Abstract
Integrins are cell adhesion receptors that communicate biochemical and mechanical signals in a bidirectional manner across the plasma membrane and thus influence most cellular functions. Intracellular signals switch integrins into a ligand-competent state as a result of elicited conformational changes in the integrin ectodomain. Binding of extracellular ligands induces, in turn, structural changes that convey distinct signals to the cell interior. The structural basis of this bidirectional signaling has been the focus of intensive study for the past 3 decades. In this perspective, we develop a new hypothesis for integrin activation based on recent crystallographic, electron microscopic, and biochemical studies.
Introduction
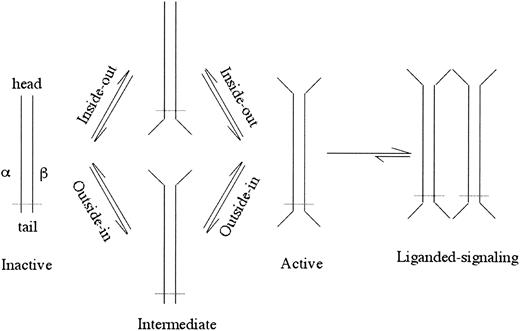
Integrins are αβ heterodimeric cell surface receptors, about 24 nm long, that assemble into a ligand-binding “head” sitting on 2 membrane-spanning legs.1,2 Integrins require activation to bind physiologic ligands (for a review, see Hynes3 ). Activation is normally induced from within the cell through the cytoplasmic and transmembrane (TM) regions (“inside-out” activation)4 and can be mimicked by deleting the TM or cytoplasmic segments (for a review, see Arnaout5 ; Figure 1). In this case, activating intracellular signals transition the integrin through conformational changes to the active state.6 This process is fast (< 1 second), temperature dependent,7 and reversible.8,9 Activation is also associated with rapid (within seconds) changes in the lateral mobility of integrins in the plasma membrane that act in concert with affinity modulation to regulate cell adhesion.10,11 Binding energetics suggest that once bound to a physiologic ligand, dissociation is energetically expensive.7 High-affinity ligand-mimic peptides,12,13 Mn2+,14 certain monoclonal antibodies (mAbs), and gain-of-function mutations in the extracellular segment (for a review, see Arnaout5 ) can mimic inside-out activation (outside-in activation; Figure 1), reflecting a dynamic link between the integrin's head and tails. A classic example of this conformational coupling is seen in electron microscopic images of an activating mAb (LIBS2) to the membrane proximal integrin leg, which causes physiologic ligand binding to the head some 16 nm away.15 The atomic basis for this bidirectional and reversible allosteric activation has remained enigmatic. New crystallographic, electron microscopic, and biochemical studies that are providing new potential solutions to this problem are reviewed here.
A schematic of inside-out or outside-in integrin activation. Opening of the head or legs/feet opens the other end of the integrin perhaps through an intermediate state(s). The ligand-occupied active integrin causes further conformational changes resulting in clustering and cell signaling. Dotted line depicts the plasma membrane.
A schematic of inside-out or outside-in integrin activation. Opening of the head or legs/feet opens the other end of the integrin perhaps through an intermediate state(s). The ligand-occupied active integrin causes further conformational changes resulting in clustering and cell signaling. Dotted line depicts the plasma membrane.
Surprising findings in the crystal structure of integrin αVβ3
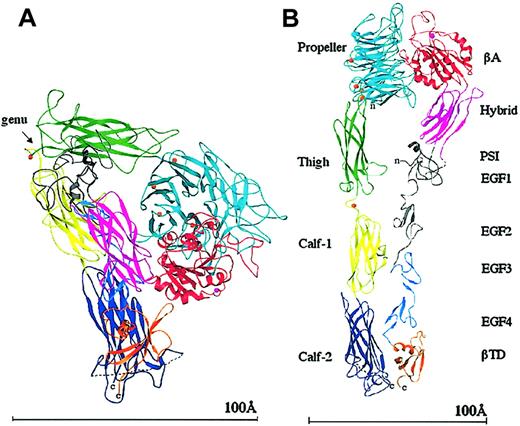
It was a big surprise when the crystal structure of the extracellular (ie, lacking the transmembrane and cytoplasmic tails) segment of integrin αVβ3 was found to be V-shaped16 (Figure 2), rather than being linear and extended as depicted in electron microscopic images. The 4-domain αV subunit and the 8-domain β3 subunit are acutely bent some 135 degrees from the linear, with the bending found in the legs, between the thigh and calf-1 domains of αV and about the second and third epidermal growth factor (EGF) repeats of β3 (at the integrin α and β knees, respectively; Figure 2). One interpretation of the severe bending of the integrin is that it is a crystal artifact.17 Another interpretation is that the bent conformation represents a constitutive and regulatable form of the integrin.16 This view is supported by electron microscopic images of negatively stained native αIIbβ3 on the cell surface,2 and more recently by electron microscopic data showing that extracellular αVβ3 is bent in solution.18 A second surprising finding in the structure was the striking resemblance in the tertiary and quaternary arrangements of the integrin head to G-proteins; the integrin head is formed by a β propeller from αV and a VWFA Rossmann fold (βA) from β3 (βA is similar to the ligand-binding αA- or αI domain found earlier in the α subunits of 9 integrins19,20 ). The integrin's βA and propeller domains were found to be the structural equivalents of the nucleotide binding (Gα) and propeller (Gβ) domains of G-proteins. This striking resemblance suggested that integrin activation might, like G-proteins, also involve tertiary and quaternary changes in the head region. The third surprising finding was that the metal ion-dependent ligand-binding site (MIDAS) of the βA domain was unoccupied by a metal ion due to an invariant glutamate side chain that partially occupies this site; instead, a metal ion was found at an adjacent site to MIDAS (ADMIDAS), linking the α1 and α7 helices of βA. The lack of a MIDAS cation and the presence of an ADMIDAS cation in βA contrasts with the situation in the integrin-αA domain, which exists in inactive “closed” and liganded “open” conformations,20,21 both with a metal ion at MIDAS but lacking the ADMIDAS coordination site.
Ribbon diagram of the structure of the unliganded extracellular αVβ3. The protein is bent at a flexible region (the genu, arrow; A). When extended at the genu,16 the structure assumes the more familiar shape seen previously by electron microscopy (B). The 12 domains are labeled. The PSI, EGF1, and EGF2 domains are not visible (indicated in gray); the tracing shown for PSI is approximate and the translated EGF3 and EGF4 domains have been used to show the approximate location of EGF1 and EGF2. The 4 metal ions (orange spheres) at the bottom of the propeller, the metal ion at the genu (orange), and the ADMIDAS ion (magenta) are shown. The dotted line in calf-2 represents the disordered loop containing the proteolytic cleavage site. A short dotted line also connects PSI and the hybrid domains, visible in panel B. The “n” and “c” indicate the amino and carboxyl terminus, respectively. Reprinted from Arnaout et al58 with permission.
Ribbon diagram of the structure of the unliganded extracellular αVβ3. The protein is bent at a flexible region (the genu, arrow; A). When extended at the genu,16 the structure assumes the more familiar shape seen previously by electron microscopy (B). The 12 domains are labeled. The PSI, EGF1, and EGF2 domains are not visible (indicated in gray); the tracing shown for PSI is approximate and the translated EGF3 and EGF4 domains have been used to show the approximate location of EGF1 and EGF2. The 4 metal ions (orange spheres) at the bottom of the propeller, the metal ion at the genu (orange), and the ADMIDAS ion (magenta) are shown. The dotted line in calf-2 represents the disordered loop containing the proteolytic cleavage site. A short dotted line also connects PSI and the hybrid domains, visible in panel B. The “n” and “c” indicate the amino and carboxyl terminus, respectively. Reprinted from Arnaout et al58 with permission.
A switchblade model of integrin activation
The discovery of the bent integrin conformation together with previous electron microscopic images of an extended form suggested that, as in the case of αA, interconversion between these 2 conformations may have important functional consequences.16 In one model, it was proposed that in response to an inside-out signal, integrins extend at the knees, snapping from the bent structure that is inactive to the straight active conformation, like the opening of a pocket knife (switchblade or jack-knife model).18 Inactivity of the bent form was inferred from the fact that it was produced in the presence of the generally inhibitory cation Ca2+ and from predictions that the head of the integrin would face down toward the membrane in this conformation and is therefore unable to bind ligands.22 Evidence has been gathered in support of the 2-state “switchblade” model. First, artificially constrained recombinant integrins, where a disulfide bridge clasps the membrane-proximal α and β legs together, bind immobilized ligand only when unclasped23 ; in the bent crystal structure, the α and β legs are very close together, potentially hampering access to activating mAbs.22 Second, estimation of the Stokes radius of extracellular αVβ3 by molecular sieve chromatography revealed that in buffers containing Ca2+ or Mn2+, the Stokes radius is 5.7 nm and 6.0 nm, respectively, and the respective electron microscopic images revealed a bent conformation except for a minority of the Mn2+-bound molecules (∼20%) that were extended. When a cyclic RGDfV (Arg-Gly-Asp-[D]Phe-Val) pentapeptide was added in Ca2+ or Mn2+ buffers, the Stokes radius shifted to about 6.4 nm and the vast majority of the molecules were now seen extended by electron microscopy. Third, a double mutation designed to hold the head against the legs via a cysteine bridge resulted in an inactive integrin unless the receptor was reduced with dithiothreitol (DTT).18
Crystal structures of αVβ3 in Mn2+ and in complex with RGD
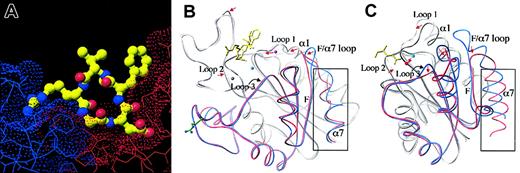
In the meantime, we determined 2 additional crystal structures of extracellular αVβ3. One was generated by cocrystallization in the presence of 5 mM Mn2+,24 and the second after allowing the high-affinity ligand mimetic cyclic (RGDf[N-Me]V) to diffuse into existing αVβ3 crystals grown in the presence of Mn2+.24 The former structure was virtually identical to that generated earlier in the presence of Ca2+, including the lack of a MIDAS cation in βA and the presence of a cation-occupied ADMIDAS. In the αVβ3-RGD structure, RGD occupies a shallow crevice between the propeller and βA domains in the integrin head (Figure 3A), with the arginine and aspartic acid residues exclusively contacting the propeller and βA domains, respectively. RGD binding is associated with tertiary changes, mostly affecting βA, and small quaternary changes in the propeller/βA interface. The tertiary changes reshape the 3 loops surrounding the βA MIDAS (Figure 3B). These changes are triggered by a similar movement of the N-terminal α1 helix to that seen when αA is liganded (compare panels B and C in Figure 3). In βA, this movement is accompanied by 2 major changes. (1) Due to its shift toward the RGD ligand, the coordination of ADMIDAS cation changes; the F-α7 loop flips so that this cation no longer interacts with the top of the C-terminal α7 helix, thus severing the ionic bond between α1 and α7 helices. (2) The invariant glutamic acid side chain from loop 2, which prevents occupation of MIDAS in unliganded αVβ3, changes its position to coordinate a third metal ion at a novel ligand-associated metal ion-binding site (LIMBS). Displacement of this glutamate now allows for occupation of MIDAS with a metal ion, that is, the βA MIDAS only binds a metal ion in the presence of the high-affinity ligand mimic. This contrasts with the situation in αAwhere MIDAS is metal bound even in the inactive “closed” conformation. Ligand-dependent occupation of the βA MIDAS is consistent with recent biochemical analysis of 45Ca binding to integrin α4β1, which identified a moderate affinity (median effective dose [ED50] about 50-100 μM) site seen only when a high-affinity ligand is present.25 In isolated αA, movement of the α1 helix reorients the metal ion at MIDAS and the flip in the F-α7 loop leads to a major 10-Å downward movement of the α7 helix (Figure 3C), not seen in the liganded βAstructure (Figure 3B). The functional end result, however, is essentially the same; the metal ion at MIDAS can now be ligated by an acidic residue from ligand. Similar changes in metal coordination, associated with the conversion of the molecule from the inactive to the active state, are found in G-proteins; these are also linked to large tertiary changes in 2 switch regions.26 But these changes are not accompanied by movement of the C-terminal helix, as in βA, indicating that this movement is not essential for switching of the homologous G-proteins to the active state.
The ligand binding site of αVβ3 and the unliganded and liganded conformations of βA and αA domains. (A) Surface representation of the RGD ligand-binding site in the head section of αVβ3. The ligand arginine-binding pocket is located in the αV propeller (blue) and the aspartic acid–binding pocket in βA (red). The ligand peptide is shown as a ball-and-stick model (carbons are shown in green, amides in blue, and oxygens in red). (B) Superposition of the β3A domain in the unliganded and liganded conformations. Ligand is in yellow. The 3 metal ions present in the liganded form (LIMBS, MIDAS, and ADMIDAS) are shown in gray, cyan, and magenta, respectively. (C) Superposition of CD11b A domain in the “closed” and “open” forms. Ligand is in yellow. The MIDAS cation is in cyan. In both panels B and C, the C-terminal βαβαβα structures are highlighted in color (blue for unliganded and red for liganded), and the remaining parts of the molecules are in gray. The C-terminal α helix (α7) is boxed. In panel B, superpositions are generated for the whole molecule with TOP,59 whereas in panel C, superpositions are based on the central β sheet only.
The ligand binding site of αVβ3 and the unliganded and liganded conformations of βA and αA domains. (A) Surface representation of the RGD ligand-binding site in the head section of αVβ3. The ligand arginine-binding pocket is located in the αV propeller (blue) and the aspartic acid–binding pocket in βA (red). The ligand peptide is shown as a ball-and-stick model (carbons are shown in green, amides in blue, and oxygens in red). (B) Superposition of the β3A domain in the unliganded and liganded conformations. Ligand is in yellow. The 3 metal ions present in the liganded form (LIMBS, MIDAS, and ADMIDAS) are shown in gray, cyan, and magenta, respectively. (C) Superposition of CD11b A domain in the “closed” and “open” forms. Ligand is in yellow. The MIDAS cation is in cyan. In both panels B and C, the C-terminal βαβαβα structures are highlighted in color (blue for unliganded and red for liganded), and the remaining parts of the molecules are in gray. The C-terminal α helix (α7) is boxed. In panel B, superpositions are generated for the whole molecule with TOP,59 whereas in panel C, superpositions are based on the central β sheet only.
As suggested,24 the structural changes seen in the αVβ3-RGD complex may correspond to outside-in activation of membrane-bound12,13 or recombinant extracellular integrins27 induced by short RGD ligand mimetic peptides. These induce binding of physiologic ligands in the integrin head and expression of neoepitopes in the legs.15,28,29 Biochemical studies using mAbs and limited proteolysis identified the α1 helix of βA and the preceding and adjacent ligand-specificity loops,25,30-34 the propeller/thigh and propeller/βA interfaces,35,36 and the disordered PSI/hybrid/EGF1 interface35,37 as regions that become exposed only in the active state (for a review, see Arnaout5 ). The congruence of the activation-sensitive regions as defined biochemically in integrins activated from “inside-out” and crystallographically by comparing the structures of the unliganded and liganded αVβ3 suggests that the tertiary changes induced by RGD in our crystals mirror those induced by inside-out activation. The switchblade model argues based on comparisons of negatively stained electron microscopic images of extracellular αVβ3,18 that in addition to these changes, there is a downward movement of the C-terminal helix of βA in the liganded structure, which is prevented by crystal contacts.
The missing link: the integrin “ankles” or TM segments
The impact of the TM segment on protein movements leading to activation is yet to be determined, and it is in this context that the switchblade model faces some challenges. In a recent study, the 20-Å cryoelectron microscopic structure of the native inactive αIIbβ3 was described.38 Although the electron microscopic images are not consistent with the bent structure of the crystallized protein, they do show a rather collapsed structure that is perhaps more similar to, and retains features of, the crystallized protein. In the “best fit” model, the membrane-proximal calf-2 domain is almost parallel to the plasma membrane; thus the ligand-binding face is facing away from the membrane and able to potentially engage ligands. In this structure, movements at additional “joints” besides the knees are necessary for the integrin to snap to the extended linear form.38 Second, inspection of detailed mAb epitope maps of active and inactive αIIbβ3 on platelet membranes led to the conclusion that both states can assume a compact form consistent with the bent conformation, that is, the structure does not need to linearize to become active.39 The inactivating effect of “freezing” an integrin in its bent unliganded state by artificial disufides linking the head and legs, which supports the switchblade model, may not be that surprising; this modification probably blocks the tertiary and quaternary changes needed for activation that occur even in protein crystals. It may thus well be that extension at the knees is not a necessary feature of activation per se, but a postbinding event linked to outside-in signaling. Activation and extension can be dissociated; extracellular α5β1 tied at its legs assumed a straight conformation by electron microscopy yet was inactive.23 Also, the binding of cyclic peptide ligand mimics to αIIbβ3 in intact platelets triggers different conformational changes in the receptor, as reflected by expression of different LIBS epitopes,40 suggesting that various ligands initiate distinct functional consequences within the receptor. Varying degrees of extension at the knees may provide the structural basis for these findings.
Inside-out activation of the integrin heterodimer: the “deadbolt” model
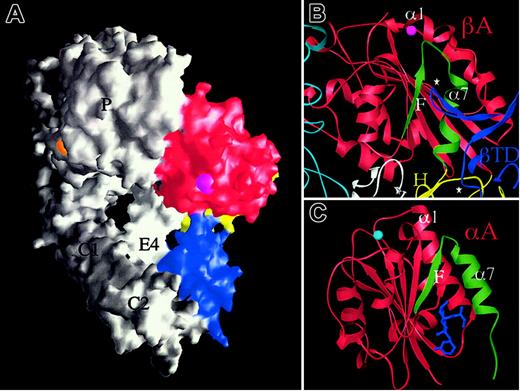
If extension at the knees is a postbinding “outside-in” signaling event, and the bent structure can be activated (stabilized) by high-affinity RGD, then how is inside-out activation triggered? The answer is currently unknown, but the bent crystal structure may offer yet another intriguing clue. The F/α7 loop is the region in βA that undergoes the most dramatic conformational change on RGD binding (Figure 3B). This loop contacts the CD loop of the βTD in unliganded αVβ3 (Figure 4A-B) and this contact is lost in the liganded structure. The contact region covers a very small surface area (∼64 Å2 ) in unliganded αVβ3 and the βTD loop has high temperature factors. Thus this contact does not contribute much stabilizing energy to the bent form in the crystal structure. The same side of the βTD (α1/A loop) also makes a small contact (∼270 Å2 of surface area) of a mixed nature with the hybrid domain that may stabilize the βTD/βA contact. The close proximity of the βTD and βA domains may produce a more substantial contact following minor rearrangement of the α and β subunits and their domain interfaces in the native membrane-bound structure. In the structurally related αA domains, integrin affinity can be regulated allosterically by modulating the structure of the F/α7 region.41,42 In addition, lovastatin, a drug that binds at the lower part of the F/α7 interface (Figure 4C), has been shown to lock the αA domain from integrin CD11a in the inactive state, by preventing the tertiary changes in its C-terminal α7 helix and preceding F/α7 loop needed for activation.43 We suggest that the elongated CD loop of the βTD may serve a similar function, acting as a regulatable “deadbolt” that locks βA in the inactive state in the native structure. In the latter state, the deadbolt would be more firmly engaged than observed in the crystallized protein, thus preventing the flip of the F/α7 loop required for the activating movement of the α1 helix. Inside-out activation transmitted through the integrin cytoplasmic tails4,44 may lead through pistonlike, seesaw,4 sliding,45 or rotational46 movements of the TM helices to unlocking of the adjacent βTD “deadbolt.” The deadbolt would slide away from βA, rendering it ligand competent. If true, one would not expect to see an extensive contact in an active integrin (or one where the cytoplasmic tails and TM segments are removed47 as in the crystallized protein) because the deadbolt is not fully inserted into its βA “lock.” Perhaps this is why the CD loop of βTD has high temperature factors; some mobility might help it re-engage the βA domain to reversibly inactivate the integrin, a motion of only 0.3 nm. A model, where the βTD and the βA domains interact over a couple of angstroms via the βTD “deadbolt” to maintain the inactive state, may act as the key that allows intracellular or membrane proximal signals to effect subjectively long range activation at the βA domain. A particularly attractive feature of this deadbolt model for reversible inside-out activation in a membrane-bound integrin is that this mechanism seems more efficient than evoking a snap opening of the molecule into the linear form. It also incorporates the unique and hitherto unexplained features of the βA domain; the ADMIDAS cation is situated right at the top of the F/α7 loop and could participate in stabilizing this loop through an ionic bridge to the α1 helix in the inactive native state. The deadbolt model does not exclude a subsequent physiologic ligand-induced switchblade movement that could be key to outside-in signaling.
The deadbolt model of inside-out integrin activation. (A) Surface representation of unliganded αVβ3 in which the βTD (blue), hybrid (yellow), and βA (red) domains are colored and the propeller (P), thigh (T), calf-1 and 2 (C1, C2), and EGF4 (E4) are labeled. The cations at ADMIDAS (magenta) and blade 6 (orange) are visible in this view. (B) Ribbon diagram showing the 2 βTD loops (⋆, blue) that contact βA (red) and the hybrid (yellow) domains. The long CD loop of the βTD contacts the top of the activation-sensitive F/α7 of βA (colored in green). The ADMIDAS cation links this loop to the top of the α1 helix in the unliganded state. Parts of the propeller (cyan), hybrid (yellow), and EGF4 (white) are shown. (C) Ribbon diagram of the inactive form of αA from integrin CD11a in complex with the allosteric inhibitor lovastatin (blue) showing the position of this drug in relation to the F/α7 region (colored in green). The MIDAS ion is in cyan. There is no ADMIDAS in this or other known αA domains.
The deadbolt model of inside-out integrin activation. (A) Surface representation of unliganded αVβ3 in which the βTD (blue), hybrid (yellow), and βA (red) domains are colored and the propeller (P), thigh (T), calf-1 and 2 (C1, C2), and EGF4 (E4) are labeled. The cations at ADMIDAS (magenta) and blade 6 (orange) are visible in this view. (B) Ribbon diagram showing the 2 βTD loops (⋆, blue) that contact βA (red) and the hybrid (yellow) domains. The long CD loop of the βTD contacts the top of the activation-sensitive F/α7 of βA (colored in green). The ADMIDAS cation links this loop to the top of the α1 helix in the unliganded state. Parts of the propeller (cyan), hybrid (yellow), and EGF4 (white) are shown. (C) Ribbon diagram of the inactive form of αA from integrin CD11a in complex with the allosteric inhibitor lovastatin (blue) showing the position of this drug in relation to the F/α7 region (colored in green). The MIDAS ion is in cyan. There is no ADMIDAS in this or other known αA domains.
Many of the existing data supportive of the switchblade model on inside-out activation may in fact be more consistent with the deadbolt model. A part of the βTD/hybrid interface is exposed when αIIbβ3 is activated; Asp393, a contact residue at the βTD/hybrid interface becomes accessible to Asp-N protease only in the active state.35 Conformational changes at the βTD/hybrid interface may act in the bent structure to unlock the deadbolt. Second, epitopes of several activating mAbs map at or adjacent to the βTD/βA interface. For example, the activating anti-β1 mAb JB1B involves the βTD CD loop of the deadbolt48 ; the epitopes of the activating anti-β1 mAbs HUTS-4, HUTS-7, HUTS-21,49 and 15/750 involve the adjacent βTD contact with the hybrid domain. The activating anti-β2 mAb, KIM185, maps to EGF4 and the N-terminus of the βTD.51 The βTD of β3 contains several epitopes for activating mAbs, including those for LIBS2, LIBS3, and LIBS6.15 In our model, these mAbs would also act by unlocking the deadbolt without necessarily straightening the integrin. Third, 4 naturally occurring activating mutations are now known in αIIbβ3: Thr562Asn52 and Cys560Phe53 (both in EGF3); and Gly579Ser53 and Cys598Tyr54 (both in EGF4). Thr562 is the last residue in EGF3; its replacement with asparagine permits an aberrant glycanation at this site, which may disrupt the EGF3/4 interface and thence the βTD/βA interface. Cys560 participates in the interdomain disulfide bridge linking EGF3 and 4; loss of this cysteine will likely have an equivalent structural effect to that caused by Thr562Asn. Cys598 forms an intradomain disulfide bond, linking the major and minor strands of EGF4; interruption of this linkage may also loosen/disrupt the βTD/βA interface. Fourth, deletion of the transmembrane/cytoplasmic segments activates integrins (for a review, see Humphries et al55 ). This is expected to loosen the grip of the deadbolt, as seen in the tailless structure of αVβ3. Finally, the rare activating anti-αIIb mAb PMI-1 binds to calf-2,56,57 in a region adjacent to the βTD and could also function in part by unlocking the deadbolt.
Because integrins are emerging therapeutic targets in many human pathologies, the ability to recognize and to target the appropriate molecular form, and to ensure this does not trigger a potentially harmful activating switch, has become a central focus of therapy. The deadbolt model predicts a defined region in the native integrin that normally locks it in a default inactive state. Drugs mimicking the deadbolt mechanism may achieve the long-sought goal of inactivating an integrin by preventing the activation switch altogether, thus avoiding potentially serious side effects.
Prepublished online as Blood First Edition Paper, April 24, 2003; DOI 10.1182/blood-2003-01-0334.
Supported by grants from the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Disease, National Institute of Allergy and Infectious Diseases, and National Heart, Lung and Blood Institute).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal