Abstract
Central to the pathophysiology of sickle cell disease are the vaso-occlusive events that lead to tissue damages and life-threatening complications. Lungs are particularly vulnerable to vaso-occlusion because of their specific vasculature. We developed a mouse model of hypoxia/reoxygenation lung injury closely mimicking the lung pathology of patients with sickle cell disease. This model involves the exposure of transgenic sickle cell (SAD) mice to hypoxia (8% oxygen) for 4, 10, and 46 hours followed by 2 hours of reoxygenation. Gene expression profiling of SAD lung tissue pointed to the specific induction of genes involved in the response to ischemic stress and microcirculation remodeling: Hspcb, Hsp86-1, Nfe2l2, Ace, and Fgf7. Hypoxia/reoxygenation also induced a marked increase in bronchoalveolar (BAL) total leukocyte and neutrophil counts, BAL total protein content, and BAL tumor necrosis factor α (TNF-α), interleukin 6 (IL-6), IL-1α, and macrophage inflammatory protein 2 (MIP-2) levels, all indicators of enhanced inflammatory response as compared with control mice. Nitric oxide (NO) was administered to SAD mice. NO (40 ppm) inhalation protected SAD mice from the histopathologic lesions of ischemic/reperfusion lung injury with corresponding normalization and/or modulation of tissue gene expression profiles. Inhaled NO (1) significantly reduced the increase in BAL total protein content, BAL total leukocyte, and neutrophil counts; (2) modulated BAL cytokine network; and (3) did not affect hemoglobin and methemoglobin levels. The present study provides evidences for the beneficial effects of inhaled NO in pulmonary injury induced by hypoxia/reoxygenation in a mouse model of sickle cell disease (SCD) and opens new avenues in drug design based on tissue gene expression profiling.
Introduction
Sickle cell disease (SCD) is caused by a single point mutation in codon 6 of the human β-globin gene that results in the substitution of valine for glutamic acid. In homozygotes, the abnormal hemoglobin (Hb) [HbS
Because many cellular and molecular events participate in the pathophysiology of the vaso-occlusive crises, it has been difficult to distinguish causative from secondary mechanisms and, therefore, devise effective therapies. Vaso-occlusive events in the microcirculation may result from the interactions between plasma factors and different cell types, including dense, dehydrated sickle cells, reticulocytes, endothelial cells, leukocytes, and platelets.4,5 For instance, endothelial cells are abnormally activated, displaying increased expression of adhesion molecules in association with a procoagulant phenotype.1-5
Transgenic mouse models of SCD offer the opportunity to study the pathophysiology of SCD and devise new therapeutic strategies.6-12 The vaso-occlusive events of human SCD can be mimicked in sickle cell mouse models by exposure to hypoxia followed by reoxygenation.6,7,12-17 The SAD mouse model of sickle cell disease expresses a “super” hemoglobin S (Hb SAD) that results in a mild sickle cell syndrome characterized by erythrocyte sickling, dehydration, and organ damages typical of SCD.6,7 On severe hypoxia (6% oxygen), death occurs for all SAD mice but not for control mice,6,7 and the rate of survival to lethal hypoxia has been reported to be improved by antisickling agents6 or by inhaled nitric oxide (NO).16
In this study, we have developed a mouse model of ischemic/reperfusion lung injury that closely mimics the pathologic changes described in patients with SCD. We then used membrane cDNA microarrays to analyze the pattern of gene expression in whole lung tissue of SAD mice with or without having been submitted to hypoxia/reoxygenation. Gene expression profiling of lung tissue pointed to the specific induction of genes involved in the response to ischemic stress and microcirculation remodeling. Because NO is a potent vasodilator and inhibitor of vascular remodeling and also affects the multistep cascade events involved in leukocyte, platelets, and endothelial activation,18-35 we have investigated whether inhaled NO, which diffuses directly from alveoli to pulmonary vascular smooth muscle, could prevent lung injury consecutive to pulmonary ischemic/reperfusion damage and studied its mechanism(s) of action.
Materials and methods
Drugs and chemicals
NaCl, KCl, ouabain, and bumetanide were purchased from Sigma Chemical (St Louis, MO). MgCl2, Mg(NO3)2, dimethylsulfoxide (DMSO), and n-butyl phthalate were purchased from Fisher Scientific (Hampton, NH). Sulfamic acid (SFa), Tris(hydroxymethyl) aminomethane (Tris) and 3(N-morpholino) propanesulfonic acid (MOPS) were purchased from Sigma Chemical. Choline chloride was purchased from Calbiochem-Boehring (San Diego, CA). Bovine serum albumin fraction V was purchased from Boehringer Mannheim (Mannheim, Germany). All solutions were prepared using double-distilled water. Atlas Mouse cDNA Expression Array Membranes were purchased from Clontech Laboratories (Palo Alto, CA; catalog no. 7741-1).
Animals and experimental design
Transgenic Hbbs/Hbbs SAD mice and C57B6/2J control mice were used (male and female, 25-20 g body weight).6,7,36 C57B6/2J mice were divided into 4 groups of 6 mice each. One group was considered as baseline in normoxia, the other mouse groups were exposed to 4, 10, or 46 hours hypoxia (8% oxygen) followed by 2 hours normoxia. SAD mice were divided into 7 groups of 6 mice each. One group was maintained in normoxia and used as baseline. The other SAD mouse groups were exposed at 4, 10, or 46 hours hypoxia (8% oxygen) with or without inhaled NO at 40 ppm followed by 2 hours reoxygenation (21% oxygen) with or without inhaled NO at 40 ppm. The choice of NO 40 ppm was based on preliminary dose-response experiments showing the absence of significant changes in methemoglobin levels (data not shown).
The gas mixture was blended using separately regulated and calibrated mass flowmeters (RDM 280; Airliquid, Paris-La-Defense, France) with high fresh gas flows of air, nitrogen, and oxygen (RDM 280; Airliquid) maintaining a gas flow constant at about 1.5 L/minute through a 5-L exposure cage. The FiO2 was kept constant at 0.21 in baseline experiments and 0.08 in hypoxia experiments using a polarographic electrode (oxygen analyzer; Ohemda 5120, Englewood, CO). NO concentration was monitored in the effluent gas by NO-NO2 analyzer (model Polytron; Dräger, Lübeck, Germany) calibrated with a 90-ppm NO gas tank (Airliquid). NO was maintained at 40 ppm, and serial measurements revealed NO2 levels 2% or less. Mice were given free access to water and food.
Lung tissue histology
Lungs were removed from mice; one lung was immediately frozen in liquid nitrogen, and the other was fixed in formalin and embedded in paraffin. Multiple (at least 5) 3-μm whole mount sections were obtained by each paraffin embedded lung and stained with hematoxylin eosin, Masson trichome, and May-Grünwald-Giemsa. Two pathologists blindly performed the analysis that consisted in the evaluation of the tissue architecture and changes for wild-type mice under hypoxia and SAD mice under hypoxia with or without inhaled NO. The pathologic criteria for quantification of the changes observed in vessels and bronchi are detailed in Table 1. Briefly, the vessels were evaluated for the presence and entity of congestion, constriction, and thrombi, whereas the bronchi were evaluated for the presence and entity of mucus and inflammatory cell infiltrate. The latter quantification was expressed as the mean number of cells per field at magnification of × 250, as resulting by the analysis of at least 4 different fields on each hematoxylin-eosin–stained whole lung section using an Olympus Provis AX 70 (Olympus, Paris, France) microscope equipped with a wide-field eye piece number 26.5.
Lung pathology of wild-type and SAD mice exposed to hypoxia/reoxygenation and effects of inhaled nitric oxide (NO)
. | Wild type + hypoxia . | . | . | . | SAD + hypoxia . | . | . | . | . | SAD + hypoxia + NO . | . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | 1 . | 2 . | 3 . | 4 . | 1 . | 2 . | 3 . | 4 . | 5 . | 1 . | 2 . | 3 . | 4 . | ||||||||||
| Vessels | |||||||||||||||||||||||
| Congestion | + | + | 0 | + | +++ | +++ | ++ | +++ | +++ | + | + | + | + | ||||||||||
| Constriction, μm | 0.027±0.009 | 0.08±0.01* | 0.025±0.005† | ||||||||||||||||||||
| Thrombi | 0 | 0 | 0 | 0 | + | + | + | 0 | + | 0 | 0 | 0 | 0 | ||||||||||
| Bronchus | |||||||||||||||||||||||
| Mucus | 0 | 0 | 0 | 0 | ++ | ++ | ++ | ++ | ++ | 0 | 0 | 0 | 0 | ||||||||||
| Inflammatory cells | 24.0±2.9 | 40.5±4.7* | 25.2±8.8 | ||||||||||||||||||||
| Lymphocyte | 12.5±3.7 | 19.8±1.9 | 15.5±6.6 | ||||||||||||||||||||
| Macrophage | 9.5±3.1 | 13.1±3.0 | 7±2.1 | ||||||||||||||||||||
| Neutrophil | 2.8±1.3 | 6.9±1.0* | 3.1±1.2 | ||||||||||||||||||||
. | Wild type + hypoxia . | . | . | . | SAD + hypoxia . | . | . | . | . | SAD + hypoxia + NO . | . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | 1 . | 2 . | 3 . | 4 . | 1 . | 2 . | 3 . | 4 . | 5 . | 1 . | 2 . | 3 . | 4 . | ||||||||||
| Vessels | |||||||||||||||||||||||
| Congestion | + | + | 0 | + | +++ | +++ | ++ | +++ | +++ | + | + | + | + | ||||||||||
| Constriction, μm | 0.027±0.009 | 0.08±0.01* | 0.025±0.005† | ||||||||||||||||||||
| Thrombi | 0 | 0 | 0 | 0 | + | + | + | 0 | + | 0 | 0 | 0 | 0 | ||||||||||
| Bronchus | |||||||||||||||||||||||
| Mucus | 0 | 0 | 0 | 0 | ++ | ++ | ++ | ++ | ++ | 0 | 0 | 0 | 0 | ||||||||||
| Inflammatory cells | 24.0±2.9 | 40.5±4.7* | 25.2±8.8 | ||||||||||||||||||||
| Lymphocyte | 12.5±3.7 | 19.8±1.9 | 15.5±6.6 | ||||||||||||||||||||
| Macrophage | 9.5±3.1 | 13.1±3.0 | 7±2.1 | ||||||||||||||||||||
| Neutrophil | 2.8±1.3 | 6.9±1.0* | 3.1±1.2 | ||||||||||||||||||||
Congestion was evaluated as the percentage of vessel area filled with red blood cells (RBCs): 0 indicates no RBCs; +, RBCs filling less than 30% of the vessel section; ++, RBCs filling between 30% and 50% of the vessel; +++, more than 50% of the vessel filled with RBCs. Constriction: 0 indicates no constriction; +, presence of empty area of less than 0.05 μm; ++, presence of an empty area around the vessel larger than 0.05 μm. Data are expressed as mean number of cells per field at ×250 and presented as means ± SDs. Thrombi: 0 indicates no thrombi; +, presence of thrombi in small vessels. Mucus: 0 indicates no mucus; +, mucus filling less than 50% of the area of the bronchus section; ++, mucus filling more than 50% of the area of the bronchus section. Data are expressed as mean number of cells per field at ×250 and presented as means ± SDs
P < .05 compared with wild-type mice
P < .05 compared with SAD mice exposed to hypoxia
Molecular analysis of lung tissue
RNA preparation. Total RNA was isolated from frozen lungs by a guanidinium-isothiocyanate-cesium chloride gradient procedure,37-42 quantified by GeneQuant 2 spectrophotometer (Pharmacia, Uppsala, Sweden), and its integrity was qualitatively controlled by agarose gel electrophoresis. cDNA array hybridization. The Atlas mouse cDNA expression array (catalog no. 7741-1) from Clontech Laboratories was used. These arrays contain 588 known genes spotted as duplicates on nylon membranes, whose list and array coordinates are available at Clontech's web site (http://www.clontech.com). The membranes were hybridized with 3 μg RNA from each of the 7 experimental groups, according to the manufacturer's specifications, exposed to phosphoimager plates for 24 to 72 hours, and acquired using the Storm Phosphorimaging System (Molecular Dynamics, Sunnyvale, CA). Image analysis and quantitation was performed with AtlasImage 2.01 software (Clontech).38 After grid assignment, the intensity value for each gene was calculated as the average of the total signal for the duplicated spots.
Filtering and normalization of cDNA array data. All signals obtained from AtlasImage 2.01 software that showed intensity values lower than twice the background level were eliminated from further analysis. Then all signals with intensity values higher than twice the background were globally normalized, dividing each value by the median of the values within arrays, then multiplied by 100 and logarithmic (log2) transformed using R statistical software (R Foundation for Statistical Computing, Vienna, Austria) package.43
Selection of differentially expressed genes. Differentially expressed genes were selected by pair-wise comparison between the data from control SAD mice (baseline) and SAD mice exposed to hypoxia/reoxygenation at the different time points with or without NO treatment. To this purpose, log2-transformed values were entered in a xy-scatterplot, and the least-squares regression line was calculated for each comparison. The standard deviation of the distances between the points and the least-squares regression line was calculated. An area of standard deviation multiplied with 1.96 at both sides of the least-squares regression line was drawn in the xy-scatter to establish a 95% confidence interval using R statistical software package.43 Genes located outside this area were identified as differentially expressed.38,39,42,43
Validation of differential expression of selected genes by quantitative RT-PCR. We confirmed the gene expression levels for selected genes using SYBR-Green-based real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR).39-42 The 5 genes for which qRT-PCR was performed included 4 genes showing modulated expression in cDNA array hybridization (Ace, Hsp86-1, Hspcb, Nfe2l2) and MIP-2, whose soluble counterpart was detected in bronchoalveolar (BAL) fluid. β-Actin was used as internal reference in each reaction, that contained 1X SYBR Green Master Mix buffer (Applied Biosystems, Weiterstadt, Germany), 200 nM forward and reverse gene-specific primers, and 40 ng cDNA. Before quantitative measurements of experimental samples, each primer pair was validated by its ability to produce a single prominent amplicon (peak) in dissociation analysis. Assays were performed in triplicate using an Applied Biosystems Model 7700 instrument. The accumulation of PCR product was measured in real time as the increase in SYBR green fluorescence. cDNA aliquots were quantified for target genes using the threshold cycle [C(t)] method normalized for the house-keeping gene β-actin. After normalization, quantitative gene expression in hypoxic mice groups were compared with corresponding normoxia mice groups set arbitrarily at 1, either for wild-type and SAD mice. The primers used were Ace sense, GGAAGAGCAGAATCAGCGGAA, antisense, TTCCACGAACCTGTCAGCCTT; Hsp86-1 sense, AGGAGTCTGATGATAAACCT, antisense, TCATTAGTGATGTCATCAGG; Hspcb sense, TGACGATGTTCTTGCGGATG, antisense, CTCAACTTTATCCGCGGTGT; Nfe2l2 sense, CTTGGGCCACTTAAAAGACGAG, antisense, TTGCCATCTCTGGTTTGCTG; MIP-2 sense, TGCGCTGTCAATGCCTGAA, antisense, TTTTGACCGCCCTTGAGAGTG; β-actin sense, AGAAGCTGTGCTATGTTGCCC, antisense, GGAACCGCTCATTGCCAAT.
Bronchoalvear (BAL) fluid and cytokine content
BAL fluids were collected by instilling and withdrawing 5 mL sterile phosphate-buffered saline (PBS), 4 times via intratracheal cannula.29,30,44 Cells were recovered by centrifugation and counted by microcytometry. The percentage of neutrophils was determined by cytospin centrifugation, fixation, and staining.29,30,44 Remaining BAL samples were centrifuged at 1500g for 10 minutes at 4°C. The supernatants were used to measure total protein content30,44 and quantify the following cytokines: tumor necrosis factor α (TNF-α), interleukin 1α (IL-1α), IL-6, and macrophage inflammatory protein-2 (MIP-2), by commercial enzyme-linked immunosorbent assay (ELISA; R&D Systems Europe, Abingdon, United Kingdom; Amersham, Oxford, United Kingdom), according to the manufacturer's instructions.
Hematologic parameters
The hematologic parameters were measured as described.6,7,36 Density distribution curves were obtained according to Danon and Marikovsky.6,7,36 The density values, defined as the 20% densest fraction of cells (D20), were determined for each curve. The remaining cells were washed 4 additional times with choline washing solution (172 mM choline chloride, 1 mM MgCl2, and 10 mM Tris-MOPS, pH 7.4 at 4°C) for measurements of internal Na+ and K+ contents by atomic absorption spectrometry.36 The methemoglobin content was calculated from the optical spectrum recorded at the end of the oxygen equilibrium measurements. P50 values were calculated by linear regression from the Hill equation for oxygen saturation levels between 40% and 60%.18,34
Statistical analysis for soluble and hematologic parameters
Data were treated by the 2-way analysis of variance (ANOVA) algorithm for repeated measures between treatment schedules. A difference with a P value less than .05 was considered significant.
Results
SAD mice exposed to hypoxia/reoxygenation recapitulate the pathophysiology of lung sickle cell vaso-occlusive events
We first asked whether the SAD transgenic mouse submitted to hypoxia/reoxygenation could be a pertinent model of the specific lung pathology observed during acute vaso-occlusive sickle cell events. The main foreseen advantages of the use of SAD mouse in the present study were as follows: (1) the animals do not have an added thalassemic syndrome, and (2) SAD mice exhibit very little pulmonary defect in young adults at baseline. Accordingly, one can expect to observe meaningful lesions on acute hypoxia-reoxygenation, not obscured by previous damage as observed in aged SAD mice or in more severe sickle cell mouse models.
Histopathologic analysis. The results of the histopathologic analysis are detailed in Table 1, and a representative example is shown in Figure 1.
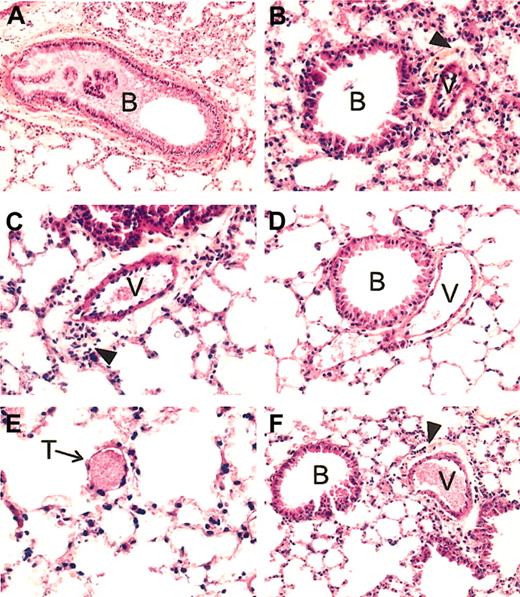
Lung histopathology of SAD mice exposed to hypoxia/reoxygenation and effect of inhaled NO. In the left panels are shown representative examples of SAD mouse lungs exposed to hypoxia/reoxygenation for 46 hours. (A) More than 50% of the bronchus lumen (B) is filled with mucus; (C), a constricted vessel (V) is evident for the large surrounding clear area containing many inflammatory cells; (E), a small vessel filled with a thrombus (T) is shown. (B) Representative section of a SAD mouse lung exposed to hypoxia/reoxygenation and inhaled NO. Here the bronchus lumen (B) is devoid of mucus, the constriction of the vessel (V) is much less intense than that seen in panel C, as evident by the clear area around the vessel whose thickness is not superior to that of the vessel wall, associated with a mild inflammatory infiltrate. Panel D exemplifies the lung histology of SAD mice under normoxia. The lumen of the bronchus (B) is devoid of mucus; the neighboring vessel (V) is not constricted and contains few red blood cells. There is no inflammatory infiltrate. (F) Example of lung histopathology of wild-type mice exposed to 46 hours hypoxia/reoxygenation. The lumen of the bronchus (B) is devoid of mucus; the neighboring vessel (V), filled with erythrocytes, is constricted, with a mild inflammatory infiltrate (arrowhead) in the clear area surrounding the vessel. All tissue sections are stained with hematoxylin and eosin, and the original magnification is × 250 (with the exception of panel A, × 200).
Lung histopathology of SAD mice exposed to hypoxia/reoxygenation and effect of inhaled NO. In the left panels are shown representative examples of SAD mouse lungs exposed to hypoxia/reoxygenation for 46 hours. (A) More than 50% of the bronchus lumen (B) is filled with mucus; (C), a constricted vessel (V) is evident for the large surrounding clear area containing many inflammatory cells; (E), a small vessel filled with a thrombus (T) is shown. (B) Representative section of a SAD mouse lung exposed to hypoxia/reoxygenation and inhaled NO. Here the bronchus lumen (B) is devoid of mucus, the constriction of the vessel (V) is much less intense than that seen in panel C, as evident by the clear area around the vessel whose thickness is not superior to that of the vessel wall, associated with a mild inflammatory infiltrate. Panel D exemplifies the lung histology of SAD mice under normoxia. The lumen of the bronchus (B) is devoid of mucus; the neighboring vessel (V) is not constricted and contains few red blood cells. There is no inflammatory infiltrate. (F) Example of lung histopathology of wild-type mice exposed to 46 hours hypoxia/reoxygenation. The lumen of the bronchus (B) is devoid of mucus; the neighboring vessel (V), filled with erythrocytes, is constricted, with a mild inflammatory infiltrate (arrowhead) in the clear area surrounding the vessel. All tissue sections are stained with hematoxylin and eosin, and the original magnification is × 250 (with the exception of panel A, × 200).
In wild-type mice, hypoxia/reoxygenation resulted in a mild inflammatory response, characterized by (1) mild vascular congestion, (2) mild vascular constriction (0.027 ± 0.009 μm; n = 4), (3) absence of thrombi, (4) mucus filling less than 30% of the bronchi section area, and (5) mild inflammatory infiltrate (24 ± 2.9 cells/high-power field, n = 4) composed mainly of lymphocytes and macrophages associated with few neutrophils (2.8 ± 1.3 cells/high-power field; n = 4). The transgenic SAD mice exposed to hypoxia/reoxygenation showed lung injury typical of the ischemic/reperfusion syndrome observed in human SCD vaso-occlusive events. Histophathologic changes were compatible with severe inflammatory response with extensive ischemic tissue changes, including (1) severe vascular congestion, (2) severe vascular constriction (0.08 ± 0.01 μm; n = 5), (3) presence of thrombi in small vessels, (4) mucus filling more than 50% of the bronchus section area, and (5) severe lymphomonocytic inflammatory infiltrate (40.5 ± 4.7 cells/high-power field; n = 5) associated with a significant amount of neutrophils (6.9 ± 1 cells/high-power field; n = 5) (Table 1, Figure 1).
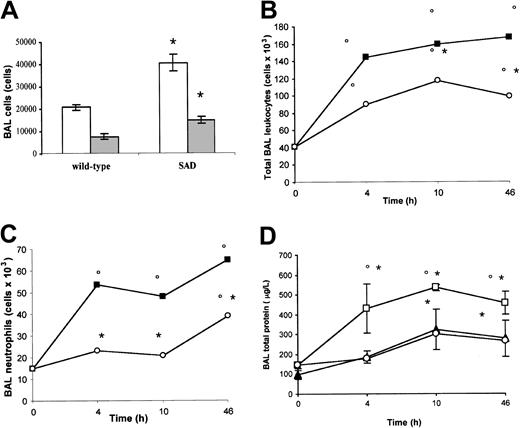
Analysis of cells and protein content in BAL. Under normoxia (21% oxygen), total leukocyte and neutrophil counts in BAL were significantly increased in SAD mice compared with wild-type mice, suggesting the existence of a basic chronic lung inflammatory state (Figure 2A).
BAL cells and proteins in normoxia, hypoxia/reoxygenation and effects of inhaled NO. (A) the BAL total leukocyte (white bars) and neutrophil (gray bars) counts in wild-type and transgenic sickle SAD mice under normoxia conditions (n = 12 in each group) are reported. (B-D) BAL total leukocyte count, neutrophil count, and total protein content in wild-type mice (closed triangles), in SAD mice exposed to hypoxia/reoxygenation alone (open boxes), and in SAD mice breathing inhaled NO (open circles) (n = 6 in each group). Values are expressed as means + SEMs. *P < .05 compared with baseline, °P < .05 hypoxia/reoxygenation versus hypoxia/reoxygenation plus NO, as determined by ANOVA with post hoc Bonferroni corrected t test.
BAL cells and proteins in normoxia, hypoxia/reoxygenation and effects of inhaled NO. (A) the BAL total leukocyte (white bars) and neutrophil (gray bars) counts in wild-type and transgenic sickle SAD mice under normoxia conditions (n = 12 in each group) are reported. (B-D) BAL total leukocyte count, neutrophil count, and total protein content in wild-type mice (closed triangles), in SAD mice exposed to hypoxia/reoxygenation alone (open boxes), and in SAD mice breathing inhaled NO (open circles) (n = 6 in each group). Values are expressed as means + SEMs. *P < .05 compared with baseline, °P < .05 hypoxia/reoxygenation versus hypoxia/reoxygenation plus NO, as determined by ANOVA with post hoc Bonferroni corrected t test.
Hypoxia/reoxygenation of SAD mice resulted in a marked increase in BAL total leukocyte and neutrophil counts after 4, 10, and 46 hours (Figure 2B-C). In wild-type mice, hypoxia/reoxygenation resulted in a significant increase in BAL total leukocyte count at 10 and 46 hours together with a significant increase in BAL neutrophil count at 46 hours (Figure 2B-C). The BAL cellular increase was significantly lower in wild-type mice compared with SAD mice under the same hypoxia/reoxygenation conditions (P < .05). Hypoxia/reoxygenation also resulted in a significant increase in BAL total protein content, indicating a dysfunction in microvascular permeability (Figure 2D).35 In wild-type mice, hypoxia/reoxygenation resulted in a significant increase in BAL total protein at 10 and 46 hours (Figure 2D). The BAL protein increase was significantly lower in wild-type mice than in SAD mice under the same hypoxia/reoxygenation conditions (Figure 2D, P < .05).
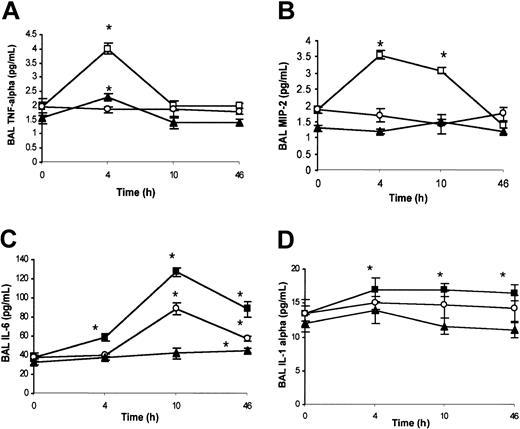
Analysis of chemokines in BAL. Under ambient condition (21% oxygen), no significant differences were detected by ELISA analysis for BAL TNF-α, MIP-2, IL-6, and IL-1α content between wild-type and SAD mice (data not shown). SAD mice exposed to hypoxia/reoxygenation showed a peak of BAL TNF-α levels at 4 hours, decreasing to baseline levels at 10 and 46 hours (Figure 3A). In wild-type mice, hypoxia/reoxygenation induced a peak of BAL TNF-α at 4 hours, returning to baseline at 10 and 46 hours (Figure 3A). The increase in BAL TNF-α levels in wild-type mice was lower than in SAD mice (Figure 3A). In SAD mice, BAL MIP-2 was increased at 4 and 10 hours after hypoxia/reoxygenation, followed by a decline to baseline levels at 46 hours (Figure 3B). The concentration of MIP-2 in the BAL fluid was not modified in wild-type mice exposed to hypoxia/reoxygenation (Figure 3B). These results were confirmed by quantitative RT-PCR analysis (described in “SAD mice exposed to hypoxia/reoxygenation show induction of lung-specific stress response and vascular remodeling genes” and Figure 6E). In SAD mice exposed to hypoxia/reoxygenation, IL-6 was increased in BAL fluid at 4, 10, and 46 hours, with a peak at 10 hours (Figure 3C). In wild-type mice, hypoxia/reoxygenation yielded an increase in BAL IL-6 levels at 46 hours to values lower than in SAD mice (Figure 3C). BAL IL-1α was significantly increased in both wild-type and SAD mice exposed to hypoxia/reoxygenation at 4 to 10 and 46 hours; the increase in BAL IL-1α was greater in SAD mice than in wild-type mice (Figure 3D).
BAL cytokines in hypoxia/reoxygenation and effects of inhaled NO. The content of BAL TNF-α (A), macrophage inflammatory protein-2 (MIP-2) (B), IL-6 (C), and IL-1α (D) is shown for wild-type mice (closed triangles), SAD mice exposed to hypoxia/reoxygenation alone (open boxes), and SAD mice breathing inhaled NO (open circles) (n = 6 in each group). Values are expressed as means + SEMs. *P < .05 compared with baseline, °P < .05 hypoxia/reoxygenation versus hypoxia/reoxygenation plus NO, as determined by ANOVA with post hoc Bonferroni corrected t test.
BAL cytokines in hypoxia/reoxygenation and effects of inhaled NO. The content of BAL TNF-α (A), macrophage inflammatory protein-2 (MIP-2) (B), IL-6 (C), and IL-1α (D) is shown for wild-type mice (closed triangles), SAD mice exposed to hypoxia/reoxygenation alone (open boxes), and SAD mice breathing inhaled NO (open circles) (n = 6 in each group). Values are expressed as means + SEMs. *P < .05 compared with baseline, °P < .05 hypoxia/reoxygenation versus hypoxia/reoxygenation plus NO, as determined by ANOVA with post hoc Bonferroni corrected t test.
Quantitative RT-PCR expression profiles of Hspcb, Hsp86-1, Nfe2l2, Ace, and Mip-2. The lines represent log2-transformed gene expression ratios between SAD mice exposed to different hypoxia times (4, 10, and 46 hours) plus 2 hours of reoxygenation alone (black diamond) or with NO 40 ppm (white square) (n = 6 for each condition); wild-type mice exposed to hypoxia/reoxygenation alone (white triangle) (n = 6) compared with an untreated SAD mouse normoxic group (time 0, n = 6).
Quantitative RT-PCR expression profiles of Hspcb, Hsp86-1, Nfe2l2, Ace, and Mip-2. The lines represent log2-transformed gene expression ratios between SAD mice exposed to different hypoxia times (4, 10, and 46 hours) plus 2 hours of reoxygenation alone (black diamond) or with NO 40 ppm (white square) (n = 6 for each condition); wild-type mice exposed to hypoxia/reoxygenation alone (white triangle) (n = 6) compared with an untreated SAD mouse normoxic group (time 0, n = 6).
Together, these data provide convincing evidence that the SAD mouse exposed to acute hypoxia/reoxygenation represents an experimental model that closely mimics the phenomena observed in vaso-occlusive lung events occurring in patients with SCD.
SAD mice exposed to hypoxia/reoxygenation show induction of lung-specific stress response and vascular remodeling genes
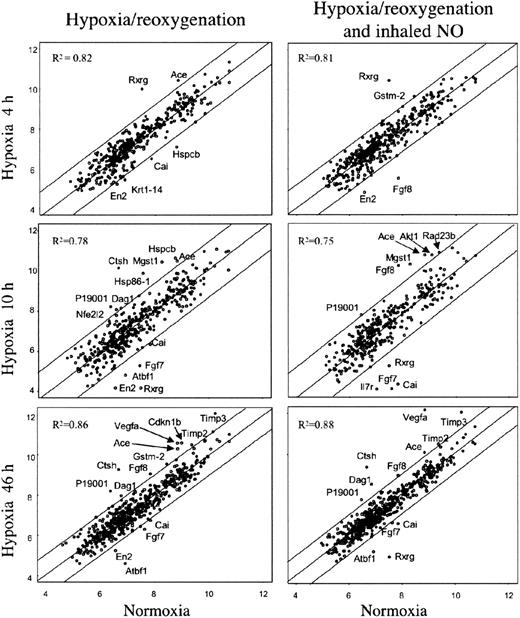
In an effort to identify specific genes associated with pathophysiologic changes related to hypoxia/reoxygenation of SAD mice, we analyzed the expression profiles of lung tissue samples using mouse cDNA arrays containing 588 known genes. The results of cDNA arrays are shown in Figure 4 and detailed in Table 2.38,41,42 A total of 23 genes were found to be differentially expressed at the various time points in lungs of SAD mice exposed to either hypoxia-reoxygenation or hypoxia-reoxygenation plus NO. In summary, Timp2, Timp3, Mgst1, type I cytoskeletal keratin 19, Rxrg, and Vegfa genes showed differential expression associated with different hypoxia time points with no or minor influence by NO treatment (Table 2). Krt1-14 gene showed a down-regulation by hypoxia that was rescued by NO treatment (Table 2). The expression of the remaining 16 genes showed variations influenced by both hypoxia and NO treatment (Table 2).
Gene expression in lung of SAD mice exposed to variable time hypoxia followed by 2 hours reoxygenation and effects of inhaled NO. On the left, the xy-scatterplots display the pair-wise comparison between gene expression levels in lungs of SAD mice control group (x axis) and that of SAD mice exposed to the indicated times of hypoxia followed by 2 hours reoxygenation (y axis). On the right, the xy-scatterplots display the pair-wise comparison between gene expression levels in lungs of SAD mice control group (x axis) and that in lungs of SAD mice exposed at the indicated times hypoxia followed by 2 hours reoxygenation breathing NO (y axis). Both x and y values represent the log2 expression level of the same genes in the different experimental conditions (groups). The central line is the least squares regression line (Y = b + ax), and the upper and lower lines are obtained by the respective addition and subtraction to the regression line equation of the standard deviation of residuals multiplied by 1.96. The Pearson correlation value (R2) for each comparison is indicated. Spots outside the upper and lower lines represent the genes upor down-regulated with respect to SAD baseline, respectively (described in “Materials and methods”).
Gene expression in lung of SAD mice exposed to variable time hypoxia followed by 2 hours reoxygenation and effects of inhaled NO. On the left, the xy-scatterplots display the pair-wise comparison between gene expression levels in lungs of SAD mice control group (x axis) and that of SAD mice exposed to the indicated times of hypoxia followed by 2 hours reoxygenation (y axis). On the right, the xy-scatterplots display the pair-wise comparison between gene expression levels in lungs of SAD mice control group (x axis) and that in lungs of SAD mice exposed at the indicated times hypoxia followed by 2 hours reoxygenation breathing NO (y axis). Both x and y values represent the log2 expression level of the same genes in the different experimental conditions (groups). The central line is the least squares regression line (Y = b + ax), and the upper and lower lines are obtained by the respective addition and subtraction to the regression line equation of the standard deviation of residuals multiplied by 1.96. The Pearson correlation value (R2) for each comparison is indicated. Spots outside the upper and lower lines represent the genes upor down-regulated with respect to SAD baseline, respectively (described in “Materials and methods”).
Genes modulated in lungs of SAD mice exposed to hypoxia/reoxygenation and effects of inhaled nitric oxide (NO)
. | . | . | Hypoxia . | . | . | Hypoxia plus NO . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GenBank accession no. . | Symbol . | Description . | 4 h . | 10 h . | 46 h . | 4 h . | 10 h . | 46 h . | ||||
| M36829 | Hspcb | Heat shock protein 1, beta | 0.3 | 3.5 | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| M13806 | Krt1-14 | Keratin complex 1, acidic, gene 14 | 0.4 | 0.4 | 0.4 | 1.0 | 1.0 | 1.0 | ||||
| J05186 | Cai | Calcium binding protein, intestinal | 0.4 | 0.4 | 0.4 | 1.0 | 0.1 | 0.4 | ||||
| L12705 | En2 | Engrailed 2 | 0.4 | 0.1 | 0.4 | 0.3 | 1.0 | 1.0 | ||||
| Z22703 | Fgf7 | Fibroblast growth factor 7 | 2.0 | 0.2 | 0.4 | 1.0 | 0.1 | 0.5 | ||||
| U10440 | Cdkn1b | Cyclin-dependent kinase inhibitor 1B (P27) | 1.0 | 0.5 | 2.6 | 1.0 | 1.0 | 1.0 | ||||
| D12482 | Fgf8 | Fibroblast growth factor 8 | 1.0 | 1.0 | 2.3 | 0.2 | 5.6 | 2.2 | ||||
| U06119 | Ctsh | Cathepsin H | 1.0 | 11.0 | 6.0 | 1.0 | 1.0 | 6.6 | ||||
| M36830 | Hsp86-1 | Heat shock protein, 1 | 1.0 | 3.8 | 1.0 | 1.0 | 1.0 | 1.5 | ||||
| U20532 | Nfe212 | Nuclear factor erythroid derived 2 like 2 | 1.0 | 2.3 | 1.0 | 1.0 | 0.9 | 1.0 | ||||
| M95200 | Vegfa | Vascular endothelial growth factor A | 1.0 | 1.0 | 2.7 | 1.0 | 1.0 | 10.0 | ||||
| J04696 | Gstm2 | Glutathione S-transferase, mu 2 | 1.0 | 2.0 | 2.1 | 2.3 | 1.0 | 1.0 | ||||
| L19622 | Timp3 | Tissue inhibitor of metalloproteinase 3 | 1.0 | 1.0 | 3.5 | 1.0 | 1.0 | 3.5 | ||||
| U43512 | Dag1 | Dystroglycan 1 | 2.0 | 2.1 | 2.1 | 1.0 | 0.5 | 3.2 | ||||
| D26046 | Atbf1 | AT motif binding factor 1 | 2.0 | 0.2 | 0.2 | 1.0 | 2.0 | 0.3 | ||||
| J04946 | Ace | Angiotensin converting enzyme | 3.0 | 3.0 | 2.3 | 1.0 | 4.0 | 2.34 | ||||
| M84819 | Rxrg | Retinoid X receptor gamma | 5.7 | 0.1 | 1.0 | 7.7 | 0.2 | 0.2 | ||||
| X62622 | Timp2 | Tissue inhibitor of metalloproteinase 2 | 1.0 | 1.0 | 2.2 | 1.0 | 1.0 | 2.2 | ||||
| M29697 | Il7r | Interleukin 7 receptor | 1.0 | 1.0 | 1.0 | 1.0 | 0.1 | 1.0 | ||||
| J03752 | Mgst1 | Microsomal glutathione S-transferase 1 | 1.0 | 4.0 | 1.0 | 1.0 | 4.2 | 1.0 | ||||
| X92411 | Rad23b | RAD23b homolog (S. cerevisiae) | 1.0 | 1.0 | 1.0 | 1.0 | 2.88 | 1.0 | ||||
| P19001 | none | Type I cytoskeletal keratin 19 | 1.0 | 3.5 | 3.5 | 1.0 | 2.5 | 2.5 | ||||
| M94335 | Akt1 | Thymoma viral proto-oncogene 1 | 1.0 | 1.0 | 1.0 | 1.0 | 3.32 | 1.0 | ||||
. | . | . | Hypoxia . | . | . | Hypoxia plus NO . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GenBank accession no. . | Symbol . | Description . | 4 h . | 10 h . | 46 h . | 4 h . | 10 h . | 46 h . | ||||
| M36829 | Hspcb | Heat shock protein 1, beta | 0.3 | 3.5 | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| M13806 | Krt1-14 | Keratin complex 1, acidic, gene 14 | 0.4 | 0.4 | 0.4 | 1.0 | 1.0 | 1.0 | ||||
| J05186 | Cai | Calcium binding protein, intestinal | 0.4 | 0.4 | 0.4 | 1.0 | 0.1 | 0.4 | ||||
| L12705 | En2 | Engrailed 2 | 0.4 | 0.1 | 0.4 | 0.3 | 1.0 | 1.0 | ||||
| Z22703 | Fgf7 | Fibroblast growth factor 7 | 2.0 | 0.2 | 0.4 | 1.0 | 0.1 | 0.5 | ||||
| U10440 | Cdkn1b | Cyclin-dependent kinase inhibitor 1B (P27) | 1.0 | 0.5 | 2.6 | 1.0 | 1.0 | 1.0 | ||||
| D12482 | Fgf8 | Fibroblast growth factor 8 | 1.0 | 1.0 | 2.3 | 0.2 | 5.6 | 2.2 | ||||
| U06119 | Ctsh | Cathepsin H | 1.0 | 11.0 | 6.0 | 1.0 | 1.0 | 6.6 | ||||
| M36830 | Hsp86-1 | Heat shock protein, 1 | 1.0 | 3.8 | 1.0 | 1.0 | 1.0 | 1.5 | ||||
| U20532 | Nfe212 | Nuclear factor erythroid derived 2 like 2 | 1.0 | 2.3 | 1.0 | 1.0 | 0.9 | 1.0 | ||||
| M95200 | Vegfa | Vascular endothelial growth factor A | 1.0 | 1.0 | 2.7 | 1.0 | 1.0 | 10.0 | ||||
| J04696 | Gstm2 | Glutathione S-transferase, mu 2 | 1.0 | 2.0 | 2.1 | 2.3 | 1.0 | 1.0 | ||||
| L19622 | Timp3 | Tissue inhibitor of metalloproteinase 3 | 1.0 | 1.0 | 3.5 | 1.0 | 1.0 | 3.5 | ||||
| U43512 | Dag1 | Dystroglycan 1 | 2.0 | 2.1 | 2.1 | 1.0 | 0.5 | 3.2 | ||||
| D26046 | Atbf1 | AT motif binding factor 1 | 2.0 | 0.2 | 0.2 | 1.0 | 2.0 | 0.3 | ||||
| J04946 | Ace | Angiotensin converting enzyme | 3.0 | 3.0 | 2.3 | 1.0 | 4.0 | 2.34 | ||||
| M84819 | Rxrg | Retinoid X receptor gamma | 5.7 | 0.1 | 1.0 | 7.7 | 0.2 | 0.2 | ||||
| X62622 | Timp2 | Tissue inhibitor of metalloproteinase 2 | 1.0 | 1.0 | 2.2 | 1.0 | 1.0 | 2.2 | ||||
| M29697 | Il7r | Interleukin 7 receptor | 1.0 | 1.0 | 1.0 | 1.0 | 0.1 | 1.0 | ||||
| J03752 | Mgst1 | Microsomal glutathione S-transferase 1 | 1.0 | 4.0 | 1.0 | 1.0 | 4.2 | 1.0 | ||||
| X92411 | Rad23b | RAD23b homolog (S. cerevisiae) | 1.0 | 1.0 | 1.0 | 1.0 | 2.88 | 1.0 | ||||
| P19001 | none | Type I cytoskeletal keratin 19 | 1.0 | 3.5 | 3.5 | 1.0 | 2.5 | 2.5 | ||||
| M94335 | Akt1 | Thymoma viral proto-oncogene 1 | 1.0 | 1.0 | 1.0 | 1.0 | 3.32 | 1.0 | ||||
The values are presented as the gene expression ratio between any given time point and normoxic SAD mice
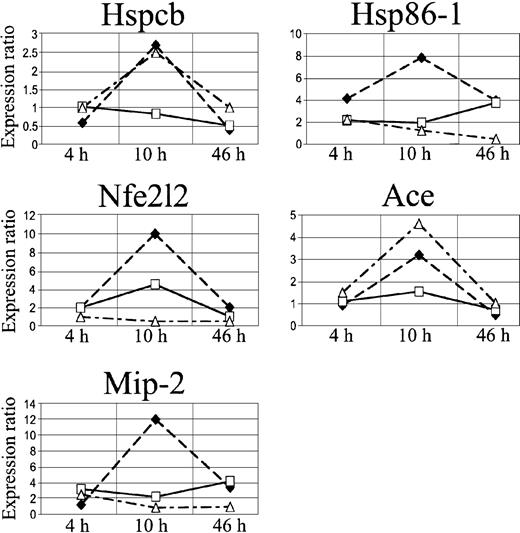
We focused our attention on 3 stress genes, heat shock protein 1 beta (Hspcb), heat shock protein 1 (Hsp86-1), and nuclear factor erythroid derived 2 like 2 (Nfe2l2), and on 2 genes involved in microcirculation remodeling, angiotensin-converting enzyme (Ace) and fibroblast growth factor 7 (Fgf7) (Figure 6, Table 2).
In SAD mice, Hspcb, Hsp86-1, and Nfe2l2 RNA levels were increased by 3.5-, 3.8-, and 2.3-fold, respectively, 10 hours after hypoxia/reoxygenation and returned to baseline levels after 46 hours (Figure 5A-B, D). Ace RNA levels increased by 3-fold, starting 4 hours after the induction of hypoxia, and remained significantly elevated 10 and 46 hours after hypoxia/reoxygenation (Figure 5C). The Fgf7 gene was up-regulated at 4 hours and down-regulated after 10 and 46 hours (Figure 5E).
cDNA array expression profiles of Hspcb, Hsp86-1, Nfe2l2, Ace, and Fgf7. The lines represent log2-transformed gene expression ratios between SAD mice exposed to different hypoxia times (4, 10, and 46 hours) plus 2 hours of reoxygenation (black diamond) or breathing NO 40 ppm (white square), compared with an untreated SAD mouse normoxic group.
cDNA array expression profiles of Hspcb, Hsp86-1, Nfe2l2, Ace, and Fgf7. The lines represent log2-transformed gene expression ratios between SAD mice exposed to different hypoxia times (4, 10, and 46 hours) plus 2 hours of reoxygenation (black diamond) or breathing NO 40 ppm (white square), compared with an untreated SAD mouse normoxic group.
Quantitative reverse transcription–PCR (qRT-PCR) was used as an independent method to validate the differences in expression levels of the Hspcb, Hsp86-1, Ace, and Nfe2l2 genes previously identified by cDNA array. qRT-PCR confirmed the results (Figure 6). The same genes were also analyzed by qRT-PCR in the 4 groups of wild-type mice exposed to hypoxia/reoxygenation, showing increased Hspcb mRNA level at 10 hours, returning to baseline at 46 hours, and unmodified Hsp86-1 mRNA level at the different time points (Figure 6A-B). The expression level of Ace, after an initial latent period of 4 hours, showed a marked increase at 10 to 46 hours, whereas Nfe2l2 expression was unchanged at the various time points analyzed (Figure 6C-D).
SAD mice inhaling NO show alleviation of hypoxia/reoxygenation-mediated lung injury with normalization of lung gene expression profiles
Inhaled NO ameliorated the lung vaso-occlusive syndrome of SCD. In fact, in SAD mice inhaled NO reduced the inflammatory response and subsequent lung tissue damages related to hypoxia/reoxygenation stimulus as supported by the observed (1) mild vascular congestion, (2) mild vascular constriction (0.025 ± 0.005 μm; n = 4), (3) absence of thrombi, (4) absence of mucus in the bronchus section, and (5) mild inflammatory infiltrate (total cells, 25.2 ± 8.8; neutrophils, 3.1 ± 1.2 cells/high power field; n = 4) (Figure 1, Table 1). In addition, in SAD mice submitted to hypoxia/reoxygenation, inhaled NO normalized the expression of the Hspcb, Hsp86-1, and Nfe2l2 genes, whereas Fgf7 gene expression was unmodified at 4 hours and down-regulated at 10 to 46 hours; the up-regulation of the Ace gene was delayed to 10 hours after hypoxia/reoxygenation (Figures 5,6) (Table 2).
We then set out to examine the mechanism(s) by which NO administration is beneficial in SAD mice submitted to hypoxia/reoxygenation.
NO inhibits neutrophil migration to the broncho-alveolar space. Inhaled NO significantly reduced the hypoxia/reoxygenation-mediated increase in BAL total leukocyte counts and prevented the increase in BAL neutrophil counts at 4 and 10 hours. In addition, the increase in BAL neutrophil counts observed at 46 hours remained lower than that in hypoxia/reoxygenation SAD mice not treated with NO (Figure 2B-C). Furthermore, inhaled NO significantly reduced the hypoxia/reoxygenation-related increase in BAL total protein content (Figure 2D). These results suggest that the inflammatory response to hypoxia/reoxygenation is amplified in SAD mice, compared with wild-type mice, and that inhaled NO inhibits the migration of neutrophils from pulmonary vasculature into the broncho-alveolar space and reduces the lung microvascular dysfunction (Figure 2B-C).
NO controls the inflammatory cytokine storm in the lung. Inhaled NO prevented the increase in BAL TNF-α and MIP-2 levels (Figure 3A-B). This latter result was confirmed by qRT-PCR analysis (Figure 6E). The IL-6 temporal expression was similar in both SAD mouse groups subjected to hypoxia/reoxygenation with or without NO. At 10 to 46 hours, the IL-6 levels were significantly lower in hypoxic SAD mice breathing NO (Figure 3C). No significant difference in IL-1α levels was observed at the different time points between hypoxia/reoxygenation SAD mice with or without inhalation of NO (Figure 3D).
NO ameliorates SAD red blood cell dehydration without change in O2 Hb affinity and methemoglobin formation
We asked whether NO may affect SAD mice submitted to hypoxia/reoxygenation primarily by the modification of the affinity of HbS for oxygen. We also investigated whether methemoglobin was produced at therapeutic levels of inhaled NO.
The effects of inhaled NO on hematologic parameters were evaluated at the different time points, 4, 10, and 46 hours, in SAD mice exposed to hypoxia (8% oxygen) breathing or not breathing NO (40 ppm). Hypoxia with or without NO affected neither hematocrit (Hct), Hb, methemoglobin levels nor reticulocyte counts at the various time points studied (Table 3).
Effects of inhaled nitric oxide (NO) on hematologic parameters and cell cation content in SAD mice exposed to hypoxia
. | . | Hypoxia, 4 h . | . | Hypoxia, 10 h . | . | Hypoxia, 46 h . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
. | Baseline . | HY . | HY + NO 40 ppm . | HY . | HY + NO 40 ppm . | HY . | HY + NO 40 ppm . | |||
| Hct, % | 43.0±1.1 | 42.7±1.5 | 43.8±0.2 | 42.7±1.1 | 42.9±1.2 | 43.3±1.5 | 42.2±1.4 | |||
| Hb, g/dL | 12.9±0.4 | 12.5±0.4 | 12.2±1.5 | 13.1±0.5 | 11.9±0.9 | 13.1±0.3 | 12.5±0.4 | |||
| MetHb, % | <1 | <1 | <1 | <1 | <1 | <1 | <1 | |||
| Reticulocytes, % | 6.3±0.4 | 5.8±1.2 | 6.1±0.9 | 7.4±2.1 | 6.9±1.5 | 7.2±0.5 | 5.9±1.4 | |||
| Cell Na, mM/kg Hb | 38.9±4.9 | — | — | — | — | 40.1±5.7 | 36.0±7.3 | |||
| Cell K, mM/kg Hb | 340±22.3 | — | — | — | — | 283±13.7* | 309.1±17.2*† | |||
. | . | Hypoxia, 4 h . | . | Hypoxia, 10 h . | . | Hypoxia, 46 h . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
. | Baseline . | HY . | HY + NO 40 ppm . | HY . | HY + NO 40 ppm . | HY . | HY + NO 40 ppm . | |||
| Hct, % | 43.0±1.1 | 42.7±1.5 | 43.8±0.2 | 42.7±1.1 | 42.9±1.2 | 43.3±1.5 | 42.2±1.4 | |||
| Hb, g/dL | 12.9±0.4 | 12.5±0.4 | 12.2±1.5 | 13.1±0.5 | 11.9±0.9 | 13.1±0.3 | 12.5±0.4 | |||
| MetHb, % | <1 | <1 | <1 | <1 | <1 | <1 | <1 | |||
| Reticulocytes, % | 6.3±0.4 | 5.8±1.2 | 6.1±0.9 | 7.4±2.1 | 6.9±1.5 | 7.2±0.5 | 5.9±1.4 | |||
| Cell Na, mM/kg Hb | 38.9±4.9 | — | — | — | — | 40.1±5.7 | 36.0±7.3 | |||
| Cell K, mM/kg Hb | 340±22.3 | — | — | — | — | 283±13.7* | 309.1±17.2*† | |||
Data are presented as means ± SEM of 6 experiments. HY indicates hypoxia (8% oxygen); MetHb, methemoglobin; and —, not done
P<.05 compared with baseline, as determined by ANOVA with post hoc Bonferroni corrected t test
P<.05 compared with hypoxia group, as determined by ANOVA with post hoc Bonferroni corrected t test
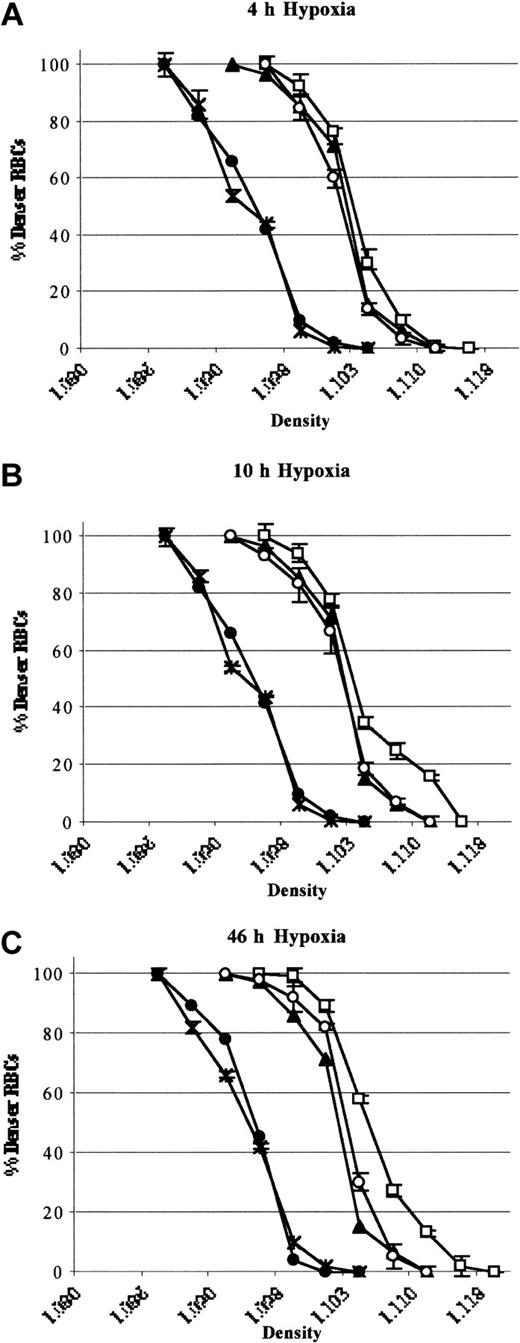
In SAD mice exposed to 46 hours hypoxia, inhaled NO did not significantly modify the erythrocyte oxygen affinity (P50 hypoxia, 49.2 ± 2 versus P50 hypoxia plus NO, 50.3 ± 1.4, n = 6; P = NS).34 A shift in the density curve of red blood cells toward higher density values (D20) was initiated after 4 hours of hypoxia and became significant after 46 hours (P < .05, n = 6; Figure 7). This result indicated a time dependency of hypoxia in exacerbating the dehydration of red blood cells of SAD mice (Figure 7). Inhaled NO partially but significantly prevented the shift in the red cell density profiles (D20), indicating a reduction in hypoxia-induced red cell dehydration (P < .05, Figure 7). Hypoxia caused a significant reduction in red blood cell K+ content (46 hours), which was partially but significantly prevented by inhaled NO (Table 3). Under normoxia, inhaled NO (40 ppm) did not affect SAD red blood cell density profiles and cell K+ content (data not shown). In wild-type mice, hypoxia did not affect the red cell density profiles either in presence or absence of inhaled NO (Figure 7). These data indicate that NO does not directly change the affinity of HbS for O2.
Red cell density profiles. Red cell density profiles in wild-type mice exposed to hypoxia/reoxygenation (close circles; 4 hours D20, 1.098 ± 0.002; 10 hours D20, 1.097 ± 0.003; 46 hours D20, 1.097 ± 0.002 n = 6), SAD mouse groups exposed to hypoxia /reoxygenation (open boxes; 4 hours D20, 1.106 ± 0.001; 10 hours D20, 1.107 ± 0.003; 46 hours D20, 1.111 ± 0.001 n = 6), and SAD mice exposed to hypoxia/reoxygenation plus inhaled NO (open circles; 4 hours D20, 1.104 ± 0.001; 10 hours D20, 1.104 ± 0.002; 46 hours D20, 1.104 ± 0.001; n = 6) compared with normoxia mouse groups (SAD, close triangles, D20, 1.102 ± 0.001; n = 6; wild-type, black asterisk; D20, 1.098 ± 0.001; n = 6). Data are presented as means ± SEMs (n = 6).
Red cell density profiles. Red cell density profiles in wild-type mice exposed to hypoxia/reoxygenation (close circles; 4 hours D20, 1.098 ± 0.002; 10 hours D20, 1.097 ± 0.003; 46 hours D20, 1.097 ± 0.002 n = 6), SAD mouse groups exposed to hypoxia /reoxygenation (open boxes; 4 hours D20, 1.106 ± 0.001; 10 hours D20, 1.107 ± 0.003; 46 hours D20, 1.111 ± 0.001 n = 6), and SAD mice exposed to hypoxia/reoxygenation plus inhaled NO (open circles; 4 hours D20, 1.104 ± 0.001; 10 hours D20, 1.104 ± 0.002; 46 hours D20, 1.104 ± 0.001; n = 6) compared with normoxia mouse groups (SAD, close triangles, D20, 1.102 ± 0.001; n = 6; wild-type, black asterisk; D20, 1.098 ± 0.001; n = 6). Data are presented as means ± SEMs (n = 6).
Discussion
Our data demonstrate that, in transgenic SAD mice submitted to hypoxia/reoxygenation, inhaled NO prevents histopathologic lung damage, attenuates the inflammatory response, and modulates genes involved in ischemic/reperfusion injury and microcirculation remodeling and ameliorates red cell dehydration.
The choice of the transgenic sickle cell SAD mouse, which exhibits a mild form of SCD compared with transgenic knock-out mice expressing exclusively human βS globins, enabled us to study the specific effects of hypoxia/reoxygenation damage. Knock-out mice for βS globin already display severe tissue inflammation and pathology, so that hypoxia/reoxygenation may only worsen the baseline state, thus resulting in ambiguous data gathering.
Recent studies have focused on inhaled NO for the treatment of tissue damage in various ischemic syndromes, including cardiovascular disease, pulmonary hypertension, and acute lung distress syndrome.19-26 The possible therapeutic role of inhaled NO has been studied in different animal models of lung injury induced by ischemic/reperfusion.19-26 Inhaled NO prevents leukocyte migration and reduces the permeability of the peripheral microvasculature.21,30,44 In association with surfactant, inhaled NO alleviates alveoli's edema and reduces bronchoalveolar leukocytes and neutrophils infiltration in animal models of ischemic lung injury.29,30 The beneficial effects of inhaled NO in maintaining endothelial integrity have also been proposed in lung transplantation, which triggers per se ischemic/reperfusion injury.29,30,32
The beneficial role of inhaled NO in SCD has been recently reported in the treatment of acute vaso-occlusive crisis in a placebo-controlled randomized clinical trial, although the mechanism of action in SCD remained unknown.18 Plasma NO metabolites are decreased in patients with SCD during either vaso-occlusive crisis associated with severe pain or acute chest syndrome.27,28 A decrease in exhaled NO has been reported in patients with sickle cell disease, suggesting a role for NO in the pathogenesis of the pulmonary complications.28
In the present paper, we report that transgenic SAD mice exposed to hypoxia/reoxygenation show lung tissue pathologic lesions similar to those observed in human patients experiencing vaso-occlusive events. These lesions are characterized by an amplified inflammatory response, vessel contraction, congestion, and microvascular clogging. Inhaled NO significantly ameliorates the lung histopathology as supported by the observed reduction in inflammatory response and microvascular damage.
Being hypoxia is a cellular stress, different cell pathways might be affected by it, including expression of vasoactive substances and matrix proteins involved in remodeling microvasculature and surrounding tissue.21,45,46 Thus, inhaled NO may act directly as vasodilator and/or indirectly by switching off hypoxia-induced genes.45 Our results indicate that hypoxia/reoxygenation up-regulates Hspcb, Hsp86-1, and Nfe2l2 genes in SAD mice and that these molecular changes are prevented by inhaled NO. In wild-type mice subjected to hypoxia/reoxygenation, the up-regulation of Hspcb is similar to that seen in hypoxic SAD mice, whereas Hsp86-1 and Nfe2l2 expression is unaltered, resembling what is observed in hypoxic SAD mice breathing NO. This finding suggests that in both wild-type and SAD mouse strains Hspcb is induced by hypoxia/reoxygenation stress, whereas the up-regulation of Hsp86-1 and Nfe2l2 genes is related to the sickle cell phenotype during hypoxia/reoxygenation stress. Hspcb, Hsp86-1, and Nfe2l2 genes are molecular chaperons essential for cell survival.46-49 In mice, the expression of Hspcb, Hsp86-1 varies with respect to each other in response to different stress, such as hypoxia and/or inflammation.45,46 Hsp86-1 has been reported to be up-regulated, with no changes in Hspcb, in relation to inflammatory stimulus in a mouse model of osteoarthritis48 ; whereas Hspcb has been found to be up-regulated in an animal model of kidney ischemia.49 Lung hsp90 protein expression has been recently reported to be modulated by hypoxia in another transgenic sickle cell mouse model, lacking in lungs of control mice.15 Because the pulmonary circulation is an efficient filter and responds to hypoxia with vasoconstriction, the sickled red blood cells may be trapped in the lung microcirculation and not yet be present in the peripheral circulation, thereby promoting an amplified inflammatory response and ischemic tissue damage. Thus, in SAD mice, inhaled NO reduces ischemic/reperfusion lung injury at least in part by preventing the activation of injury-sensitive transcription factors and by attenuating the expression of stress-inducible genes that include Hspcb, Hsp86-1, and Nfe2l2.
Accumulating evidences on ischemic/reperfusion syndromes, suggest that the inhibition of ACE may reduce postischemic injury and prevent microcirculation remodeling.50,51 In SAD mice hypoxia/reoxygenation induced Ace up-regulation, which was delayed by inhaled NO. In wild-type mice hypoxia induces a persistent up-regulation of Ace, similarly to that reported in other animal models.50,51 The pathophysiologic role of Ace has been studied in Ace-deficient mice exposed to chronic hypoxia (4 weeks), which show a lower level of pulmonary vascular remodeling compared with healthy controls.50 Studies of the canine vascular pulmonary system have shown that the pulmonary microvascular endothelial cells51 are the most abundant source of hypoxia-sensitive Ace activity. Thus, inhaled NO in SAD mice may attenuate postischemic microvascular remodeling, which plays an important role in the development of hypoxic pulmonary hypertension,50,51 by modulating Ace expression.
Similarly to Ace, Fgf7 is a mitogen factor for type II alveolar epithelial and microvascular cells, and it is up-regulated during ischemia.52,53 In SAD mice, hypoxia/reoxygenation triggers Fgf7 up-regulation, whereas inhaled NO delayed Fgf7 up-regulation and, presumably, the subsequent structural remodeling.
The present results also indicate that under room air condition SAD mice display increased BAL total leukocyte and neutrophil counts, suggesting a basic chronic pulmonary inflammatory state, that becomes amplified in response to hypoxia/reoxygenation, as suggested by the further and persistent increase in BAL total leukocyte and neutrophil counts and in BAL total protein content. Hypoxia/reoxygenation in wild-type mice causes an increase in BAL total leukocyte and neutrophil count that is delayed and lower than in SAD mice, indicating that the rapid and amplified inflammatory response is related to SCD. Inhaled NO causes a significant reduction in BAL total leukocyte and neutrophil counts and in BAL total protein as well as in the lung inflammatory infiltration, suggesting a role for NO in inhibiting the leukocyte recruitment during the ischemic/reperfusion lung injury.
The neutrophil recruitment in hypoxia/reoxygenation-injured lungs of SAD mice may be linked to the activation of BAL cytokine network by activated lung macrophages and airway epithelial cells.54-57 The temporal sequence of TNF-α, IL-1α, and IL-6 changes in BAL of SAD mice exposed to hypoxia/reoxygenation suggests that TNF-α may be the early stimulus (4 hours) and IL-6 the late stimulus (10-46 hours) for neutrophil recruitment and activation, both cytokines synergizing with IL-1α. The present study suggests that MIP-2 may contribute to the in vivo accumulation of neutrophils in SAD mouse ischemic/reperfusion lung injury, primarily by acting as a chemotactic factor.54-57 The differences found between mRNA and BAL protein peaks have been previously reported in other animal models and may be related to preformed sources of this chemokine.54-57 Thus, in SAD mice, the acute hypoxic/reoxygenation lung injury stimulates first TNF-α, IL-1α, and MIP-2 from epithelial cells and resident alveolar macrophages. Next, IL-6, IL-1α, and MIP-2 synergize in contributing to the expansion of the lesion by recruiting mono-nuclear phagocytes. Inhaled NO may modulate in vivo the hypoxia/reoxygenation-induced cytokines and attenuate the inflammatory response, as supported by the observed changes in the cytokine network, the reduction in BAL neutrophil counts, the reduction in total protein content, and the amelioration of lung histopathology. In wild-type mice, hypoxia/reoxygenation did not activate the BAL cytokine network except for an increase in BAL TNF-α at 4 hours, indicating a lack of major inflammatory response that supports a crucial role of sickle cells in the pathogenesis of ischemic/reperfusion lung injury.
It has been reported that in the transgenic SAD mouse model exposed to severe hypoxia (6% O2, for 1 hour), inhaled NO (20 ppm) resulted in improvement of the survival rate, with no changes in methemoglobin levels and in oxyhemoglobin dissociation curves; the reported increase in methemoglobin was significant starting from the dose of 60 ppm inhaled NO.16 In patients with sickle cell disease, inhaled NO (80 ppm) does not modify the erythrocyte oxygen affinity, even when associated with a modest increase in methemoglobin.32,33 Here, we found that inhaled NO (40 ppm) ameliorates red blood cell dehydration in SAD mice exposed to 46 hours of hypoxia, with an increase in red blood cell K+ content. We previously reported that hypoxia-induced red blood cell dehydration in SAD mice is mostly mediated by the Ca2+-activated K+ channel and prevented by its specific inhibitor, clotrimazole.36 In another transgenic sickle cell mouse model, arginine diet supplementation, used to increase nitric oxide production, causes a decrease in the proportion of dense red blood cells associated with a reduction in the Ca2+-activated K+ channel activity.58 Thus, it is likely that inhaled NO ameliorates SAD red blood cell hypoxia/reoxygenation-induced dehydration through the modulation of Ca2+-activated K+ channel function.
In conclusion, the pulmonary beneficial effects of inhaled NO may be elective on the microcirculation and reperfused tissue in which endothelial NO production is reduced, providing protection during ischemia/reperfusion injury. The present study provides solid experimental evidences for a protective effect of inhaled NO in attenuating transgenic sickle cell SAD mouse injury induced by hypoxia/reoxygenation and provides the rationale for clinical trials with inhaled NO or NO donors in sickle cell patients with hypoxia-related lung injury.
Prepublished online as Blood First Edition Paper, April 10, 2003; DOI 10.1182/blood-2002-07-2135.
Supported by Verona University (MIUR 60%) (L.d.F.); Fondazione Cassa di Risparmio di Verona (Bando 2001) (A.S. and R.C.); Associazione Italiana Ricerca sul Cancro, Milan, Italy (A.S.); Italian Ministry of University and Research (grant 2001067845).
L.d.F. and A.B. contributed equally to this study.
Presented at the 43rd annual meeting of the American Society of Hematology, Orlando, FL, December 7-11, 2001.59
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Prof Carlo Brugnara, Harvard Medical School, Boston, MA, for the helpful discussion.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal