Abstract
Myelodysplastic and leukemic stem cell clones that evolve in children and adults with Fanconi anemia universally bear complex cytogenetic abnormalities. The abnormalities are generally recurring deletions or chromosomal loss and involve precisely the same chromosomes with the same frequency as has been described in marrow cells from patients with secondary acute leukemia induced by alkylating agents. Reasoning that acquired Fanconi anemia protein dysfunction might contribute to cytogenetic instability in secondary acute myelogenous leukemia (AML) cells, we analyzed leukemic cells bearing characteristic complex cytogenetic defects obtained from a 68-year-old man whose lymphoblasts showed no evidence of Fanconi anemia. Unlike the lymphoblasts, this myeloid leukemia cell line (UoC-M1) was hypersensitive to mitomycin-C (MMC) and diepoxybutane (DEB) and exhibited a marked decrease in nuclear FANCA, FANCG, and FANCD2-L. Retroviral transduction of FANCA significantly reduced MMC sensitivity but FANCF, FANCG, and FANCC did not. Overexpression of FANCA restored levels of both FANCA and FANCG, whereas overexpression of FANCG or FANCC did not restore FANCA levels. The molecular mass of cytoplasmic FANCA, FANCG, FANCC, and nuclear FANCD2 were normal. All exons of FANCA and FANCG were sequenced, and no mutations were found. We conclude that perturbations of as yet unidentified factors that govern the binding activity or intracellular localization of FANCA may promote cytogenetic instability and clonal progression in patients with AML who do not have Fanconi anemia. (Blood. 2003;102: 7-16)
Introduction
A variety of molecular defects have been discovered in patients with myeloproliferative syndromes and acute myelogenous leukemia, many of which result in the activation of autonomous signal transduction pathways for growth and survival.1-5 Although oncogenic mutations in such pathways are sufficient to induce myeloproliferative disorders in transgenic mice,2,3 the development of acute leukemia requires additional mutations that ultimately interfere with myeloid differentiation.6 The risk of this second type of mutation would be necessarily enhanced by cytogenetic instability, a manifestation of which in the leukemic clone would be multiple cytogenetic defects. That such complex cytogenetic defects universally occur in the myeloid leukemic clones of children with Fanconi anemia, a cytogenetic instability syndrome, supports this notion. In fact, because the clonal abnormalities found in this clinical context exactly recapitulate those found in patients with secondary myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML),7,8 and high-risk MDS,9 we hypothesized that acquired Fanconi anemia (FA) protein dysfunction might serve as a progression factor for the evolution of cytogenetically unstable AML clones, even in patients without hereditary evidence of Fanconi anemia. To test this notion, we examined a variety of AML cells and cell lines, including a cell line from a 68-year-old patient with cytogenetic defects characteristic of secondary AML. The patient's lymphoblasts were normal, but the leukemic cell population exhibited hypersensitivity to mitomycin C, a feature of Fanconi anemia. Nuclear FANCC, FANCG, and FANCA levels were markedly reduced, and retroviral complementation indicated that the leukemic cells exhibited a secondary defect of FANCA function. Because lymphoblasts from this same patient showed normal levels of nuclear Fanconi proteins, we propose that this acquired defect of FANCA function occurred during the course of leukemic clonal evolution and may have been the cause of the cytogenetic instability.
Patient, materials, and methods
Patient clinical history
A brief clinical history from this individual has been previously reported.10 There was no family history of Fanconi anemia or acute myelogenous leukemia. The patient had not been exposed to alkylating agents or chemical leukemogens and had no café-au-lait spots or developmental defects of skeleton, kidney, or nervous tissue.
Origins and maintenance of cell lines
The establishment of UoC-M1, a spontaneously immortalized, factor-independent, myeloid leukemia cell line has been previously reported.10 The cell line 4081 was obtained by Epstein-Barr virus (EBV)–mediated immortalization of peripheral blood, mononuclear cells from the same patient as UoC-M1 and is maintained in RPMI-1640 supplemented with 15% fetal calf serum (FCS; Hyclone Labs, Logan, UT), penicillin, and streptomycin. The human, cervical adenocarcinoma cell line HeLa was obtained from the American Type Culture Collection (ATCC, Manassas, VA). Human FANCE cDNA was kindly provided by James Hejna (Oregon Health & Science University, Portland). The FANCE coding sequence (GenBank no.AF265210, bp 181-1815) was subcloned into the EcoRI/Xhol sites of pcDNA3.1+ (Invitrogen Life Technologies, Carlsbad, CA). This plasmid was stably transfected into subconfluent HeLa cells by the calcium phosphate method followed by selection in G418. The FA-C lymphoblastoid cell line HSC536N was generously contributed by Manuel Buchwald (Hospital for Sick Children, University of Toronto). The factor-dependent, myeloid cell line Mo7e11 was a gift from Hal E. Broxmeyer (Indiana University, Bloomington).
Breakage analysis
Cultures were treated with 40 ng/mL mitomycin C (MMC) or 200 ng/mL diepoxybutane (DEB) for 72 hours in the dark, at 37°C. Cultures were then harvested after a 2-hour exposure to 0.25 μg/mL Colcemid (Sigma, St Louis, MO). After a 10-minute treatment with 75 mM KCl, the cells were fixed with a 3:1 mixture of methanol and acetic acid, respectively. Wet-mount slides were prepared, air-dried, and treated with Wright stain. At least 50 metaphase cells from each culture were scored for DNA breaks and radial formations per experiment.
Quantitative real-time RT-PCR
Quantitative real-time reverse-transcription polymerase chain reaction (QRT-PCR) analysis was performed in triplicate (or more in some instances) for each primer set and in each cell line. The overall number of UoC-M1 amplification cycles per primer set was calibrated against an internal control of 18S cDNA. The cycle number was then standardized against the cycle number for the same primer pair in a concurrently amplified reaction in the myeloid cell line Mo7e. For each FA gene, the amplification threshold obtained with Mo7e was defined as a fold value of 1. Total cellular RNA was isolated using TriReagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. cDNA synthesis was performed with 1 μg total RNA and Superscript II reverse transcriptase (Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. QRT-PCR was then performed using an ABI Prism 7700 Sequence Detection System (ABI, Foster City, CA). Amplification was performed using the TaqMan PCR system (ABI) per the manufacturer's instructions. Specific primer and probe sequences were predicted using ABI Primer Express software (ABI). All oligomers used were obtained from Integrated DNA Technologies (Coralville, IA). The sequences are as follows: hFANCA (forward primer), 5′-CGTCATGGAGGCCAGCATAT-3′; hFANCA (reverse primer), 5′-CACGGGAGTCAGGGACTTTG-3′; hFANCA (dual-labeled probe), 5′-FAM-CCCGCCCTGCTCACACCTCG-TAM-3′; hFANCC (forward), 5′-TGGCTCCAGACACTGAAGCA-3′; hFANCC (reverse), 5′-AACAGGAACCAGCTCTCAAAGG-3′; hFANCC (dual-labeled probe), 5′-FAM-AAGCAGTTGAAGACCAGACTCATGGGTCCT-TAM-3′; hFANCD2 (forward), 5′-GGGATTCCCAGCACGCT-3′; hFANCD2 (reverse), 5′-TCGGAGGCTTGAAAGGACAT-3′; hFANCD2 (dual-labeled probe), 5′-FAM-CTCAGTGACCTACTGATAGAGAATACTTCACTCACTGTCC-TAM-3′; hFANCE (forward), 5′-GACCAGCCTGTCATGGCAGT-3′; hFANCE (reverse), 5′-TCCTGGATAGCTTTGGGCAA-3′; hFANCE (dual-labeled probe), 5′-FAM-AAGACTGGCGAGGACGGTTCGAATC-TAM-3′; hFANCF (forward), 5′-TTCCTGAAGGTGATAGCGGTG-3′; hFANCF (reverse), 5′-CTCCCCTCTCCAGGTGATTTG-3′; hFANCF (dual-labeled probe), 5′-FAM-CTGTTGCAGCCGCCTTTGTCTCGT-TAM-3′; hFANCG (forward), 5′-CTCCCTGCAGCTGTTCCTGT-3′; hFANCG (reverse), 5′-GCCTGATCCTCTGTGAAACCC-3′; hFANCG (dual-labeled probe), 5′-FAM-CCTTGGAGCTGACTGTCACCTGCAACT-TAM-3′; h18S (forward), 5′-CGGCTACCACATCCAAGGAA-3′; h18S (reverse), 5′-GGGCCTCGA AAGAGTCCTGT-3′; and 18S (dual-labeled probe), 5′-VIC-CAGCAGGCGCGCAAATTACCCA-TAM-3′.
FANCD2 and FANCE antibody production
A 14-amino acid polypeptide (RDLQGEEIKSQNSQ) was synthesized corresponding to amino acids 1392 to 1405 of FANCD2 (1471 total). For FANCE, a hydrophilic 14-amino acid polypeptide (EEENRDSQQPGKRR) was synthesized corresponding to amino acids 188 to 201 (536 total). Additional cysteine residues were added to the N-termini to facilitate conjugation to carrier protein. These peptides were used to raise polyclonal antisera against FANCD2 and FANCE (ProSci, Poway, CA). FANCD2 antiserum was characterized by immunoblot analysis of HeLa, HeLa/FANCD2, and PD20L lysates using antiserum blocked or unblocked with peptide prior to immunoblotting. PD20 is an FA-D2 cell line and was generously contributed by Markus Grompe (Oregon Health & Science University, Portland). FANCE antisera were screened against wild-type HeLa cells and HeLa cells overexpressing FANCE protein. Positive antisera were affinity purified with the peptides described earlier bound to a HiTrap N-hydroxy-succinimide (NHS)–activated column (Amersham Pharmacia Biotech AB, Piscataway, NJ) according to the manufacturer's recommendations.
Cell fractionation and immunoblot analysis
Whole cell extracts were obtained as previously described.12,13 Subcellular fractions were prepared as previously described.14 4081 cell extracts in 50 mM Tris (tris(hydroxymethyl)aminomethane) pH 7.4, 150 mM NaCl, 1% Triton X-100 were precleared with Sepharose CL-4B (Amersaham-Pharmacia) and then depleted of ubiquitin with a monoclonal mouse antiubiquitin antibody (catalog no. 13-1600; Zymed Laboratories, San Francisco, CA) with overnight rocking at 4°C. Antibody-bound ubiquitin was removed by incubating with Protein A-Sepharose, and the resulting extracts were analyzed. Proteins were resolved via denaturing, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (7.5%) and immobilized to nitrocellulose membranes (Bio-Rad, Hercules, CA) by electroblotting as previously described.15 Polyclonal antibodies raised against FANCD2 (described in “FANCD2 and FANCE antibody production”), FANCC,14 FANCA, and FANCG (the latter 2 were kindly provided by Alan D'Andrea, Dana-Farber Cancer Institute, Boston, MA) were exposed to blocked filters at 4°C overnight in a solution of 5% (wt/vol) powdered milk (Nestle, Solon, OH) in Tris-buffered saline (TBS; 10 mM Tris-base, 150 mM NaCl, pH 8.0). The following day, filters were washed 6 times in a solution of 50 μL TWEEN-20 (BioRad) per liter of TBS. Goat antirabbit-horseradish peroxidase (HRP) secondary antibodies (BioRad) were then added in a 5% solution of powdered milk in TBS with rocking for 1 hour at room temperature. For BRCA2 analysis, extracts were prepared with Nonidet P-40 (NP-40) buffer (1% NP-40, 20 mM Tris-HCl, pH 8.0, 137 mM NaCl, and 10% glycerol) containing protease inhibitors as previously described.14 Samples were electrophoresed on NuPAGE 3% to 8% Tris-Acetate gels (Invitrogen, Carlsbad, CA) for 3 hours and transferred to nitrocellulose (Bio-Rad) via electroblotting (90 V constant for 5 hours at 4°C). Blots were incubated with either polyclonal FANCD2 antiserum (described in “FANCD2 and FANCE antibody production”) or polyclonal anti-BRCA2 antiserum (Ab-2; Oncogene Research, San Diego, CA).
Retroviral transduction
The retroviral constructs used in these studies16,17 were kindly provided by Alan D'Andrea, MD (Dana-Farber Cancer Institute). Each vector contains a puromycin-resistance gene. High-titer retroviral supernatants containing 1 of the 4 Fanconi anemia cDNAs (pMMP-FAA IRES/PURO, pMMP-FAC IRES/PURO, pMMP-FAF IRES/PURO, and pMMP-FAG IRES/PURO) were obtained. The resulting puromycin-resistant, retrovirally transduced lines were subsequently identified as UoC-M1/FA, UoC-M1/FC, UoC-M1/FF, and UoC-M1/FG.
Statistical analysis
The statistical significance of differential chromosomal breakage and radial formation after exposure to MMC or DEB was determined via contingency table analysis (chi-square) on pooled data from multiple, independent experiments. Two different MMC sensitivity comparisons were (1) between UoC-M1 controls and cell lines retrovirally transduced with constructs encoding 4 different Fanconi genes (UoC-M1/FA, UoC-M1/FC, UoC-M1/FF, and UoC-M1/FG) in pair-wise comparisons, and (2) between UoC-M1 controls and an EBV-transformed lymphoblastoid cell line from the same patient (4081). Yates correction for continuity was used.18 In each analysis, the null hypothesis (Ho) was defined as no difference between the data sets compared. Cutoffs were established wherein differential sensitivities were required to meet a P < .05 to be labeled as significant. The standard deviation (SD) was calculated on the basis of pooled data from all experiments for each cell type tested.
Gene dosage
For FANCA, the gene dosage, multiplex assay was based on that previously described by Morgan et al.19 A multiplex assay has also been designed for FANCG using exons 1 and 14. Primer sequences have been previously published19 with the following exceptions: FANCA, exon 19/20, forward, 5′-GAAACACCGGTCACCGTCTGTG-3′; FANCA, exon 19/20, reverse, 5′-AGATCCACGATTCTTCGCATTGTC-3′; FANCG, exon 1, forward, 5′-CCTTCCTAAGTCCGCTTCTG-3′; FANCG, exon 1, reverse, 5′-CACATGAGGGAGGGGTTGTCAC-3′; FANCG, exon 14, forward, 5′-CCAGCTTTCTGCCAATCTTT-3′; FANCG, exon 14, reverse, 5′-CACAGAGAGACAGCCCACTG-3′; NCX1, exon 1, forward, 5′-CACTGTTCTCTGTTCCCAGG-3′; NCX1, exon 1, reverse, 5′-TGACATGTTTCAAAAGTGGC-3′. The sizes in base-pairs (bp) of the expected amplicons from each primer set are the following: FANCA, exon 5, 250 bp; FANCA, exon 11, 301 bp; FANCA, exon 17, 207 bp; FANCA, exon 19/20, 348 bp; FANCA, exon 31, 308 bp; FANCG, exon 1, 234 bp; FANCG, exon 14, 214 bp; MPZ, exon 1, 385 bp; NCX1, exon 1, 226 bp. Only 22 PCR cycles were performed to keep the reaction within the exponential phase.20 An aliquot of the PCR product (3 μL) was added to 16.5 μL formamide and 0.5 μL internal lane size standard (Genescan-500 Rox; ABI). The samples were denatured for 5 minutes at 94°C, snap-cooled on ice, and transferred to an ABI 310 capillary-based system (ABI). Data were then analyzed by means of Genescan software to obtain electrophoretograms for each sample.
Fanconi gene sequencing
Sequences of interest were amplified using PCR, and the products were purified from 1% agarose gels using a commercial kit (Qiagen, Valencia, CA). Purified product (5 μL) was then added to 4 μL commercial sequencing kit (ABI), 1.6 pmol either the forward of reverse primer, and using a Perkin-Elmer 480 thermocycler 25 amplification cycles of 96°C for 30 seconds, 50°C for 15 seconds, and 60°C for 4 minutes were carried out. Salt precipitation was achieved by adding 10 μL product to 37 μL of 0.5 mM MgCl2/70% EtOH and leaving to stand for 10 to 15 minutes after which the mixture was centrifuged at 13 000 rpm for 20 minutes. The supernatant was removed, and the pellet was left to air dry for 20 minutes before being resuspended in 12 μL Template Suppression Agent (ABI), denatured (5 minutes at 94°C), and snap-cooled on ice. Sequence data were obtained using an ABI 310 capillary-based system and analyzed using Sequence Navigator software (ABI).
Haplotype analysis
To verify the genetic identity of the UoC-M1 myeloid and 4081 lymphoblastoid cell lines, we constructed haplotypes using PCR-based genomic mapping panels for human chromosomes 7 and 8 (ABI). Two Centre d'Etude du Polymorphisme Humain (CEPH) reference DNAs were used as standards for individual allele size calculations (CEPH 1347-02 and 1347-15) (http://www.cephb.fr/cephdb/). PCR reactions were subjected to capillary-gel electrophoresis in an ABI-Prism 310 machine (ABI) to resolve and detect individual amplicons. Alleles were labeled according to the calculated amplicon sizes (GeneScan version 3.1 and Genotyper version 2.5; ABI) obtained for each primer set as compared with concurrently amplified and electrophoresed CEPH genomic DNA controls (CEPH pedigree 1347) (http://www.cephb.fr/cephdb/) and a concurrently electrophoresed Genescan-400 Rox size standard (ABI). An estimate of the overall haplotype frequency was obtained by multiplying the individual allele frequencies reported for each marker as indicated in the CEPH marker database (http://www.cephb.fr/cephdb/).
Results
UoC-M1 cells are hypersensitive to MMC
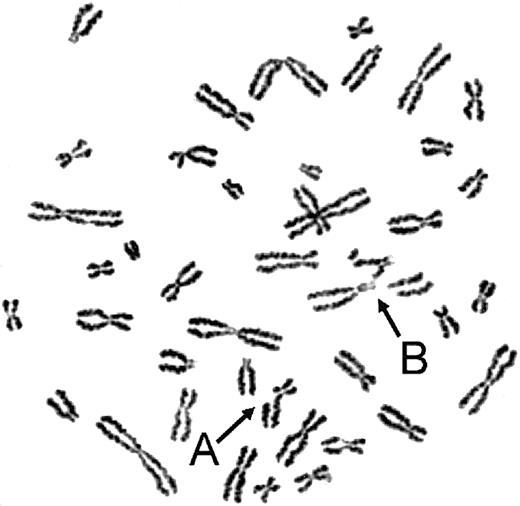
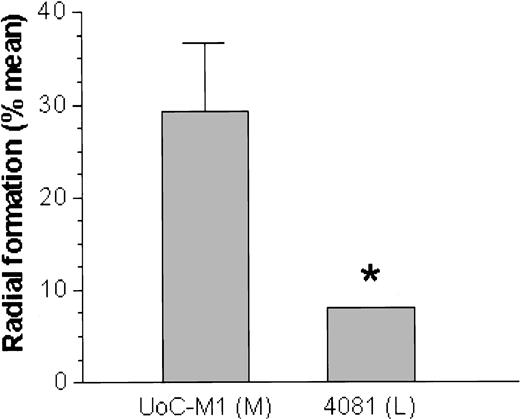
On exposure to the bifunctional alkylating agents MMC or DEB, the myeloid cell line UoC-M1 demonstrated extensive chromosomal breakage and unusual end-to-end chromosomal fusions termed radial forms (Figure 1). Even in the absence of clastogens, the cells occasionally exhibited chromosomal breakage (data not shown). These experiments were performed against an independent myeloid leukemic cell line (FA-AML1), and the sample identities were reconfirmed by virtue of their distinct G-banded karyotypes (data not shown). Although the percentage of clastogen-induced radial formation in UoC-M1 varied across multiple experiments, it never fell below the diagnostic cutoff used in our laboratory (20%). The fraction of metaphases exhibiting radial forms was 29.3% ± 7.6% (n = 300; range, 20%-38%) (Figure 2, column 1). To rule out the possibility that UoC-M1 cells represent material from a person that truly had hereditary Fanconi anemia (the patient's advanced age and unexceptional prior medical and family history make this unlikely), we obtained EBV-immortalized lymphoblastoid cells derived from the same individual (cell line 4081) and subjected them to MMC/DEB sensitivity testing. As shown in Figure 2, the percentage of radial formation in 4081 (8%) is significantly lower than that found in UoC-M1 (chi-square value of 8.995, 1 degree of freedom, P = .003) and within the normal range (< 10%). As increased cellular sensitivity to chromosomal damage following exposure to MMC and DEB represents the current diagnostic laboratory standard for Fanconi anemia,21 we next sought to determine whether the clastogen sensitivity demonstrated by UoC-M1 could be the result of acquired abnormalities in the Fanconi anemia pathway.
UoC-M1 is sensitive to chromosomal breakage and radial formation following MMC treatment. Arrows indicate representative chromosomal anomalies in UoC-M1: (A) single chromatid break and (B) complex chromosomal rearrangement with at least 2 chromosomal breaks and one end-to-end fusion (radial). Such findings in clastogen-treated cells represent the clinical diagnostic laboratory standard for Fanconi anemia. UoC-M1 also demonstrated occasional, low-level chromosomal breakage without clastogen treatment, although most of the untreated cells had no evidence of instability. No radial forms or cells with multiple chromosomal breaks were ever noted in untreated specimens (of more than 500 total metaphases analyzed), although cells with single breaks were occasionally noted (maximum of 20% in an untreated specimen that when clastogen treated in a simultaneously prepared, parallel flask demonstrated 26% breakage with up to 3 breaks per cell and 38% radial formation; data not shown).
UoC-M1 is sensitive to chromosomal breakage and radial formation following MMC treatment. Arrows indicate representative chromosomal anomalies in UoC-M1: (A) single chromatid break and (B) complex chromosomal rearrangement with at least 2 chromosomal breaks and one end-to-end fusion (radial). Such findings in clastogen-treated cells represent the clinical diagnostic laboratory standard for Fanconi anemia. UoC-M1 also demonstrated occasional, low-level chromosomal breakage without clastogen treatment, although most of the untreated cells had no evidence of instability. No radial forms or cells with multiple chromosomal breaks were ever noted in untreated specimens (of more than 500 total metaphases analyzed), although cells with single breaks were occasionally noted (maximum of 20% in an untreated specimen that when clastogen treated in a simultaneously prepared, parallel flask demonstrated 26% breakage with up to 3 breaks per cell and 38% radial formation; data not shown).
A lymphoblastoid cell line from the same patient is resistant to MMC. The myeloid cell line (UoC-M1) was exposed to MMC as was the lymphoblastoid cell line (4081) derived from the same patient. Although the UoC-M1 cell line consistently demonstrated radial formation in excess of 20% (mean = 29%) in multiple experiments (left bar), the 4081 cell line was resistant (P = .003) to chromosomal breaks and radial formation after exposure to MMC (right bar). Treatment with DEB showed a similar difference. Error bars represent ± SD. *P = .003; M indicates myeloid; L, lymphoid.
A lymphoblastoid cell line from the same patient is resistant to MMC. The myeloid cell line (UoC-M1) was exposed to MMC as was the lymphoblastoid cell line (4081) derived from the same patient. Although the UoC-M1 cell line consistently demonstrated radial formation in excess of 20% (mean = 29%) in multiple experiments (left bar), the 4081 cell line was resistant (P = .003) to chromosomal breaks and radial formation after exposure to MMC (right bar). Treatment with DEB showed a similar difference. Error bars represent ± SD. *P = .003; M indicates myeloid; L, lymphoid.
FA gene transcripts are quantitatively normal
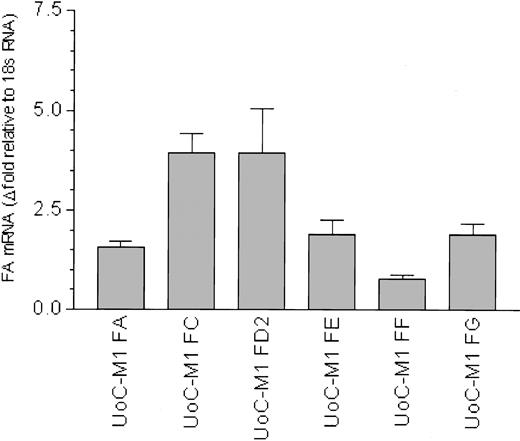
To date, 7 FA genes have been cloned, and the sequences of the representative cDNAs for the FANCA,22,23 FANCC,24 FANCD2,25 FANCE,26 FANCF,27 FANCG,28 and FANCD1 (BRCA2)29 genes are available. QRT-PCR was used to quantify transcripts from each of these genes relative to an internal amplification standard (18S RNA) and also compared with the myeloid cell line Mo7e (Figure 3). Mo7e was used as a control for 3 reasons: (1) it is not MMC hypersensitive, (2) it is in the same myeloid lineage as UoC-M1 (megakaryoblastoid),11 and (3) it shares a myeloid leukemic phenotype with UoC-M1.11 Although FA mRNA levels vary slightly between UoC-M1 and Mo7e (Figure 3, columns 1-6), the RNA levels for FANCA, FANCC, FANCD2, FANCE, and FANCG were equivalent to or more abundant than those observed in Mo7e. An exception to this was the observed 2-fold reduction in FANCF mRNA (Figure 3, column 5), a reduction we attribute to monosomy 11p found in UoC-M1 cell,10 as FANCF is located on human chromosome band 11p15.27 Levels of FANCA, FANCC, and FANCG mRNA (Figure 3, columns 1, 2, and 6) are slightly increased relative to Mo7e. We conclude that the MMC hypersensitivity of UoC-M1 cells does not derive from reduced transcription of Fanconi genes.
Fanconi anemia gene transcripts are not reduced in UoC-M1. QRT-PCR reactions were performed in at least triplicate. Error bars represent standard deviations. Fold values were obtained by externally standardizing against identical amplifications in Mo7e and by internally standardizing against 18S RNA in each cell line. Compared with Mo7e, UoC-M1 does not exhibit reduced levels of mRNA for FANCA, FANCC, FANCD2, FANCE, and FANCG. An approximate 2-fold reduction in FANCF was observed. This finding was attributed to monosomy 11p in UoC-M1,10 as FANCF is encoded on chromosome 11.27
Fanconi anemia gene transcripts are not reduced in UoC-M1. QRT-PCR reactions were performed in at least triplicate. Error bars represent standard deviations. Fold values were obtained by externally standardizing against identical amplifications in Mo7e and by internally standardizing against 18S RNA in each cell line. Compared with Mo7e, UoC-M1 does not exhibit reduced levels of mRNA for FANCA, FANCC, FANCD2, FANCE, and FANCG. An approximate 2-fold reduction in FANCF was observed. This finding was attributed to monosomy 11p in UoC-M1,10 as FANCF is encoded on chromosome 11.27
Nuclear FA proteins are markedly reduced
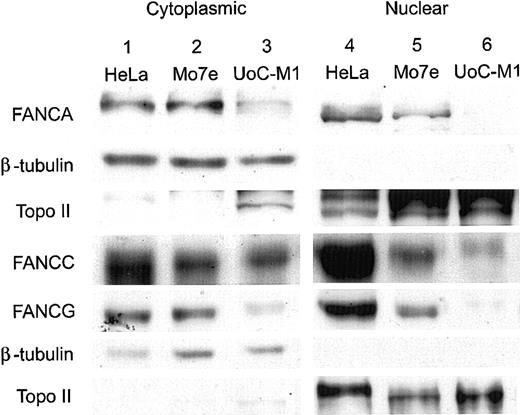
As shown in Figure 4, compared with the human, cervical carcinoma cell line HeLa (lane 1) and the factor-dependent, myeloid cell line Mo7e (lane 2), UoC-M1 (lane 3) had a mild reduction in the levels of cytoplasmic FANCA, FANCC, and FANCG proteins (rows 1, 4, and 5, respectively). FANCE levels in UoC-M1 were comparable to controls (data not shown). However, nuclear fractions (lanes 4-6) demonstrated a reduction of FANCC in UoC-M1 (row 4, lane 6) and a near absence of nuclear FANCA (row 1, lane 6) and FANCG proteins (row 5, lane 6). This observed reduction confirmed earlier experiments performed with whole cell extracts (data not shown) and further indicated that the most substantial reductions in UoC-M1 cells were due to losses in the nuclear compartment, the compartment in which Fanconi protein complexes function to effect MMC resistance.13 Purity of subcellular fractions was confirmed by reprobing stripped filters with antisera against beta-tubulin (Roche Diagnostics, Indianapolis, IN) (rows 2 and 6) and topoisomerase II (NeoMarkers, Fremont, CA) (rows 3 and 7). To investigate the significance of nuclear FA protein loss in UoC-M1, a downstream component of the FA protein pathway (FANCD2-L) was assayed via immunoblot analysis.
Nuclear FA proteins are reduced in UoC-M1 cells. Immunoblots of cytoplasmic and nuclear components were probed with antisera for the FANCA, FANCC, and FANCG proteins. Compared with the human, cervical carcinoma cell line HeLa (lane 1) and the factor-dependent, myeloid cell line Mo7e (lane 2), UoC-M1 (lane 3) has a mild reduction in the levels of cytoplasmic FANCA, FANCC, and FANCG proteins (rows 1, 4, and 5, respectively). Nuclear fractions (lanes 4-6) demonstrated a reduction of FANCC in UoC-M1 (row 4, lane 6) and a nearly complete absence of nuclear FANCA (row 1, lane 6) and FANCG proteins (row 5, lane 6). Purity of nuclear fractions was confirmed by reprobing the filters with antisera against beta-tubulin (rows 2 and 6) and topoisomerase II (rows 3 and 7).
Nuclear FA proteins are reduced in UoC-M1 cells. Immunoblots of cytoplasmic and nuclear components were probed with antisera for the FANCA, FANCC, and FANCG proteins. Compared with the human, cervical carcinoma cell line HeLa (lane 1) and the factor-dependent, myeloid cell line Mo7e (lane 2), UoC-M1 (lane 3) has a mild reduction in the levels of cytoplasmic FANCA, FANCC, and FANCG proteins (rows 1, 4, and 5, respectively). Nuclear fractions (lanes 4-6) demonstrated a reduction of FANCC in UoC-M1 (row 4, lane 6) and a nearly complete absence of nuclear FANCA (row 1, lane 6) and FANCG proteins (row 5, lane 6). Purity of nuclear fractions was confirmed by reprobing the filters with antisera against beta-tubulin (rows 2 and 6) and topoisomerase II (rows 3 and 7).
FANCD2 is not mono-ubiquitinylated in UoC-M1 cells
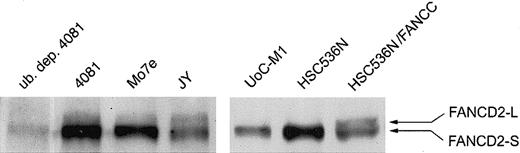
A nuclear multimeric complex of the FANCA, FANCC, FANCF, FANCE, and FANCG proteins is required for the monoubiquitinylation of FANCD2, a posttranslational change enhanced by DNA damage.13 This posttranslational modification of FANCD2 is thought to be required for normal cellular responses to certain types of genotoxic stress.13 If the reduction of nuclear FANCA and FANCG in UoCM1 cells is the cause of MMC hypersensitivity, then FANCD2-L should be undetectable. As shown in Figure 5, UoC-M1 (lane 5) does not demonstrate the mono-ubiquitinylated form of FANCD2 (FANCD2-L for long form). FANCD2-L is also absent in lane 6 containing an extract from HSC536N, an EBV-transformed lymphoblastoid line from an FA-C patient, but is present in the FANCC complemented FA-C cells (lane 7). The 4081 lymphoblastoid cell line obtained from the same patient as UoC-M1 exhibits FANCD2-L formation (lane 2). The FANCD2-L band disappeared in lysates depleted of ubiquitin (lane 1). Lanes 3 (the Mo7e myeloid leukemic cell line) and 4 (JY, an EBV-transformed lymphoblastoid cell line from a healthy volunteer) both demonstrate FANCD2-L. Additionally, as truncating mutations of BRCA2 have recently been shown to cause FA complementation group D1 (FA-D1),29 we investigated BRCA2 via immunoblot analysis. Full-length BRCA2 was detected in UoC-M1 cells at levels comparable to HeLa controls but not in concurrently analyzed FANCD1 cells or the BRCA2 mutant cell line DU145,30 ruling out the possibility that BRCA2 loss has occurred in UoC-M1 (data not shown). To further establish a linkage between MMC sensitivity in UoC-M1 cells and the absence of nuclear FA proteins and absence of FANCD2-L, we transduced UoC-M1 cells with each of 4 retroviral vectors encoding FANCA, FANCC, FANCF, and FANCG.
UoC-M1 does not monoubiquitinylate FANCD2. A lymphoblastoid cell line control from an FA-C patient (HSC536N) does not exhibit the monoubiquitinylated form of FANCD2 (FANCD2-L), whereas retroviral correction of the FANCC defect in these cells (HSC536N/FANCC) restores FANCD2-L. UoC-M1 is incapable of forming FANCD2-L. The 4081 lymphoblastoid cell line (from the same patient as the UoC-M1 myeloid cell line) demonstrates FANCD2-L that is capable of being immunodepleted with a monoclonal antiubiquitin antibody (lane 1; ub. dep. 4081). Both Mo7e (a factor-dependent myeloid leukemia cell line) and JY (an EBV-transformed lymphoblastoid line from a healthy volunteer) demonstrate cellular resistance to MMC and FANCD2-L.
UoC-M1 does not monoubiquitinylate FANCD2. A lymphoblastoid cell line control from an FA-C patient (HSC536N) does not exhibit the monoubiquitinylated form of FANCD2 (FANCD2-L), whereas retroviral correction of the FANCC defect in these cells (HSC536N/FANCC) restores FANCD2-L. UoC-M1 is incapable of forming FANCD2-L. The 4081 lymphoblastoid cell line (from the same patient as the UoC-M1 myeloid cell line) demonstrates FANCD2-L that is capable of being immunodepleted with a monoclonal antiubiquitin antibody (lane 1; ub. dep. 4081). Both Mo7e (a factor-dependent myeloid leukemia cell line) and JY (an EBV-transformed lymphoblastoid line from a healthy volunteer) demonstrate cellular resistance to MMC and FANCD2-L.
Functional complementation with a FANCA cDNA
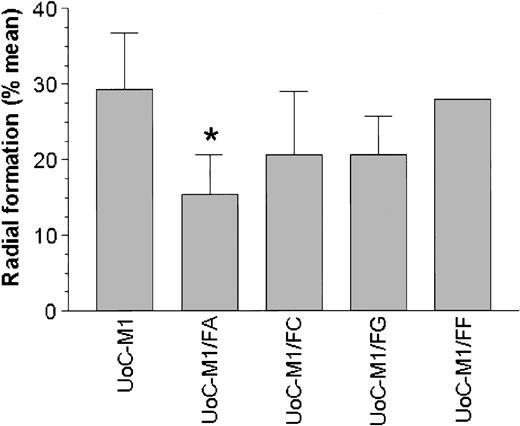
UoC-M1 cells were retrovirally transduced with the coding sequences for the FANCA, FANCC, FANCF, or FANCG cDNA and were positively selected with puromycin. MMC sensitivity was determined in each resulting cell line (UoC-M1/FA, UoC-M1/FC, UoC-M1/FF, and UoC-M1/FG). As shown in Figure 6, only transduction with the cDNA encoding FANCA (cell line UoC-M1/FA) resulted in a significant reduction in MMC sensitivity in UoC-M1 (chi-square value of 10.058 with 1 degree of freedom, P = .002). Transduction with constructs encoding FANCC (UoC-M1/FC) and FANCG (UoC-M1/FG) failed to significantly reduce MMC sensitivity (P = .059, and P = .064, respectively). In 2 independent experiments, transduction of UoC-M1 with the FANCF vector (UoC-M1/FF) did not influence MMC sensitivity (average radial formation of 32% over 100 total cells). The data in UoC-M1/FA support the notion that an anomaly of FANCA function is directly related to MMC sensitivity in UoC-M1. We next assured that each transgene was highly expressed in transduced cells (Figure 7).
Introduction of a FANCA cDNA reduces MMC sensitivity in UoC-M1. MMC resistance was evaluated in transduced UoCM-1 cells using cytogenetic analysis of radial formation after treatment with MMC. Transduction with a FANCA cDNA (UoC-M1/FA) reduced MMC resistance significantly (*P = .002). Neither FANCC nor FANCG transduction (UoC-M1/FC and UoC-M1/FG) significantly altered MMC sensitivity (P = .059 and P = .064, respectively). UoC-M1/FF demonstrated continued MMC sensitivity in 2 independent experiments with 38% and 26% radial formation in each (of 50 metaphases analyzed per experiment). Values shown are means ± SD.
Introduction of a FANCA cDNA reduces MMC sensitivity in UoC-M1. MMC resistance was evaluated in transduced UoCM-1 cells using cytogenetic analysis of radial formation after treatment with MMC. Transduction with a FANCA cDNA (UoC-M1/FA) reduced MMC resistance significantly (*P = .002). Neither FANCC nor FANCG transduction (UoC-M1/FC and UoC-M1/FG) significantly altered MMC sensitivity (P = .059 and P = .064, respectively). UoC-M1/FF demonstrated continued MMC sensitivity in 2 independent experiments with 38% and 26% radial formation in each (of 50 metaphases analyzed per experiment). Values shown are means ± SD.
FANCA transduction enhances FA protein complex stability and restores nuclear FANCA levels. (A) Whole cell lysates. All 4 rows depict data obtained from the same filter, cut horizontally at 80 kDa to allow FANCA to be interrogated on the top portion and FANCC (then FANCG after stripping the FANCC signal) on the bottom portion. Retroviral transduction with each of 3 FA cDNA constructs resulted in increased levels of their respective proteins (the results from independently obtained duplicates are shown for each transduced cell line). Transduction with FANCA (UoC-M1/FA) also led to an increase in cellular levels of FANCG, whereas the reverse was not observed (ie, transduction with FANCG [UoC-M1/FG] did not increase cellular levels of FANCA). Row 4 is a constant, nonspecific fragment detected by the FANCG antibody and demonstrates equal loading in all lanes. (B) Nuclear extracts from 3 transduced lines (UoC-M1/FA, UoC-M1/FC, and UoC-M1/FG), and the myeloid cell line Mo7e probed with anti-FANCA (row 1) and anti-FANCG (row 2) antisera. Transduction with FANCA restored nuclear levels of FANCA in UoC-M1/FA (row 1, column 2), whereas transduction with FANCG did not increase nuclear levels of FANCG (UoC-M1/FG) (row 2, column 5). (C) Nuclear levels of FANCA and FANCG are not reduced in the lymphoblastoid cell line 4081. The cell line JY is an EBV-transformed lymphoblastoid cell line established from a healthy adult volunteer and also demonstrates cellular resistance to MMC. Lane 1 is a whole cell extract (WCL) from HeLa cells.
FANCA transduction enhances FA protein complex stability and restores nuclear FANCA levels. (A) Whole cell lysates. All 4 rows depict data obtained from the same filter, cut horizontally at 80 kDa to allow FANCA to be interrogated on the top portion and FANCC (then FANCG after stripping the FANCC signal) on the bottom portion. Retroviral transduction with each of 3 FA cDNA constructs resulted in increased levels of their respective proteins (the results from independently obtained duplicates are shown for each transduced cell line). Transduction with FANCA (UoC-M1/FA) also led to an increase in cellular levels of FANCG, whereas the reverse was not observed (ie, transduction with FANCG [UoC-M1/FG] did not increase cellular levels of FANCA). Row 4 is a constant, nonspecific fragment detected by the FANCG antibody and demonstrates equal loading in all lanes. (B) Nuclear extracts from 3 transduced lines (UoC-M1/FA, UoC-M1/FC, and UoC-M1/FG), and the myeloid cell line Mo7e probed with anti-FANCA (row 1) and anti-FANCG (row 2) antisera. Transduction with FANCA restored nuclear levels of FANCA in UoC-M1/FA (row 1, column 2), whereas transduction with FANCG did not increase nuclear levels of FANCG (UoC-M1/FG) (row 2, column 5). (C) Nuclear levels of FANCA and FANCG are not reduced in the lymphoblastoid cell line 4081. The cell line JY is an EBV-transformed lymphoblastoid cell line established from a healthy adult volunteer and also demonstrates cellular resistance to MMC. Lane 1 is a whole cell extract (WCL) from HeLa cells.
Expression of FANCA stabilizes FANCG
Transduction with retroviral vectors profoundly increased levels of the proper FA protein in UoC-M1 cells (Figure 7A). Transduction with FANCC (UoC-M1/FC) had no affect on levels of FANCA or FANCG. Of importance is the observation that transduction with FANCA (UoC-M1/FA) not only resulted in an increase in cellular levels of FANCA (row 1, lanes 3-4) but also of FANCG (row 3, lanes 3-4). However, transduction with FANCG (UoC-M1/FG), although greatly increasing cellular levels of FANCG (row 3, lanes 7-8), did not result in an increase in FANCA (row 1, lanes 7-8). Because these proteins are known to stabilize each other,31,32 the results point to FANCA as the dysfunctional FA protein in UoC-M1 cells. Row 4 depicts a constant, nonspecific fragment detected by the FANCG antibody and demonstrated consistent loading across all lanes.
Additional support for FANCA dysfunction evolved from studies on nuclear localization of FANCA. As shown in Figure 7B, transduction of UoC-M1 with FANCA resulted in FANCA accumulation in the nucleus of UoC-M1/FA (row 1, lane 3), whereas transduction with FANCC (UoC-M1/FC) (row 1, lane 4) and FANCG (UoC-M1/FG) (row 1, lane 5) did not increase nuclear levels of FANCA. Transduction with FANCG failed to increase nuclear levels of this protein in any of the 3 lines (row 2, lanes 3-5). Taken together, these results suggest that a defect largely involving FANCA function was present in UoC-M1.
FANCA gene harbors no mutations
The most common form of FANCA mutation is deletion of one or more exons.19 We, therefore, subjected DNA from UoC-M1 to quantitative multiplex-PCR–based gene dosage analysis and sequencing. As shown in Table 1, relative to the myelin protein zero (MPZ) gene on 1q2233 or the NCX1 sodium channel exchanger gene on 2p22-p23,34 6 exons of FANCA (exons 5, 11, 17, 19/20, and 31) and 3 exons of FANCG (exons 1, 2, and 10) did not show significant dosage disparities. Therefore, the genomic regions covered by these intervals are intact and contain a proper copy number of FANCA and FANCG.
Gene dosage analysis for FANCA and FANCG in UoC-M1
. | FANCA . | . | . | . | . | . | FANCG . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment . | Control . | Exon 5 . | Exon 11 . | Exon 17 . | Exon 19/20 . | Exon 31 . | Control . | Exon 1 . | Exon 2 . | Exon 10 . | ||||||||
| 1 | MPZI | 1.60 | 0.84 | 0.92 | 0.80 | 0.94 | MPZI | 1.26 | 1.26 | 1.60 | ||||||||
| 2 | MPZI | 0.87 | 0.82 | ND | ND | ND | NCXI | 1.13 | 0.82 | 1.03 | ||||||||
| 3 | NCXI | 1.03 | 0.61 | ND | ND | ND | ND | ND | ND | ND | ||||||||
| 4 | NCXI | ND | 0.97 | ND | ND | ND | ND | ND | ND | ND | ||||||||
| Average dosage quotient | 1.17 | 0.81 | 0.92 | 0.80 | 0.94 | 1.19 | 1.04 | 1.31 | ||||||||||
. | FANCA . | . | . | . | . | . | FANCG . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment . | Control . | Exon 5 . | Exon 11 . | Exon 17 . | Exon 19/20 . | Exon 31 . | Control . | Exon 1 . | Exon 2 . | Exon 10 . | ||||||||
| 1 | MPZI | 1.60 | 0.84 | 0.92 | 0.80 | 0.94 | MPZI | 1.26 | 1.26 | 1.60 | ||||||||
| 2 | MPZI | 0.87 | 0.82 | ND | ND | ND | NCXI | 1.13 | 0.82 | 1.03 | ||||||||
| 3 | NCXI | 1.03 | 0.61 | ND | ND | ND | ND | ND | ND | ND | ||||||||
| 4 | NCXI | ND | 0.97 | ND | ND | ND | ND | ND | ND | ND | ||||||||
| Average dosage quotient | 1.17 | 0.81 | 0.92 | 0.80 | 0.94 | 1.19 | 1.04 | 1.31 | ||||||||||
ND indicates not done.
We further analyzed the FANCA and FANCG genes by performing DNA sequencing of all exons, approximately 30 bp of sequence upstream and downstream of each exon and all known splice sites. Sequence analysis of FANCG in UoC-M1 was normal with no anomalies noted. A heterozygous, silent transition polymorphism (Gly1290Ala) was noted in exon 14 of FANCA. FANCA was otherwise unremarkable, including the 2 putative bipartite nuclear localization signals (NLSs),23 that were intact (data not shown).
Genetic analysis of a lymphoblastoid cell line from the same individual
QRT-PCR in 4081 (standardized against a lymphoblastoid cell line established from a healthy individual, JY) indicated that RNA for each of the 6 cloned FA genes (as before) was present (data not shown). As shown in Figure 7C, immunoblot analysis of nuclear fractions from 4081 indicates nuclear abundance of both FANCA and FANCG, clearly demonstrating that the function of FANCA in 4081 cells was normal.
It became important to demonstrate that UoC-M1 and 4081 cells were genetically identical. We constructed haplotypes using highly variable dinucleotide repeats from chromosomes 7 and 8. As shown in Table 2, UoC-M1 and 4081 genetically agree at 12 of 12 markers (11 markers from chromosome 7 and 1 marker from chromosome 8). On the basis of the frequencies reported for each of these alleles in the CEPH database, the overall frequency of the indicated haplotype is conservatively estimated to be 5 × 10-17, confirming a common genetic identity between UoC-M1 and 4081. Of note, only a single allele for chromosome 7 markers in UoC-M1 is indicated, because UoC-M1 cells are monosomic for chromosome 7.10
Haplotype analysis in UoC-M1 and 4081 indicates they are derived from the same individual
. | Alleles . | . | . | |
|---|---|---|---|---|
| Marker . | UoC-M1 . | 4081 . | Band . | |
| D7S531 | 295 | 295, 280 | 7p22.3-7p22.2 | |
| D7S493 | 206 | 206, 229 | 7p21.3-7p15.2 | |
| D7S484 | 114 | 114, 114 | 7p22.1-7p14.2 | |
| D7S519 | 267 | 267, 271 | 7p13-7p12.3 | |
| D7S502 | 300 | 300, 304 | 7q11.21-7q11.22 | |
| D7S669 | 190 | 190, 181 | 7q11.22-7q21.11 | |
| D7S486 | 221 | 221, 231 | 7q31.31 | |
| D7S640 | 134 | 134, 120 | 7q31-7q33 | |
| D7S661 | 313 | 313, 319 | 7q34-7q35 | |
| D7S798 | 71 | 71, 76 | 7q35 | |
| D7S2465 | 333 | 333, 335 | 7q35 | |
| D8S550 | 193, 204 | 193, 204 | 8pter-8qter | |
. | Alleles . | . | . | |
|---|---|---|---|---|
| Marker . | UoC-M1 . | 4081 . | Band . | |
| D7S531 | 295 | 295, 280 | 7p22.3-7p22.2 | |
| D7S493 | 206 | 206, 229 | 7p21.3-7p15.2 | |
| D7S484 | 114 | 114, 114 | 7p22.1-7p14.2 | |
| D7S519 | 267 | 267, 271 | 7p13-7p12.3 | |
| D7S502 | 300 | 300, 304 | 7q11.21-7q11.22 | |
| D7S669 | 190 | 190, 181 | 7q11.22-7q21.11 | |
| D7S486 | 221 | 221, 231 | 7q31.31 | |
| D7S640 | 134 | 134, 120 | 7q31-7q33 | |
| D7S661 | 313 | 313, 319 | 7q34-7q35 | |
| D7S798 | 71 | 71, 76 | 7q35 | |
| D7S2465 | 333 | 333, 335 | 7q35 | |
| D8S550 | 193, 204 | 193, 204 | 8pter-8qter | |
The molecular mass (bp) of amplimers at each polymorphic marker (column 1) is given in columns 2 and 3. Column 4 lists the chromosomal location.
Note: Only a single allele for chromosome 7 makers in UoC-M1 is indicated, as UoC-M1 has complex cytogenetics, including functional monosomy 7.10
Discussion
FA is a rare, autosomal-recessive disorder characterized by bone marrow failure and various developmental defects, including skeletal, skin pigmentation, and visceral anomalies.35,36 The disease demonstrates genetic heterogeneity. Eight complementation groups have been defined by somatic cell fusion studies (FA-A through FA-G, including FA-D1 and FA-D2) with each complementation group corresponding to a distinct gene.25,37 The majority of all patients (60%-65%) fall within group FA-A. Seven of the FA genes have been cloned.22-29 Apart from BRCA2 (the FANCD1 gene), the cloned sequences bear homologies neither to other known genes nor to one another. The FA genes clearly protect cells from genotoxic stress. Mutant cells exhibit spontaneous chromosomal breakage38 and increased sensitivity to the development of cytogenetic anomalies on in vitro treatment with cross-linking agents39 such as MMC40 and DEB.41
Additional functions for FA proteins have also been described, including modulation of cytokine-mediated signaling,42-50 responsiveness to oxidative stress,51-57 and chromatin remodeling.58 Moreover, at least 2 members of the FA genes (FANCC and FANCD2) are multifunctional. For example, selective mutations of FANCC can impair some functions of FANCC but not others.49 The same is true for FANCD2, which plays a dual role in mediating resistance to chemical cross-linking agents (for which FANCD2 monoubiquitinylation is required) and ionizing radiation (for which FANCD2 monoubiquitinylation and serine phosphorylation are required).59 However, although distinct FA proteins likely have individual cellular activities, the universal phenotypic feature of bone marrow failure across all complementation groups suggests that these proteins share important hematopoietic activities by acting together in either a multimeric complex, a sequential linear pathway, or both.
Garcia-Higuera et al13 suggest that FA proteins function in a linear pathway to effect protection from or repair of DNA damage. Indeed, there exists an upstream multimeric complex made up of at least the FANCA, FANCC, FANCE, FANCF, and FANCG proteins.31,32,60-65 This complex is required to permit a posttranslational modification (monoubiquitinylation) of the FANCD2 gene product.13 The FANCD2 protein has been shown to coimmunoprecipitate with BRCA1 and further, to colocalize with BRCA1 to damage-induced nuclear foci.13 The ability to form such foci is absent in cell lines from multiple FA complementation groups but is restored on correction of these lines with appropriate cDNAs.13 That BRCA2 was recently identified as the FANCD1 gene provides further evidence of association between the Fanconi and BRCA pathways.29
The hematopoietic defect in FA involves initial bone marrow failure followed by myelodysplasia and AML.66 Hematopoietic defects account for most of the deaths in patients with FA. Only rare cases have been reported into their fourth decades. A single set of siblings diagnosed in their 50s has been reported; one with anemia and the other hematologically asymptomatic (although MMC sensitive).67 Furthermore, no case presenting initially as MDS or AML without bone marrow failure (BMF) has been reported in a patient older than 28 years.68
Xie et al69 suggested, based on observations from coimmunoprecipitation studies, that AML cells from non-FA patients may exhibit abnormal FA protein complex formation, but the functional significance of the observations was unclear. Specifically, MMC sensitivity was not reported in all cells tested, FA genes were not sequenced, complementation studies were not performed, FANCD2-S/L status was not examined (the identification of FANCD2 was reported after the publication of the report by Xie et al69 ), and nonleukemic cells from the patients with AML were not analyzed. In the studies outlined herein, all of these standards were met, and the results demonstrate that dysfunction of FANCA can indeed occur as an acquired defect in AML cells and that it has unambiguous functional significance.
First, the UoC-M1 myeloid cell line is hypersensitive to MMC and DEB, but lymphoblasts from the same patient (4081) are not. Second, nuclear FANCA and FANCG protein levels were undetectable in UoC-M1 cells but were abundant in 4081 cells. Third, 4081 cells contained FANCD2-L and -S, but UoC-M1 cells contained only FANCD2-S. Fourth, full-length BRCA2 (recently shown to be the FANCD1 gene29 and known to function distal to FANCD270 ) was present in UoC-M1. Fifth, transduction of UoC-M1 cells with a FANCA but not FANCG cDNA increased both FANCA and FANCG levels and induced MMC resistance. The capacity of FANCA to increase FANCG levels reflects the FANCG-stabilizing activity of FANCA.31,32 That FANCG did not influence FANCA points to FANCA dysfunction in UoC-M1 cells.
As mRNA encoded by 5 cloned FA genes (FANCA, FANCC, FANCD2, FANCE, and FANCG) was present in UoC-M1 at equal or greater levels than in Mo7e controls, gene dosage analysis for several exons of both FANCA and FANCG indicated standard copy number, and because the sequences of all exons of FANCA and FANCG were normal, we propose that cofactors for nuclear localization of FANCA or for FANCG binding of FANCA are dysfunctional in UoC-M1 cells, a deficiency that could be overcome by overexpression of FANCA but not by FANCG or FANCC. We have not yet identified what factors account for FANCA dysfunction in UoC-M1 cells, but it does not seem to be a result of structural or transcriptional regulatory mutations of FANCA or FANCG genes. Because of the capacity of FA proteins to bind to cellular chaperones,50,71 as well as to each other,13,31,32,60-65,72-76 it is possible that factors governing intracellular (ie, nuclear) localization may be dysfunctional in these cells. It has been reported that abnormalities in both complementation groups FA-B and FA-C lead to reductions of the FANCA/FANCG complex.31 However, as retroviral transduction with a FANCC construct does not significantly reduce the MMC sensitivity of UoC-M1, it is unlikely that a defect of FANCC exists in these cells. It is, therefore, remotely possible that the apparent abnormality of FANCA function derives from a defect in FANCB, a gene that has not yet been unambiguously identified. Should a FANCB defect be found in UoC-M1 cells, the 4081 cells might be either heterozygous or wild type for FANCB. It is also possible that further investigation into the observed FANCA defect in UoC-M1 may reveal dysfunction of a novel protein corresponding to no known FA complementation group. Because most known FA cases fall into the complementation groups already defined, it is likely that biallelic inactivation of this protein would result in embryonic lethality and that ascertainment of a role in disease pathogenesis could be achieved only by observing somatic dysfunction through loss of both genes in noncarriers or by loss of heterozygosity (LOH) in carriers. Such has clearly been the case with other disorders.77-79
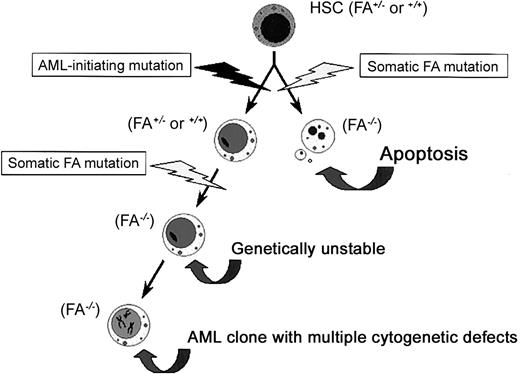
These studies support a model of clonal evolution in this patient in which acquired FANCA dysfunction occurred in an already initiated clone that ultimately gave rise to the cytogenetically unstable UoC-M1 cells but did not give rise to the 4081 lymphoblasts (Figure 8). There are 2 alternative explanations for our results. First, one might propose that the cells we studied acquired the FANCA defect during propagation of this immortalized line. However, the cytogenetic defects described in the original patient material were highly complex and were the same as they are currently in the cells of this line, indicating no new cytogenetic evolution.10 Secondly, this patient could theoretically represent the oldest ever reported case of FA with lymphoid mosaicism. Mosaicism has been shown to arise in vivo by a variety of mechanisms, including reversion of frameshift mutations by subsequent nucleotide insertions or deletions.80 A case reported recently in a young adult with FA demonstrated the occurrence of leukemia, although the patient had high-level mosaicism (arising via mitotic recombination between alleles in a compound heterozygote).81 The leukemic clone derived from the non-reverted stem cell population.81 If such a circumstance is also considered in the patient we report here, mosaicism might have occurred early in life, leading to subsequent correction of a hematopoietic defect with late-arising clonal evolution in a preserved, non-revertant stem cell. We believe this circumstance to be unlikely in view of his age, clinical stability, and absence of other stigmata of FA in the patient or any family member, as well as that no mutations were found within the patient's FANCA and FANCG genes. It is also unusual for patients with mosaicism to be fully hematologically healthy, as this individual was known to have been prior to the onset of AML. Consequently, it is most likely that our studies indicate that in some adult patients with AML, likely those with complex cytogenetic defects, acquired FA dysfunction may serve as a progression factor for clonal evolution.
Acquired dysfunction of FA proteins, a potential progression factor in secondary AML/MDS. We propose that FANCA dysfunction was acquired by an evolving clone of initiated hematopoietic stem cells. As shown on the right, an early FA-inactivating event would render the cell hypersensitive to apoptotic stimuli, resulting in apoptosis. However, as shown on the left, if a similar event occurred in an already initiated cell, the cell could resist apoptotic cues. The FA defect would then contribute to genetic instability and lead to multiple cytogenetic defects in the evolving clone. HSC indicates hematopoietic stem cell; FA, any FA protein or factor that enhances FA protein function.
Acquired dysfunction of FA proteins, a potential progression factor in secondary AML/MDS. We propose that FANCA dysfunction was acquired by an evolving clone of initiated hematopoietic stem cells. As shown on the right, an early FA-inactivating event would render the cell hypersensitive to apoptotic stimuli, resulting in apoptosis. However, as shown on the left, if a similar event occurred in an already initiated cell, the cell could resist apoptotic cues. The FA defect would then contribute to genetic instability and lead to multiple cytogenetic defects in the evolving clone. HSC indicates hematopoietic stem cell; FA, any FA protein or factor that enhances FA protein function.
The activities of the FA proteins also speak to the timing of somatic mutations affecting their function. We argue that loss of FA protein function is likely a progression factor and not an initiation factor for new clones. This notion derives from work from our lab and others,42,43,45,48-50,82,83 demonstrating that FA gene dysfunction results in an apoptotic phenotype in hematopoietic cells, hardly a selectable attribute unless an initiating mutation predates the dysfunction. The proposition that FA mutations are not selectable, absent an initiating mutation, is supported by the finding that obligate FA heterozygotes do not appear to be at increased risk of AML, MDS, or cancer.84 The apoptotic phenotype of nonleukemic FA cells not only accounts for bone marrow failure in FA42 but also may exert a particular selective-pressure that contributes to the evolution of malignant stem cells by forcing the emergence of apoptosis-resistant cells.8 This type of adaptive mutation could represent a perfect initiating factor, protecting the cells from the apoptotic consequences of FA protein dysfunction but permitting subsequent FA gene mutations to cause genomic instability and MMC hypersensitivity.8 As such, loss of FA protein function may serve as an important progression factor for leukemic clones, especially the types bearing multiple, recurring cytogenetic defects. A comprehensive examination of FA protein function in primary and secondary MDS/AML cells should shed further light on these questions and is clearly warranted at this time.
Prepublished online as Blood First Edition Paper, March 13, 2003; DOI 10.1182/blood-2002-09-2781.
Supported by the National Institutes of Health T-32 HL07781 (M.W.L.) and P01 HL48546 (M.W.L., T.A.C., C.A.R., S.A.S., S.B.O., and G.C.B.); National Cancer Institute PO1 CA40046 (M.M.L.B. and R.A.L.); the Cancer Research United Kingdom (M.T. and S.V.H.); and the Fanconi Anemia Research Fund (M.W.L. and G.C.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Mathew J. Thayer, Michael C. Heinrich, and Michael Riscoe for helpful discussions.

![Figure 7. FANCA transduction enhances FA protein complex stability and restores nuclear FANCA levels. (A) Whole cell lysates. All 4 rows depict data obtained from the same filter, cut horizontally at 80 kDa to allow FANCA to be interrogated on the top portion and FANCC (then FANCG after stripping the FANCC signal) on the bottom portion. Retroviral transduction with each of 3 FA cDNA constructs resulted in increased levels of their respective proteins (the results from independently obtained duplicates are shown for each transduced cell line). Transduction with FANCA (UoC-M1/FA) also led to an increase in cellular levels of FANCG, whereas the reverse was not observed (ie, transduction with FANCG [UoC-M1/FG] did not increase cellular levels of FANCA). Row 4 is a constant, nonspecific fragment detected by the FANCG antibody and demonstrates equal loading in all lanes. (B) Nuclear extracts from 3 transduced lines (UoC-M1/FA, UoC-M1/FC, and UoC-M1/FG), and the myeloid cell line Mo7e probed with anti-FANCA (row 1) and anti-FANCG (row 2) antisera. Transduction with FANCA restored nuclear levels of FANCA in UoC-M1/FA (row 1, column 2), whereas transduction with FANCG did not increase nuclear levels of FANCG (UoC-M1/FG) (row 2, column 5). (C) Nuclear levels of FANCA and FANCG are not reduced in the lymphoblastoid cell line 4081. The cell line JY is an EBV-transformed lymphoblastoid cell line established from a healthy adult volunteer and also demonstrates cellular resistance to MMC. Lane 1 is a whole cell extract (WCL) from HeLa cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/1/10.1182_blood-2002-09-2781/5/m_h81334572007.jpeg?Expires=1769452231&Signature=gB7eEuf5Hu0vd3FqYI-T3HcMYSnpbOlADsm-kFiPcmRvCTlyETIxSBygd9FkiWPOdXDXqGds8WgJS5WQyKiF18aGsdNeBpbs~I4PrMiuY-bCSnNVz7GnnXCnGYEL~9VgzMk6HMvhSwkCWSZwmlS1le-Ug40iOBxsmD94N5upj2gYozNu7sLuTTurqp3wLBPZsQDbtWquFL-0XUshKA~eHlh~AzaovtUkFb92ZPHLpZGzswj6Bg2xuJOZgurTQHjrABGLZ~oGAMdTHAkIXESOHaiE3m2N7MQuiWm4sXuyCS4UkP1O6eqFGjJi3m-i6Fk5dy4GZonPYbTYolXfBJ57zA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal