading buffer containing 2% SDS did not change these results (data not shown). Together, these data indicate that the abundance of PML-RARα protein is less than that of endogenous PML and RARα in both nonleukemic marrow and splenic tumors derived from both mouse models.
Expression of PML-RARα protein in leukemic spleen cells. Western blots were performed for RARα and actin on transiently transfected U937 cells (harvested 4 hours after transfection) and on independent tumors from each model. Lane 1 represents U937 cells electroporated with an empty expression plasmid; lane 2, U937 cells expressing the RARα portion of human PML-RARα (RARα exons 3-9); and lane 3, U937 cells expressing full-length human PML-RARα (importantly, high-level expression of PML-RARα in these cells is highly toxic, causing the death of most U937 cells within 24 hours of transfection27 ). Lanes 4-6 represent hCG-PR tumors; lanes 7-8, mCG+/PR (ΔPGK-neo) tumors. The positions of RARα (∼55 kDa) and full-length PML-RARα (∼120 kDa) are marked. A band of approximately 50 kDa marked * represents an unknown protein recognized by the RARα antibody in hCG–PML-RARα tumors. It could potentially represent an altered PML-RARα protein.
Expression of PML-RARα protein in leukemic spleen cells. Western blots were performed for RARα and actin on transiently transfected U937 cells (harvested 4 hours after transfection) and on independent tumors from each model. Lane 1 represents U937 cells electroporated with an empty expression plasmid; lane 2, U937 cells expressing the RARα portion of human PML-RARα (RARα exons 3-9); and lane 3, U937 cells expressing full-length human PML-RARα (importantly, high-level expression of PML-RARα in these cells is highly toxic, causing the death of most U937 cells within 24 hours of transfection27 ). Lanes 4-6 represent hCG-PR tumors; lanes 7-8, mCG+/PR (ΔPGK-neo) tumors. The positions of RARα (∼55 kDa) and full-length PML-RARα (∼120 kDa) are marked. A band of approximately 50 kDa marked * represents an unknown protein recognized by the RARα antibody in hCG–PML-RARα tumors. It could potentially represent an altered PML-RARα protein.
An interstitial deletion of chromosome 2 is found in a small fraction of knock-in tumor cells
We analyzed 9 cryopreserved tumor samples with FISH using a chromosome 2–specific painting probe and by posthybridization DAPI (46′-diamidino-2-phenylindole-2 HCl)–produced G-like banding (Table 2). Six tumors from mCG+/PR mice (13437, 13441, 13487, 13644, 13646, and 13659) had a homogenous, near diploid cell population with a chromosome number ranging from 38 to 46; most of the cells from these 6 tumors contained 40 chromosomes. Among these, tumor 13437 had fewer cells with the normal chromosome number (2n = 40). Tumors 13498 and 13499 showed wider variations in chromosome number, which ranged from 39 to 50 and 36 to 62, respectively. Only one tumor (13843) consisted of 2 subpopulations with near-diploid and near-tetraploid chromosome numbers (ranging from 40-47 to 71-81 chromosomes, respectively). All lines were examined for alterations of chromosome 2, which is a recurrent alteration in tumors derived from hCG–PML-RARα transgenic animals.9 An interstitial deletion of chromosome 2, similar to the deletion previously described for transgenic hCG–PML-RARα × hCG–RARα-PML tumors9 was detected in tumors 13843 and 13499. The deleted material from chromosome 2 was not translocated to any other chromosome. No other gross chromosomal abnormalities were identified.
Molecular cytogenetic analysis of mCG+/PR splenic tumors
Tumor . | Chromosomal modal number . | Chromosome number range . | % of metaphases with 2n ≠ 40 or 4n ≠ 80 . | Chromosome 2 . |
|---|---|---|---|---|
| 13437 | 40 | 39-44 | 60 | N/N |
| 13441 | 40 | 39-42 | 36 | N/N |
| 13487 | 40 | 39-42 | 36 | N/N |
| 13498 | 40 | 39-50 | 32 | N/N |
| 13499 | 40 | 36-62 | 52 | N/del(2) |
| 13843 | 40 | 40-47 | 50 | N/del(2) |
| 13843 | 80 | 71-81 | 56 | N/N/del(2)/del(2) |
| 13644 | 40 | 38-41 | 20 | N/N |
| 13646 | 40 | 39-41 | 12 | N/N |
| 13659 | 40 | 38-46 | 36 | N/N |
Tumor . | Chromosomal modal number . | Chromosome number range . | % of metaphases with 2n ≠ 40 or 4n ≠ 80 . | Chromosome 2 . |
|---|---|---|---|---|
| 13437 | 40 | 39-44 | 60 | N/N |
| 13441 | 40 | 39-42 | 36 | N/N |
| 13487 | 40 | 39-42 | 36 | N/N |
| 13498 | 40 | 39-50 | 32 | N/N |
| 13499 | 40 | 36-62 | 52 | N/del(2) |
| 13843 | 40 | 40-47 | 50 | N/del(2) |
| 13843 | 80 | 71-81 | 56 | N/N/del(2)/del(2) |
| 13644 | 40 | 38-41 | 20 | N/N |
| 13646 | 40 | 39-41 | 12 | N/N |
| 13659 | 40 | 38-46 | 36 | N/N |
N indicates normal; del(2) refers to the interstitial deletion of chromosome 2 previously described.8
Discussion
In this report, we describe mice that express PML-RARα under control of the endogenous murine cathepsin G locus. Mice heterozygous or homozygous for this mutation (which also creates a null cathepsin G allele) are viable and fertile, but all display a myeloproliferative syndrome early in life. After a long latent period, mice bearing this mutation develop a fatal APL-like syndrome that is characterized by the accumulation of early myeloid cells in the bone marrow, spleen, and liver, as well as sensitivity to ATRA in vitro. In contrast to the low penetrance of APL (15%-20%) observed in transgenic mice expressing PML-RARα under control of a human cathepsin G targeting cassette, the penetrance of APL in the knock-in mice was more than 90%. The latency of the 2 models was similar, however, suggesting that both require additional genetic events for APL progression. Although we predicted that the knock-in model would express PML-RARα at high levels in early myeloid cells, expression was actually much lower than that observed in the previous transgenic model. These results suggest that there may be an optimal level of PML-RARα expression that facilitates APL development and that this level is lower than that previously predicted.
The most striking feature of the knock-in model was the very high penetrance of APL. In previously reported experiments, we and others have demonstrated that an identical PML-RARα cDNA, when expressed in transgenic mice under the control of human cathepsin G regulatory sequences, results in a myeloproliferative syndrome in all of the mice, but APL development in only 15% to 20%, and only after a latent period of 6 to 14 months.1,2,4,5 Similarly, mice that express PML-RARα under control of the MRP8 promoter (which is expressed in both early and late myeloid cells) have a low frequency of APL development also characterized by long latency.3 Our previously reported hCG transgenic mouse model exhibited relatively low levels of expression of PML-RARα mRNA in the bone marrow, compared with endogenous PML and/or endogenous cathepsin G1 ; none of 8 transgenic lines expressing hCG–PML-RARα expressed high levels of PML-RARα mRNA.1 In contrast, the unmanipulated 6.0-kB human cathepsin G cassette was expressed at high levels in most transgenic founders, although it did exhibit integration site–specific variegation.26
These results suggested that high-level expression of PML-RARα could not be achieved in the early myeloid cells of transgenic mice. There are at least 3 possibilities for why this might occur: (1) Insertion of the PML-RARα cDNA into the 5′-untranslated region (UT) of the cathepsin G gene disrupted a critical regulatory element. (2) A critical regulatory element(s) (eg, an enhancer or locus control region) that is required for high-level expression is missing from the 6.0-kB human cathepsin G targeting cassette. (3) PML-RARα is toxic to the early myeloid cells in which it is expressed.
To address some of these issues, we generated the knock-in model described in this report. The same PML-RARα cDNA was targeted (via homologous recombination) into the identical position of the murine cathepsin G 5′ untranslated region as in the human cathepsin G transgene. With this approach, any missing regulatory element in the cathepsin G locus would be captured. When the PGK-neo cassette was left in the mutant locus, virtually no expression of the cDNA was detected, strongly suggesting that the retained PGK-neo cassette dramatically reduced expression from the mutant locus.20 We, therefore, removed the PGK-neo cassette by expressing Cre-recombinase in the targeted embryonic stem cell line. This manipulation yielded a functional mutant allele, whose expression could readily be detected, but only with RT-PCR. The expression level from the mutant allele was dramatically less than that of the unmodified cathepsin G allele in heterozygous animals (data not shown). When expression from the knock-in allele was compared with PML-RARα expression in a transgenic hCG–PML-RARα founder line, we were surprised to find that it was much lower. We and others14 had predicted that the opposite would occur. Furthermore, full-length PML-RARα protein could not be detected in the bone marrow of nonleukemic mice, nor in tumors derived from knock-in or transgenic mice, using antibodies that can detect endogenous levels of PML and RARα. This high-penetrance model of APL is, therefore, associated with an extremely low level of PML-RARα expression in early myeloid cells.
A number of explanations could account for these unexpected results: first, the mouse strains used to generate the 2 models are different. Our transgenic model was made in C3H × C57Bl/6 mice, and the knock-in model was made in 129/SvJ × C57Bl/6 mice. The 129/SvJ component could potentially provide a susceptibility locus for APL development that has not yet been characterized. This possibility seems unlikely to us, because the PML-RARα cDNA has previously been expressed in a number of different genetic backgrounds, and the latency and penetrance of APL development in all strains has been similar.1-5 However, this remains a formal possibility, and a backcross to C57Bl/6 mice is, therefore, in progress; this experiment will require another 2 to 3 years to complete.
Second, the knock-in mutation actually results in both a gain-of-function and a loss-of-function change in each cathepsin G locus. The gain-of-function mutation is provided by the PML-RARα cDNA placed in the 5′-UT of the cathepsin G gene. This mutation also causes a loss-of-function change: a null mutation of the same cathepsin G gene. However, it does not appear that the loss-of-function mutation plays a significant role in the phenotype. We did not observe a further alteration in the latency or penetrance of APL in knock-in mice that had a null mutation of cathepsin G on the residual allele. Furthermore, because cathepsin G null mice lack any detectable alterations in myeloid development21 and do not develop AML, the cathepsin G haploinsufficiency caused by the targeting event is unlikely to have affected these results.
Third, it is possible that there is a difference in the early myeloid compartment targeted by the human cathepsin G transgene versus the knock-in cathepsin G locus. To explore this possibility, we purified Sca+Lineage– mononuclear cells from 5-fluorouracil (5-FU)–treated transgenic and knock-in bone marrow cells maintained in stem cell factor, Flt3 ligand, interleukin 3 (IL-3), and thrombopoietin (TPO) for 3 days. Analysis of these populations does not show any significant difference in PML-RARα mRNA expression measured by real time RT-PCR. Culturing these cells for 2 days in media containing stem cell factor and granulocyte colony-stimulating factor (G-CSF) induces differentiation of the immature progenitor cells into a predominantly promyelocytic population; these cells display the same relative levels of PML-RARα expression as seen in Figure 4 (Lane et al27 and A.A.L. and T.J.L., unpublished observation). This observation suggests that the knock-in mCG locus does not target expression to a significantly earlier myeloid progenitor compartment than the hCG transgene.
Fourth, the translatability of the PML-RARα mRNA produced in the knock-in mice may be greater than that of the transgenic model, in which the mRNA is produced from a long concatemer of transgenes. We cannot directly address this point, because we cannot accurately measure total PML-RARα protein levels via sensitive Western blotting techniques. However, in many previous studies of multicopy transgenic mice in which RNA and protein levels could be measured, direct correlations were observed.28-30 For this reason, we feel that this explanation for these results is unlikely.
Finally, it is possible that there is a narrow “window” of PML-RARα expression in early myeloid cells that is optimal to cause the changes that ultimately lead to the development of APL. If levels of this protein are too high in these cells, they may die or become disabled, so that they cannot contribute to APL development. A larger pool of cells expressing a smaller amount of protein may make it more likely that the critical “hits” needed for APL progression may occur in a larger fraction of mice. The slightly decreased latency seen with 2 copies of PML-RARα may reflect a narrow dose response for this protein, perhaps by increasing the proportion of early myeloid cells that are susceptible for secondary transforming events. These observations are supported by observations from Grignani et al31 and Ferrucci et al32 that have shown that PML-RARα can be toxic to hematopoietic cells when overexpressed. Furthermore, in a recent study from Minucci et al,33 splenic APL tumors derived from cells transduced with a retrovirus expressing PML-RARα and an IRES-GFP cassette yielded no detectable green fluorescent protein (GFP)–positive cells. These results support the hypothesis that cells expressing high levels of PML-RARα may be deleted in vivo.
An interstitial deletion of chromosome 2 was detected in 2 of 9 APL tumors obtained from the knock-in mice. This frequency is similar to that detected in transgenic hCG–PML-RARα mice (1 of 5) and in MRP8–PML-RARα mice (3 of 30),10 but it is significantly different (P < .05) from that seen in transgenic hCG–PML-RARα × hCG–RARα-PML mice, in which 11 of 13 tumors contained del(2).9 These results show that a high-penetrance model of APL is not necessarily associated with a high frequency of del(2) during progression. The data also support the hypothesis that it is truly the expression of the RARα-PML cDNA that facilitates the acquisition of del(2) in hCG–PML-RARα × hCG–RARα-PML mice.9 Collectively, these results suggest that there are many kinds of genetic events that can contribute to APL progression in the mouse model and that del(2) is simply one of the most obvious and frequently detected at the whole chromosome level.
The results presented in this study indirectly address the hypothesis that PML-RARα contributes to the pathogenesis of APL by acting predominantly as a dominant-negative molecule for PML and RARα. In vitro, PML-RARα can clearly act as a dominant-negative factor to inhibit both PML and RARα function when overexpressed.12-14 Furthermore, PML haploinsufficiency in hCG–PML-RARα mice appeared to increase the likelihood of APL development, suggesting that PML loss-of-function might somehow “cooperate” with PML-RARα, perhaps by enforcing a dominant-negative signal.18 However, the results presented here suggest that low levels of PML-RARα expression are more efficient at producing APL than higher levels. The data are more consistent with the hypothesis that there is an optimal pathogenetic PML-RARα “dose” that produces disease. Expression levels that are too low result in no disease, whereas levels that are too high may result in toxicity and select against the expressing cells. Levels that are most appropriate lead to alterations in expressing cells that make it more likely that they will acquire the critical progression mutations that lead to the development of APL. Further experiments designed to rigorously test this hypothesis are in progress.
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2002-12-3779.
Supported by National Institutes of Health grants CA83962 (T.J.L.), HL0399102 (P.W.), and T32 HLO 7088 (A.A.L.), and by the Buder Charitable Foundation (T.J.L.).
P.W. and A.A.L. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Kelly Schrimpf for blastocyst injections and Mieke Hoock for excellent mouse colony management. Elaine Ross and Jacque Mudd provided excellent assistance with the ES targeting work. Nancy Reidelberger provided expert editorial assistance.

![Figure 1. Targeting PML-RARα to the murine cathepsin G locus by homologous recombination and generation of transgenic mice. (A) A targeting vector consisting of a PML-RARα cDNA and PGK-neo selectable marker cassette, flanked by targeting arms from the murine cathepsin G (mCG) locus were transfected into embryonic stem cells. Coding exons of the mCG gene are represented as black boxes (not to scale). The PML-RARα cDNA and PGK-neo selectable marker cassette are represented as open boxes. Transcriptional start sites for the mCG gene and PGK-neo selectable marker cassette are indicated by arrows. The mCG 5′ untranslated region is represented by a gray box. LoxP sites used for CRE recombinase-mediated cassette excision are represented as shaded ovals. The position of an external 3′-mCG DNA probe is indicated as a hatched box under mCG exons 3 and 4. The PGK-neo cassette was subsequently removed from a homologously recombinant ES cell clone by transient transfection of targeted ES cells with a Cre recombinase expression vector. (B) The targeted mCG locus was identified in ES cells by a size shift from 3.2 kb (wild type [WT]) to 7.9 kb (targeted, +PGK-neo) following HindIII digestion of genomic DNA. Removal of the PGK-neo cassette by Cre recombinase resulted in a shift in the size of the recombinant band to 6.4 kb (a loading artifact caused the slight apparent differences in the sizes of the WT mCG bands at 3.2 kb). (C) Tail DNA from the offspring a heterozygous CGPR/+ intercross was screened by Southern blotting to identify wild-type (CG+/+), heterozygous (CG+/PR), and homozygous (CGPR/PR) animals. (D) Tail DNA from the offspring of CGPR/PR × CG+/– animals was screened by Southern blotting to identify animals that carried one PML-RARα allele together with either an intact CG gene (CG+/PR) or a CG null mutation (CG–/PR) at the other allele.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2002-12-3779/6/m_h81734865001.jpeg?Expires=1769095101&Signature=pOwPwTHhlqMRPP1HHtzDVeGYys~e9zAu~NK7slBtQD0kgNYkpoSRPioCuo6zwV9cwf8hyMEkFBKjyfFpHx9MyDs9f2gKXTz-mPXwtoRmtmm~lWKyzmD9gKVUVdoBLtTa6jebfm2ZLxE8cJ4v2pE7lQSoSqhAKMz45VdlpB1kC1gZxYZh9H-Twq1sjnPSTU-8qSV6lHMitEZGoCz5wHYPxFwF9RJLVtUDNJKgzf~mtkOH-jx3O85CEQdI3WJP0CGhbYGrHiR13Hwy9Vs613iYMPBLHCK6Qbz8lXOkcdz-8UPPCVo8A~GOWvtSPJXoMpfdaLxMzhqE00uyWLqQYKag9A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal