Abstract
In mammals, it is well documented that observable circadian rhythms are controlled by a central oscillator that is organized in transcriptional and translational feedback loops involving several clock genes. Although recent studies have demonstrated that clock genes oscillate in many peripheral tissues, their characteristics in the human immune system remain unknown. The present study investigates whether circadian clock genes function in human peripheral blood mononuclear cells. On the basis of studies derived from 3 human subjects under controlled conditions, circadian clock genes hPer1, hPer2, hPer3, and hDec1 are expressed in a circadian manner in human peripheral blood mononuclear cells (PBMCs), with the peak level occurring during the habitual time of activity. The demonstration of functional circadian machinery in human PBMCs suggests that peripheral blood cells may be useful for the investigation of human circadian rhythms and their associated disorders. (Blood. 2003;102:4143-4145)
Introduction
In humans, as in other animal species, most physiological and behavioral functions are expressed rhythmically across days and nights. These daily rhythms, referred to as circadian, are controlled by self-sustained biologic oscillators. In mammals, primary neuronal cultures as well as ablation and transplantation studies indicate that the central component of this complex oscillatory system resides in the suprachiasmatic nuclei (SCN) of the anterior hypothalamus.1,2 Molecular components of the circadian oscillator in the mouse have been found to involve transcriptional-translational feedback loops of at least 10 genes, namely, mPer1-3, mCry1-2, Clock, BMal1, Tau (casein kinase Iϵ), NPAS2, and Dec1-2. Of these, mPer1-2, mCry1-2, and Dec1-2 are negative regulators, whereas BMal1 and Clock/NPAS2 are positive regulators.3-5 In addition, Tau (casein kinase Iϵ) binds and phosphorylates period proteins (PERs), thereby posttranscriptionally regulating their stability.
Studies from rodents have shown that the molecular clock mechanisms oscillate not only in the central nervous system, but also in peripheral organs, such as the liver, kidney, and fibroblast cells.6 Given that the many immune parameters, such as cytokine synthesis and white blood cell counts, fluctuate in a circadian fashion,7 we hypothesized that a veritable circadian expression of functional clock components may be observed in human peripheral blood mononuclear cells (PBMCs) and, therefore, may play certain roles in controlling the immune circadian physiology.
In this brief report, we present the expression profile of circadian clock genes hPer1, hPer2, hPer3, and hDec1 in human PBMCs from 3 human subjects studied in a time-free environment. As human blood is widely used in clinical settings, our approach may not only provide a useful tool to elucidate the molecular mechanisms of human circadian rhythms, but also prompt the investigation of new means to diagnose and treat sleep/wake cycle and mood disturbances associated with jet lag, shift work, and a variety of medical and psychiatric disorders.
Study design
Three physically and mentally healthy young men, aged 20, 22, and 27 years, were recruited from the community.8 Each provided an informed consent in accordance with the guidelines of the Douglas Hospital Research Ethics Board (Montreal, QC, Canada). For 3 weeks prior to their admission, subjects kept regular sleep times, restricted to a single 8-hour nocturnal sleep episode. This procedure served to ensure that the circadian pacemaker was well stabilized relative to the sleep-wake schedule.
Each subject was studied individually in a time-free, light-controlled laboratory room for at least 5 consecutive days. After 3 baseline days on their habitual schedule, subjects underwent a 35-hour constant routine procedure designed to unmask the endogenous circadian oscillation of physiological parameters. This procedure consists of a regimen of enforced waking, in a semirecumbent posture, under very dim light conditions (approximately 4 lux), with minimal levels of activity and with nutritional intake divided into hourly snacks. Indeed, the expression of physiologic markers known to be controlled by the circadian pacemaker may be confounded by levels of light exposure, meals, or postural changes. Thus, sampling for approximately 1.5 days under these conditions allows us to investigate at least one complete circadian cycle with limited masking of the endogenous circadian expression.
Plasma melatonin concentration was determined by means of commercially available radioimmunoassays (Stockgrand, Surrey, United Kingdom). Every 120 minutes, PBMCs were isolated from heparinized blood samples by means of Histopaque-1077 (Sigma, St Louis, MO) gradient centrifugation, washed, and subsequently frozen at -80°C. The total RNA of each sample was extracted by means of Trizol reagent, and the cDNA was generated with Superscript II reverse transcriptase. We performed a TaqMan quantitative real-time reverse-transcription polymerase chain reaction (RT-PCR) to determine the level of hPer1, hPer2, hPer3, and hDec1 expression relative to the housekeeping gene hCDK4 by means of the standard protocol described by Applied Biosystems (ABI) (Foster City, CA).9
On the basis of the published sequences,10 the primer sets for hPer1, hPer2, hPer3, hDec1, and hCDK4 are designed as follows: hPer1 forward primer, 5′-TTCCTGACGGGCCGAAT-3′; hPer1 reverse primer, 5′-CGCTTGCAACGCAGCA-3′; hPer1 TaqMan probe, 5′-FAM-TCTACATTTCGGAGCAGGCAGCCG-Tamra-3′; hPer2 forward primer, 5′-CCACGAGAATGAAATCCGCT-3′; hPer2 reverse primer, 5′-CCCGCACCTTGACCAGG-3′; hPer2 TaqMan probe, 5′-FAM-CCACCCCTTCCGCATGACGC-Tamra-3′; hPer3 forward primer, 5′-TTTCCTAATGTCGCCGAAGAG-3′; hPer3 reverse primer, 5′-CCTGGTATGTCATGAGAATGCG-3′; hPer3 TaqMan probe, 5′-FAM-ATCTGGAGAATGATACGGCAGACACCTGA-Tamra-3′; hDEC1 forward primer, 5′-GAGAATCGGAGAAGGGCGAC-3′; hDEC1 reverse primer, 5′-GCGTCCGTGGTCACTTTTG-3′; hDEC1 TaqMan probe, 5′-FAM-TGCGCAGTGAGCAGCCGTGC-Tamra-3′; hCDK4 forward primer, 5′-GAGGCGACTGGAGGCTTTT-3′; hCDK4 reverse primer, 5′-GGATGTGGCACAGACGTCC-3′; hCDK4 TaqMan probe, 5′-FAM-AGCATCCCAATGTTGTCCGGCTGA-Tamra-3′.
A dual-harmonic regression model was used on individual curves for the expression of each gene with the use of a period search from 23.91 to 24.45 hours.11 As reported previously,12 a 12/24-hour composite model explains a greater amount of variance in the data. The circadian variation of transcriptional expression was considered significant if the 95% confidence interval (CI) describing the amplitude of the first harmonic did not include the zero axis. We also performed cross-correlation analyses between the variation in the expression of clock genes and that of the plasma melatonin secretion, a reliable circadian marker.
Results and discussion
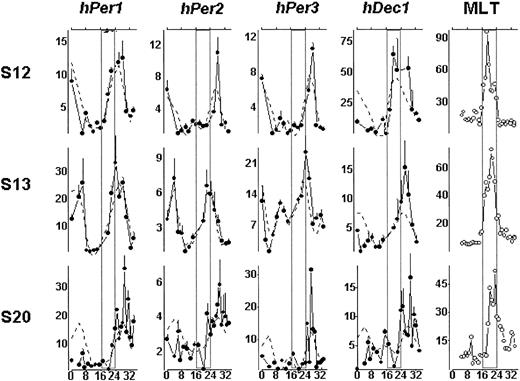
Regression analyses indicated that transcripts of all clock genes studied displayed a daily rhythm that was significantly different from zero with the exception of the expression of hDec1 for subjects S13 and S20 (Table 1). Peak clock gene expression was observed mostly during the usual time of activity and light exposure. Transcript levels of all 4 genes were found to correlate positively and significantly with the secretion of plasma melatonin, a reliable circadian marker. Peak clock gene expression followed the peak of melatonin concentration by a maximum of 9 hours (Figure 1). These results are consistent with animal studies since the oscillation of clock genes in peripheral tissues often lags several hours behind that of the SCN of the central nervous system.
Expression of clock gene mRNA in human PBMCs based on dual-harmonic regression analysis
Gene and subject . | Amplitude . | 95% CI for amplitude . | Time of fitted maximum . |
|---|---|---|---|
| hPer1 | |||
| S12 | 5.000 | 3.155-6.884 | 09:26 |
| S13 | 12.389 | 6.433-18.344 | 09:06 |
| S20 | 7.408 | 2.638-12.178 | 12:23 |
| hPer2 | |||
| S12 | 2.029 | 0.672-3.387 | 12:28 |
| S13 | 2.379 | 1.805-2.953 | 06:43 |
| S20 | 0.955 | 0.375-1.536 | 12:35 |
| hPer3 | |||
| S12 | 2.639 | 1.377-3.901 | 12:28 |
| S13 | 2.584 | 0.182-4.986 | 06:23 |
| S20 | 3.665 | 0.174-7.157 | 12:19 |
| hDec1 | |||
| S12 | 22.566 | 7.598-37.533 | 06:19 |
| S13 | 2.773 | −0.203-5.75 | 09:22 |
| S20 | 1.723 | −0.399-3.845 | 12:19 |
Gene and subject . | Amplitude . | 95% CI for amplitude . | Time of fitted maximum . |
|---|---|---|---|
| hPer1 | |||
| S12 | 5.000 | 3.155-6.884 | 09:26 |
| S13 | 12.389 | 6.433-18.344 | 09:06 |
| S20 | 7.408 | 2.638-12.178 | 12:23 |
| hPer2 | |||
| S12 | 2.029 | 0.672-3.387 | 12:28 |
| S13 | 2.379 | 1.805-2.953 | 06:43 |
| S20 | 0.955 | 0.375-1.536 | 12:35 |
| hPer3 | |||
| S12 | 2.639 | 1.377-3.901 | 12:28 |
| S13 | 2.584 | 0.182-4.986 | 06:23 |
| S20 | 3.665 | 0.174-7.157 | 12:19 |
| hDec1 | |||
| S12 | 22.566 | 7.598-37.533 | 06:19 |
| S13 | 2.773 | −0.203-5.75 | 09:22 |
| S20 | 1.723 | −0.399-3.845 | 12:19 |
Circadian variation of clock gene mRNA transcripts in human PBMCs. Transcript expression of hPer1, hPer2, hPer3, and hDec1 for each subject is shown with a solid line and the dual-harmonic regression with a dotted line. The y-axis represents the relative intensity of mRNA expression. The value of the lowest mRNA expression is designated 1, and the levels of mRNA expression at all other time points are calibrated to this value. Error bars indicate the standard deviation on the basis of the mRNA samples assayed in triplicates. The x-axis indicates the time, in hours, under the constant-routine procedure. In the melatonin (MLT) panel, the y-axis represents plasma melatonin concentration (picograms per milligram) in each subject. For the purposes of illustration, times where subjects are habitually asleep are projected as open rectangles (□).
Circadian variation of clock gene mRNA transcripts in human PBMCs. Transcript expression of hPer1, hPer2, hPer3, and hDec1 for each subject is shown with a solid line and the dual-harmonic regression with a dotted line. The y-axis represents the relative intensity of mRNA expression. The value of the lowest mRNA expression is designated 1, and the levels of mRNA expression at all other time points are calibrated to this value. Error bars indicate the standard deviation on the basis of the mRNA samples assayed in triplicates. The x-axis indicates the time, in hours, under the constant-routine procedure. In the melatonin (MLT) panel, the y-axis represents plasma melatonin concentration (picograms per milligram) in each subject. For the purposes of illustration, times where subjects are habitually asleep are projected as open rectangles (□).
Our findings of a significant oscillation of hPer mRNA levels across circadian phases in PBMCs of human subjects are consistent with the results of a prior study12 conducted under ambulatory conditions on oral mucosa and skin biopsies. One study also revealed that the expression of hPer2 in PBMCs is significantly different in the morning and the evening.13 The enforcement of constant conditions in the present study implies that the significant circadian oscillation we observe is not explained by postural changes, alteration in exposure to light, food intake, or the stress induced by needle insertion or biopsy punches. The use of the constant routine procedure also reveals that the oscillation of clock genes in PBMCs is present even in the absence of sleep/darkness episodes, an observation that supports its endogenous nature. While interindividual variability may account for the reduced regression amplitude observed for hDec1 transcripts in 2 subjects, it is possible that the period of awakening associated with the constant routine may have influenced our observations. Further investigation into the expression of clock genes under a variety of conditions will elucidate the influence of sleep and waking on clock gene expression in humans.
Although limited to a few subjects, our results support the presence of a significant and cyclic transcription of clock genes in peripheral PBMCs of healthy controls. They also demonstrate the feasibility of studying PBMCs as an accessible surrogate for the identification of rhythmic clock gene expression in humans. Experimental tools using this technique could thus be refined in humans to investigate the effects of various drugs on the sleep/wake cycle and endogenous circadian rhythms. Future studies should clarify the time relationship of clock gene expression in the SCN and PBMCs in lower mammals and extend these findings to humans.
Supported by grants from the Canadian Institutes of Health Research (D.B.B.) and the Edward Mallinckrodt, Jr Foundation (Z.S.S.).
Prepublished online as Blood First Edition Paper, July 31, 2003; DOI 10.1182/blood-2003-03-0779.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful to Dr N. M. Ng Ying Kin of the Douglas Hospital Research Center for the melatonin assays.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal