Interactions between killer immunoglobulin-like receptors (KIRs) and human leukocyte antigen (HLA) class I ligands influence development of natural killer (NK) cell repertoire and response to infection, cancer, and allogeneic tissue. As KIRs and HLA class I molecules are highly polymorphic, clinical allogeneic hematopoietic cell transplantation is predicted to frequently involve KIRmismatch, and thus to provide a unique system for study of human NK cell receptor repertoire development. Eighteen leukemia patients undergoing HLA-matched transplantation and their donors were analyzed for KIR genotype. Ten of 13HLA-identical donor-patient pairs were KIRmismatched and 3 were matched; all HLA-matched unrelated pairs were KIR mismatched. Reconstitution of recipient NK cell repertoire following transplantation was examined using flow cytometry and monoclonal antibodies specific for KIR and CD94:NKG2A. These data form 3 groups. Six to 9 months after transplantation, 8 patients (group 1) reconstituted an NK cell repertoire resembling that of their donor, and forKIR-mismatched transplants, distinct from the recipient before transplantation. In the first year after transplantation, 5 patients (group 2) exhibited a generally depressed frequency of KIR-expressing NK cells and concomitant high frequency of CD94:NKG2A expression. By 3 years after transplantation, the frequency of KIR-expressing NK cells had increased to donor values, in the 3 patients from group 2 analyzed for this period. The remaining 5 patients experienced severe clinical complications following transplantation and displayed unique features in their NK cell receptor reconstitution. These results demonstrate that a majority ofHLA-matched hematopoietic cell transplantations involveKIR mismatch and reveal differences in NK cell repertoire having potential impact for immune responsiveness and transplantation outcome.
Introduction
Natural killer (NK) cells are lymphocytes that act early in the immune response against injury and infection. They kill tumor-transformed and allogeneic cells, secrete cytokines, and contribute to inflammation.1,2 The actions of NK cells are mediated by the balance of activating and inhibitory signals sent by cell surface receptors on binding to human leukocyte antigen (HLA) class I or class I-like ligands. Although the likely role of activating NK cell receptors is in response to invasion, inhibitory receptors on NK cells discriminate healthy from diseased cells by surveying cell surfaces for expression of autologous HLA class I molecules.3-5
NK cell receptors are encoded by 2 structurally distinct families of molecules: the killer immunoglobulin-like receptors (KIRs), and the lectinlike CD94:NKG2 heterodimers. Fourteen KIR genes, located in the leukocyte receptor complex on human chromosome 19, encode inhibitory and activating receptors, some of which are specific for polymorphic epitopes of HLA-A, -B, -C, and -G.6,7 TheKIR gene family exhibits extensive variation in gene number and content, as well as allelic polymorphism.8-11 The more conserved lectinlike CD94:NKG2 molecules are encoded by genes in the natural killer cell gene complex on human chromosome 12 and interact with HLA-E, in complex with peptides derived from the leader sequences of HLA class I molecules.12-15
Within an individual, KIR and CD94:NKG2 receptors are expressed on overlapping subsets on NK cells, such that an NK cell receptor repertoire of expression is formed.16 The class I specificity of each NK cell is determined by the array of receptors present on the cell surface, rather than by a single, unique determinant.16 The NK cell repertoire is primarily determined by KIR genotype, although HLA genotype has a subtle, modifying influence.16,17 The only requirement for NK cell receptor repertoire development appears to be that every NK cell express at least one inhibitory receptor specific for autologous HLA class I, thereby ensuring self-tolerance.16
Clinical hematopoietic cell transplantation offers a unique opportunity to assess the relative effects of donor KIR genotype and host environment on the process of NK cell receptor repertoire development. As currently practiced, matching of allogeneic donors and recipients focuses on HLA polymorphisms and has no consideration of KIR polymorphism. As KIR andHLA segregate independently, and as unrelated individuals almost always have different KIR genotype, we predict that approximately 25% of transplantations between HLA-identical siblings involve KIR identity and approximately 75%KIR disparity. For transplantations involving anHLA-matched unrelated donor the frequency of KIRincompatibility is predicted to approach 100%.17-20Consequently, allogeneic transplantation provides a good system for examining the reconstitution of human NK cell repertoires under conditions of HLA and/or KIR genetic difference.
The aim of this study was to examine recovery of the NK cell receptor repertoire in 18 patients with acute myeloid leukemia (AML) and chronic myeloid leukemia (CML) following HLA-matched hematopoietic cell transplantation. As KIR genotype has been shown to be the principal determinant of KIR phenotype,17we tested the hypothesis that the NK cell receptor repertoire that emerges following HLA-matched allogeneic transplantation is determined by receptor genes in the donor cells. KIR and CD94:NKG2A expression was analyzed by flow cytometry of the donor's and recipient's NK cells prior to transplantation and for periods of approximately 1 to 3 years following transplantation. On this basis, 2 common patterns of NK cell receptor reconstitution emerged, permitting each patient to be assigned to one of 3 groups. For 8 patients (group 1), KIR expression reconstituted during the first year following transplantation and became like that of the donor and, in cases ofKIR mismatch, distinct from that of the patient before transplantation. For 5 patients (group 2), reconstitution during the first year after transplantation was characterized by low frequencies of KIR-positive NK cells and a high frequency of CD94:NKG2A expression. For 3 patients in group 2 that were studied over a longer period of time, the frequencies of KIR and CD94:NKG2A did change to become like those of the donor. The remaining 5 patients all experienced severe clinical complications and exhibited unique and sometimes transient features in their reconstituted NK cell receptor repertoires, making them distinct from either donor or recipient, as well as different from each other.
Patients, materials, and methods
Patient and donor characteristics
The study panel comprised 18 patients receiving allogeneic hematopoietic cell transplants at the Stanford University Medical Center for treatment of CML (n = 12) or AML (n = 6). Of these, 13 received bone marrow (BM) (n = 9) or granulocyte colony-stimulating factor (G-CSF)–mobilized peripheral blood progenitor cells (PBPCs) (n = 4) from HLA-matched sibling donors (matched related donors, MRDs). The remaining 5 patients received BM from matched unrelated donors (MUDs). One patient (SPN 2017), previously reported to have an unrelated donor,17 in fact received BM from an HLA-identical sibling donor. The study group included 6 men and 12 women, with a median age of 39 years (range, 21-56 years). Myeloablative regimens included a combination of busulfan and cyclophosphamide; or cyclophosphamide and/or etoposide, with or without fractionated total body irradiation (FTBI).21-24Prophylactic therapy included cyclosporine and methotrexate, with or without prednisone, for graft-versus-host disease (GVHD), heparin for prevention of veno-occlusive disease, fluconazole and acyclovir for fungal and viral infections, intravenous immunoglobulin for cytomegalovirus (CMV) infection, and additional antibiotics as needed.
Sample collection and preparation
Blood (20 mL) was drawn from donors and recipients prior to transplantation and from recipients on clinical engraftment or by day 30 after transplantation. Subsequent sample collection targeted days 30, 60, 90 and/or 100, 120, 150, 180, 270, and 1 year after transplantation. Three-year posttransplantation blood samples were collected from 5 patients. Samples scheduled for collection before day 100 were drawn within 4 days of the specified day. Later time points were collected within 2 weeks (days 120, 150, and 180) or 3 weeks (day 270) of the specified day, and 1-year posttransplantation samples were collected between days 334 and 395. Three-year samples were collected between days 1000 and 1275. In the case of missed collections, samples were drawn at interim time points whenever possible. Peripheral blood mononuclear cells (PBMCs) were isolated from anticoagulated whole blood by Ficoll-Hypaque gradient separation.
Informed consent was obtained from all participants, and patients were treated under protocols approved by the Stanford Institutional Review Board.
Genomic DNA preparation and KIR genotyping
Genomic DNA was prepared from 2 × 106 to 1 × 107 PBMCs using a QIAamp Blood Kit (QIAGEN, Valencia, CA). Generic KIR typing of genomic DNA was performed by polymerase chain reaction (PCR) amplification with primers based on conserved regions specific to each KIRlocus, as described by Uhrberg et al9 with modification, as described by Shilling et al.19 For KIRsubtyping, primers designed to discriminate allele-specific polymorphisms were paired with KIR2DL1, 2DL3,3DL1, or 3DL2 locus-specific primers as previously described.18 19
HLA class I determination and KIR epitope typing
HLA-A and HLA-B antigens, including Bw4 and Bw6, were determined serologically by the Stanford Histocompatibility Laboratory using commercially available reagents. HLA-C type was determined serologically or by PCR-SSP (sequence-specific primer) analysis of genomic DNA using C-locus SSP Unitray test kits (Pel-Freez Biological, Roger, AR), as per manufacturer's instructions. The presence of motifs encoding the class I HLA-C KIR epitopes and the HLA-Bw4 and Bw6 epitopes was determined by reverse transcription (RT)–PCR amplification, as previously described.17
Antibodies and flow cytometric analysis
Monoclonal antibodies DX27 (anti-KIR2DL2/KIR2DL3/KIR2DS2), DX9 (anti-KIR3DL1), DX31 (anti-KIR3DL2) (generously provided by L. Lanier, Cancer Research Center, University of California, San Francisco, CA), EB6 (anti-KIR2DL1/2DS1), HP-3B1 (anti-CD94), and Z199 (anti-NKG2A) (Beckman-Coulter-Immunotech, Brea, CA) were each used in combination with anti-CD3 (SK7) and anti-CD56 (NCAM16.2) (BD Biosciences, Mountain View, CA) in individual 3-color flow cytometry assays of PBMCs from each donor. After gating on the CD3−CD56+lymphocyte (ie, NK cell) subset, KIR- or CD94:NKG2A-expressing populations were identified using irrelevant, isotype-matched control antibody stains to set the lower limit of the KIR- or CD94:NKG2A-positive gate, thereby excluding background fluorescence and receptor-negative cells. Frequency and median fluorescence intensity (MFI) of NK cells binding each specific antibody were calculated for these populations. Anti-CD3 and anti-CD56 antibodies were labeled with peridinin chlorophyll protein (PerCP) and fluorescein isothiocyanate (FITC), respectively; anti-KIR, anti-NKG2A, and anti-CD94 antibodies were phycoerythrin (PE) conjugated. Flow cytometric analysis was performed on a fluorescence-activated cell sorter scan (FACScan) flow cytometer using CellQuest software (BD Biosciences).
Calculation of differences in frequency and median fluorescence intensity of KIR expression from flow cytometry data
As first described in Shilling et al,17 the difference in frequency of KIR expression by NK cells for each KIR-specific antibody stain was calculated according to the following formula: frequency (sample 1) − frequency (sample 2)/mean frequency (samples 1 and 2) = difference in frequency.
The 4 frequency differences were added to determine the sum of frequency differences. Differences in MFI were calculated using the same formula, then added to give the sum of MFI differences. All differences were considered positive. The mean was incorporated as a denominator here to normalize the data obtained for the different antibodies. The values obtained for the sum of frequency differences and the sum of MFI differences were then used to plot a single point on a 2-dimensional graph. In this analysis, the origin corresponds to identity of KIR repertoires; conversely, the further a point is away from the origin, the more disparate are the KIR repertoires.
Determination of absolute lymphocyte numbers
After transplantation, PBMCs from transplant recipients were stained with anti-CD3 (PerCP), anti-CD4 (PE), and anti-CD8 (FITC), and the frequencies of total lymphocytes that were CD3+, CD3+CD4−CD8+, CD3+CD4+CD8− were calculated. Frequencies of CD3−CD56+ NK cells were calculated from 3-color stains for KIR as described in “Antibodies and flow cytometric analysis.” These frequencies were multiplied by absolute numbers of lymphocytes, obtained from complete blood counts (CBCs), to back-calculate absolute numbers (× 103 cells per microliter) for each lymphocyte subset.
Results
Patient characteristics
That differences in KIR repertoire among healthy individuals are determined predominantly by KIR genotype predicts that recipients of hematopoietic cell transplants will reconstitute NK cell populations with donorlike repertoires of KIR expression.17 To test this hypothesis, reconstitution of NK cell receptor expression in 18 patients undergoing transplantation for treatment of CML or AML was examined. Patient characteristics, including age, sex, underlying hematologic disorder, preparative regimen, KIR compatibility, and clinical outcome, are listed in Table 1. Thirteen of the patients received BM or PBPCs from MRDs, whereas 4 patients received BM from MUDs, and one patient received marrow from an unrelated donor that wasHLA matched except for one HLA-C disparity (SPN 2067). All patients achieved complete donor chimerism within 90 days of transplantation, as measured by short terminal repeat (STR) analysis of marrow biopsies (data not shown). The KIR identity of the donor/recipient pairs was determined by PCR typing and subtyping of genomic DNA. Consistent with random segregation of parentalKIR and HLA haplotypes, 3 of the 13 (23%) MRD pairs were KIR identical; all MUD pairs were KIRdisparate (Figure 1).
Characteristics of patients undergoingHLA-matched bone marrow or peripheral blood progenitor cell transplantation
| SPN . | Age (y) . | Sex . | Diagnosis . | Transplant type . | Preparatory regimen . | Donor-patient KIR compatibility . | GVHD . | Status at last follow-up . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 | ||||||||||
| 1899 | 48 | F | CML | CP | MRD | BM | FTBI/VP16 | KIR-mismatched | None | Alive and CCR |
| 2017 | 21 | F | CML | CP | MRD | BM | FTBI/VP16 | KIR-mismatched | None | Alive and CCR |
| 1991 | 42 | F | CML | CP | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD I (skin) | Alive and CCR |
| 2025 | 29 | F | CML | CP | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD I-II (skin) | Alive and CCR |
| 2060 | 47 | F | AML | CR | MRD | BM | FTBI/VP16 | KIR-mismatched | None | Alive and CCR |
| 2036 | 38 | F | AML | CR | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD (skin) | Alive and CCR |
| 2067 | 42 | M | CML | AP | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD I (skin); cGVHD (eyes, mouth, skin) | Alive and CCR |
| 2138 | 32 | F | CML | CP | MRD | PBPC | BU/CY | KIR-mismatched | None | Alive and CCR |
| Group 2 | ||||||||||
| 1920 | 23 | M | CML | AP | MRD | BM | FTBI/VP16/CY | KIR-mismatched | None | Alive and CCR |
| 1970 | 56 | F | AML | CR2 | MRD | BM | BU/VP16/CY | KIR-mismatched | None | Died day 741, meningitis |
| 2040 | 53 | F | AML | IF | MRD | BM | BU/VP16/CY | KIR-mismatched | cGVHD (liver) | Rel day 228; died day 280 |
| 2068 | 39 | M | CML | AP | MRD | BM | BU/CY | KIR-mismatched | None | Alive and CCR |
| 2089 | 34 | F | CML | CP | MRD | PBPC | BU/CY | KIR-matched | None | Alive and CCR |
| Nongroup 1 and 2 | ||||||||||
| 1980 | 30 | F | CML | CP | MUD | BM | FTBI/CY | KIR-mismatched | None | Rel day 737; given DLI |
| 2038 | 27 | M | AML | CR1 | MRD | PBPC | FTBI/VP16 | KIR-mismatched | cGVHD (eyes, mouth, liver) | Alive and CCR |
| 2061 | 48 | M | CML | AP | MRD | PBPC | BU/CY | KIR-mismatched | cGVHD (mouth, eye); aGVHD I (skin); aGVHD IV (liver, gut) | Died day 159-aGVHD |
| 2062 | 45 | M | CML | CP | MRD | BM | BU/CY | KIR-matched | None | Rel day 180; retranspl |
| 2140 | 41 | F | AML | IF | MRD | PBPC | FTBI/VP16/CY | KIR-matched | cGVHD (extensive) | Alive and CCR |
| SPN . | Age (y) . | Sex . | Diagnosis . | Transplant type . | Preparatory regimen . | Donor-patient KIR compatibility . | GVHD . | Status at last follow-up . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 | ||||||||||
| 1899 | 48 | F | CML | CP | MRD | BM | FTBI/VP16 | KIR-mismatched | None | Alive and CCR |
| 2017 | 21 | F | CML | CP | MRD | BM | FTBI/VP16 | KIR-mismatched | None | Alive and CCR |
| 1991 | 42 | F | CML | CP | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD I (skin) | Alive and CCR |
| 2025 | 29 | F | CML | CP | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD I-II (skin) | Alive and CCR |
| 2060 | 47 | F | AML | CR | MRD | BM | FTBI/VP16 | KIR-mismatched | None | Alive and CCR |
| 2036 | 38 | F | AML | CR | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD (skin) | Alive and CCR |
| 2067 | 42 | M | CML | AP | MUD | BM | FTBI/CY | KIR-mismatched | aGVHD I (skin); cGVHD (eyes, mouth, skin) | Alive and CCR |
| 2138 | 32 | F | CML | CP | MRD | PBPC | BU/CY | KIR-mismatched | None | Alive and CCR |
| Group 2 | ||||||||||
| 1920 | 23 | M | CML | AP | MRD | BM | FTBI/VP16/CY | KIR-mismatched | None | Alive and CCR |
| 1970 | 56 | F | AML | CR2 | MRD | BM | BU/VP16/CY | KIR-mismatched | None | Died day 741, meningitis |
| 2040 | 53 | F | AML | IF | MRD | BM | BU/VP16/CY | KIR-mismatched | cGVHD (liver) | Rel day 228; died day 280 |
| 2068 | 39 | M | CML | AP | MRD | BM | BU/CY | KIR-mismatched | None | Alive and CCR |
| 2089 | 34 | F | CML | CP | MRD | PBPC | BU/CY | KIR-matched | None | Alive and CCR |
| Nongroup 1 and 2 | ||||||||||
| 1980 | 30 | F | CML | CP | MUD | BM | FTBI/CY | KIR-mismatched | None | Rel day 737; given DLI |
| 2038 | 27 | M | AML | CR1 | MRD | PBPC | FTBI/VP16 | KIR-mismatched | cGVHD (eyes, mouth, liver) | Alive and CCR |
| 2061 | 48 | M | CML | AP | MRD | PBPC | BU/CY | KIR-mismatched | cGVHD (mouth, eye); aGVHD I (skin); aGVHD IV (liver, gut) | Died day 159-aGVHD |
| 2062 | 45 | M | CML | CP | MRD | BM | BU/CY | KIR-matched | None | Rel day 180; retranspl |
| 2140 | 41 | F | AML | IF | MRD | PBPC | FTBI/VP16/CY | KIR-matched | cGVHD (extensive) | Alive and CCR |
SPN indicates Stanford patient number; F, female; M, male; CML, chronic myeloid leukemia; AML, acute myeloid leukemia; CP, chronic phase; AP, accelerated phase; CR, complete remission; IF, induction failure; MRD, matched related donor; MUD, matched unrelated donor; BM, bone marrow; PBPC, peripheral blood progenitor cell; FTBI, fractionated total body irradiation; VP16, etoposide; CY, cyclophosphamide; BU, busulfan; GVHD, graft-versus-host disease; aGVHD, acute GVHD; cGVHD, chronic GVHD; CCR, continuous complete remission; Rel, relapse; DLI, donor leukocyte infusion; retranspl, retransplanted. Patients in groups 1, 2, and nongroup 1 and 2 are separated into sections.
KIR genotyping of hematopoietic cell donors and recipients.
Donor/recipient pairs are organized according to donor type andKIR genotype: KIR-identical MRD pairs (A),KIR-disparate MRD pairs (B), and KIR-disparate MUD pairs (C). All MRD and MUD pairs are HLA identical, except patient/donor SPN 2067 (marked with *), who was disparate for one HLA-C allele. Clear boxes indicate the presence of HLA-B and C KIR epitopes and KIR genes, whereas dark gray hatched boxes indicate the absence of KIR epitopes and genes. Allelic subtypes, shown for those pairs having matching combinations ofKIR genes, are indicated by clear boxes with text. In heterozygous individuals, allele names are separated with a slash. Alleles not discriminated in subtyping are listed together, for example, as KIR2LD3*002or6; KIR3DL1*002 refers here to a set of similar alleles which includes KIR3DL1*002,*003, and *006-8.
KIR genotyping of hematopoietic cell donors and recipients.
Donor/recipient pairs are organized according to donor type andKIR genotype: KIR-identical MRD pairs (A),KIR-disparate MRD pairs (B), and KIR-disparate MUD pairs (C). All MRD and MUD pairs are HLA identical, except patient/donor SPN 2067 (marked with *), who was disparate for one HLA-C allele. Clear boxes indicate the presence of HLA-B and C KIR epitopes and KIR genes, whereas dark gray hatched boxes indicate the absence of KIR epitopes and genes. Allelic subtypes, shown for those pairs having matching combinations ofKIR genes, are indicated by clear boxes with text. In heterozygous individuals, allele names are separated with a slash. Alleles not discriminated in subtyping are listed together, for example, as KIR2LD3*002or6; KIR3DL1*002 refers here to a set of similar alleles which includes KIR3DL1*002,*003, and *006-8.
Patients have expected KIR repertoires prior to transplantation
Flow cytometric analysis of CD3−CD56+peripheral blood NK cells from each patient and donor was performed using 4 KIR-specific monoclonal antibodies that identify all KIR of well-defined, MHC class I specificity. In combination withKIR typing information, the frequency and MFI of KIR-specific antibody stains can be used as descriptors of NK cell KIR repertoire.17 For each individual, the percentage of cells binding the antibody and the median intensity of binding were determined for each of the 4 KIR-specific antibodies. From these data, summed differences in frequency and MFI were calculated for each donor-recipient pair, thereby reducing the comparisons to a single point on a 2-dimensional plot. Pairs of individuals with matching KIR repertoires yield small summed differences using these calculations, whereas disparate KIR phenotypes give higher summed differences. As seen in Figure 2, data points representing the 3 MRD pairs with identical KIR genotypes are clustered near the origin, indicating similar KIR repertoires. As a group, the KIR-disparate MRD pairs cover a wide range of summed differences, as do the MUD pairs, an expected result because they exhibit a range of differences in their KIRgenotype.
KIR repertoire comparisons of all donor-recipient pairs prior to transplantation.
Pretransplantation KIR repertoires of 18 allogeneic transplant patients and their MUD or sibling (MRD) donors are shown. Donor-recipient pairs are identified as KIR-identical, HLA-identical MRD/patient pairs (n = 3; ▪), KIR-disparate,HLA-identical MRD/patient pairs (n = 10; ■), andKIR-disparate, HLA-identical MUD/patient pairs (n = 5;  ). The KIR-disparate, HLA-C–disparate MUD patient/donor SPN 2067 is marked with an asterisk.
). The KIR-disparate, HLA-C–disparate MUD patient/donor SPN 2067 is marked with an asterisk.
KIR repertoire comparisons of all donor-recipient pairs prior to transplantation.
Pretransplantation KIR repertoires of 18 allogeneic transplant patients and their MUD or sibling (MRD) donors are shown. Donor-recipient pairs are identified as KIR-identical, HLA-identical MRD/patient pairs (n = 3; ▪), KIR-disparate,HLA-identical MRD/patient pairs (n = 10; ■), andKIR-disparate, HLA-identical MUD/patient pairs (n = 5;  ). The KIR-disparate, HLA-C–disparate MUD patient/donor SPN 2067 is marked with an asterisk.
). The KIR-disparate, HLA-C–disparate MUD patient/donor SPN 2067 is marked with an asterisk.
The data obtained from the 3 HLA- andKIR-identical pairs address the question of whether the hematopoietic malignancy suffered by one of the siblings had any effect on the repertoire of KIR expression. Although the percentage and absolute number of NK cells in the peripheral blood differed between the siblings (data not shown), the KIR repertoires of the affected siblings matched those of their healthy,KIR-identical, HLA-identical siblings (Figure 2). These observations indicate that the KIR repertoires measured in the patients prior to transplantation were an accurate reflection of their repertoires prior to the onset of malignant disease and validate comparisons with donor and posttransplantation KIR repertoire.
Distinct patterns of KIR repertoire reconstitution emerge following transplantation
Following transplantation, patients' peripheral blood was sampled at intervals over the course of 1 year, and the repertoire of KIR and CD94:NKG2A expression by NK cells determined by flow cytometry. Posttransplantation KIR repertoires were compared with those of the donor and patient prior to transplantation. KIR and CD94:NKG2 expression profiles, in conjunction with KIR repertoire comparisons, were examined to identify the pattern of NK cell receptor repertoire reconstitution following transplantation for each patient. On this basis, the patients could be divided into 3 groups.
Group 1 transplant recipients (SPN 1899, 2017, 1991, 2025, 2060, 2036, 2067, 2138)
For 8 patients (6 CML and 2 AML), the reconstituted repertoire of KIR expression 1 year after transplantation was like that of the donor and distinct from that of the patient before transplantation. Four donors were MRDs and 4 were MUDs; none of the donors wereKIR identical to their recipient. Patients in this group had good recovery following transplantation with few clinical complications, although 4 patients (SPN 1991, 2025, 2036, 2067) had grades I to II acute GVHD and one patient had chronic GVHD (SPN 2067). Myeloablative preparatory regimens for individuals in this group were FTBI, in combination with etoposide, cyclophosphamide, and/or busulfan. All individuals were alive and in continuous complete remission at the time of last follow-up (Table 1).
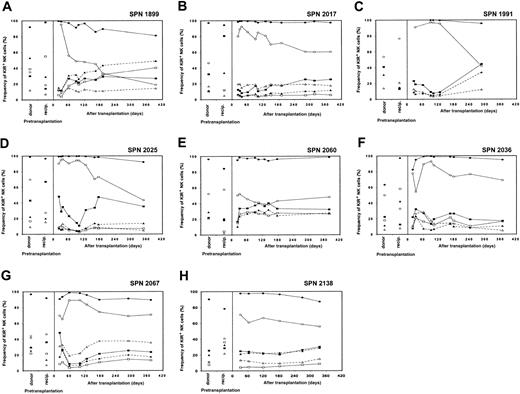
Reconstitution of NK cell receptor expression in patients from group 1 following transplantation
Recovery of NK cell receptor repertoire for each of the 8 patients is illustrated in Figure 3A-H, where each panel represents a separate patient. Posttransplantation NK cell receptor reconstitution is compared with KIR and CD94:NKG2A expression patterns in the donor and patient prior to transplantation. For each patient, few peripheral blood NK cells expressed KIR on initial engraftment, whereas the majority of NK cells were CD94:NKG2A+. The frequency of KIR expression gradually increased during the subsequent 6 to 9 months, eventually reaching levels comparable to those of the donor. Concomitant with increasing KIR expression, the proportion of NK cells expressing CD94:NKG2A gradually decreased. At 1 year after transplantation, CD94:NKG2A frequencies resembled those of the donor for 3 patients (SPN 1991, 2025, 2060), and the recipient (before transplantation) for one patient (SPN 1899), but differed from both donor and recipient pretransplantation expression levels for 4 patients (SPN 2017, 2036, 2067, 2138).
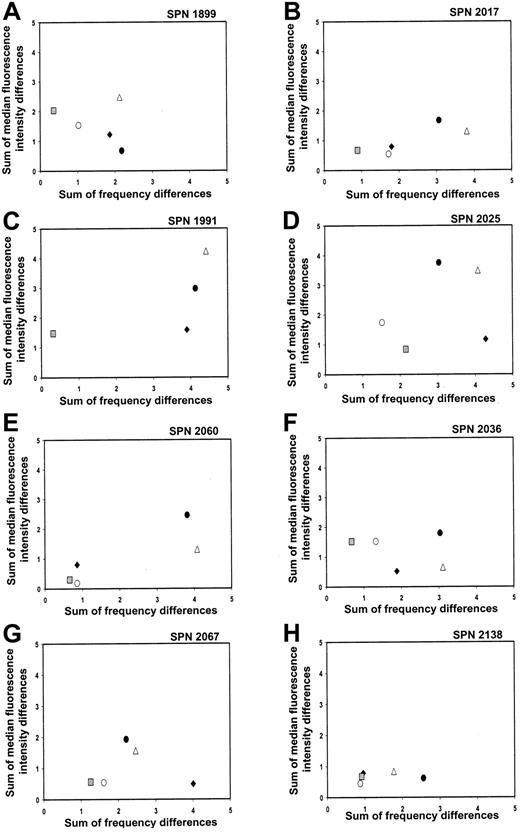
KIR repertoire comparisons for patients in group 1 during the first year after transplantation
That the 8 patients reconstituted donor-type KIR expression patterns were confirmed when the recipient's KIR repertoire at different time points prior to and following transplantation was compared with the donor's repertoire. The summed differences in the frequency of cells binding each of the 4 KIR-specific antibodies and the MFI were calculated and plotted for each donor-recipient pair at the following time points: before transplantation (solid circles), days 90 to 120 (solid diamonds), days 150 to 180 (open circles), and days 270 to 365 (gray squares) after transplantation (Figure 4A-H, where each panel represents a separate patient). Each data point in the 2-dimensional plots represents a comparison of the KIR repertoire between the donor and recipient at a specific time point. The closer a data point is to the origin, the more similar are the 2 KIR repertoires, and, conversely, the further a data point is from the origin, the more disparate are the 2 KIR repertoires under comparison.17For each patient in group 1, data points representing the donor-versus-recipient comparison before transplantation (solid circles) are away from the origin, consistent with the KIRmismatch for each donor-recipient pair. With time of sampling over the course of 1 year, the data points move progressively toward the origin, with the data point representing the 1-year posttransplantation comparison (gray squares) of recipient and donor in general being the closest (Figure 4A-H). Comparison of each patient's KIR repertoire prior to transplantation with that at 1 year after transplantation (open triangles) further demonstrates that the reconstituted posttransplantation repertoire is distinct from that of the recipient prior to transplantation. This analysis indicates that the KIR repertoire of expression for each patient in group 1 changes over a 1-year time period to resemble that of the donor.
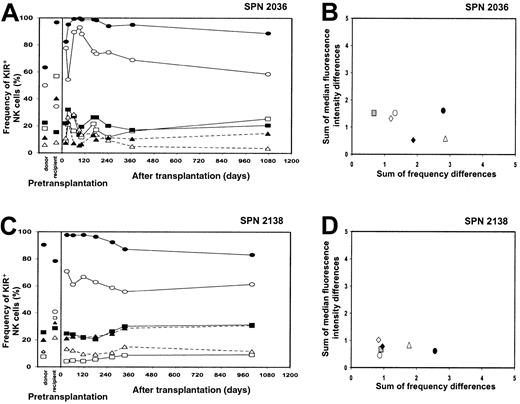
KIR expression patterns 3 years after transplantation (SPN 2036, 2138)
Peripheral blood samples were obtained from 2 of the patients in group 1 (SPN 2036, 2138) at 36 and 32 months after transplantation, respectively. NK cell receptor expression was assessed by flow cytometry and compared with that of the donor and recipient prior to and following transplantation (Figure 5A-D). The frequency of KIR- and CD94:NKG2A-positive NK cells some 3 years after transplantation were similar to that 1 year after transplantation, as shown in Figure 5A,C. This conclusion is also reached when the KIR repertoires of the donor and the recipient at the 3-year time point were compared. There was no significant change in the location of the data points representing the donor-recipient comparison 1 year after transplantation (gray squares) and the donor-recipient comparison 3 years after transplantation (open diamonds) (Figures 5B,D), suggesting that with reconstitution of the NK cell receptor repertoire in the first year following transplantation, it then becomes stable. Such stability in NK cell receptor repertoire is also a property of healthy individuals who have not received a transplant, as determined by continual assessment of KIR repertoire over a 13-month period.17
NK cell receptor expression in patients from group 1 following hematopoietic cell transplantation.
Posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for each of the 8 patients from group 1 (one panel per patient) are compared with donor and recipient pretransplantation expression. Patients in group 1 are characterized by a return to donorlike KIR expression by 6 to 9 months after transplantation. Each receptor-specific antibody is shown with a different symbol: EB6 (■; KIR2DL1/S1), DX27 (▪; KIR2DL2-3/S2), DX9 (▵; KIR3DL1), DX31 (▴; KIR3DL2), HP-3B1 (●; CD94), and Z199 (○; NKG2A).
NK cell receptor expression in patients from group 1 following hematopoietic cell transplantation.
Posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for each of the 8 patients from group 1 (one panel per patient) are compared with donor and recipient pretransplantation expression. Patients in group 1 are characterized by a return to donorlike KIR expression by 6 to 9 months after transplantation. Each receptor-specific antibody is shown with a different symbol: EB6 (■; KIR2DL1/S1), DX27 (▪; KIR2DL2-3/S2), DX9 (▵; KIR3DL1), DX31 (▴; KIR3DL2), HP-3B1 (●; CD94), and Z199 (○; NKG2A).
Changes in KIR repertoire in each patient from group 1 over a 1-year time period following hematopoietic cell transplantation.
Each panel shows comparisons between the NK cell KIR repertoires of the donor and patient at different time points prior to and following hematopoietic cell transplantation. Each panel represents a different patient, and each symbol represents a different time point under comparison: donor versus patient before transplantation (●); donor versus patient days 90 to 120 (♦); donor versus patient days 150 to 180 (○); donor versus patient days 270 to 365 (░); patient before versus day 365 after transplantation (▵).
Changes in KIR repertoire in each patient from group 1 over a 1-year time period following hematopoietic cell transplantation.
Each panel shows comparisons between the NK cell KIR repertoires of the donor and patient at different time points prior to and following hematopoietic cell transplantation. Each panel represents a different patient, and each symbol represents a different time point under comparison: donor versus patient before transplantation (●); donor versus patient days 90 to 120 (♦); donor versus patient days 150 to 180 (○); donor versus patient days 270 to 365 (░); patient before versus day 365 after transplantation (▵).
NK cell repertoire of expression for 2 group 1 patients 3 years after transplantation.
(A,C) These panels show posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for 2 patients from group 1 (SPN 2036 and 2138, respectively) followed for a 3-year time period and compared with donor and recipient pretransplantation expression. Symbols representing receptor-specific antibodies are the same as in Figure 3. (B,D) These panels compare the NK cell repertoires between donor-recipient pairs at different time points following transplantation. Symbols representing each time point under comparison are the same as in Figure 4, with the addition of the following: donor versus patient 3 years after transplantation (⋄).
NK cell repertoire of expression for 2 group 1 patients 3 years after transplantation.
(A,C) These panels show posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for 2 patients from group 1 (SPN 2036 and 2138, respectively) followed for a 3-year time period and compared with donor and recipient pretransplantation expression. Symbols representing receptor-specific antibodies are the same as in Figure 3. (B,D) These panels compare the NK cell repertoires between donor-recipient pairs at different time points following transplantation. Symbols representing each time point under comparison are the same as in Figure 4, with the addition of the following: donor versus patient 3 years after transplantation (⋄).
Group 2 transplant recipients (1920, 1970, 2040, 2068, 2089)
Reconstitution in 5 patients (3 CML, 2 AML) 1 year after transplantation was characterized by reduced frequencies of KIR-positive NK cells, whereas CD94:NKG2A expression frequencies remained high. All 5 donor-recipient pairs wereHLA-identical siblings and one was KIR identical (SPN 2089). Four patients in this group experienced few clinical complications, although one patient (SPN 1970) died of meningitis 2 years after transplantation. The fifth patient (SPN 2040) had chronic GVHD but showed a similar pattern of KIR reconstitution through day 223 after transplantation, the last time point sampled prior to AML relapse and consequent death of this patient. Myeloablative preparatory regimens for patients in this group included combinations of busulfan, etoposide, and cyclophosphamide, and only one patient (SPN 1920) received FTBI (Table 1).
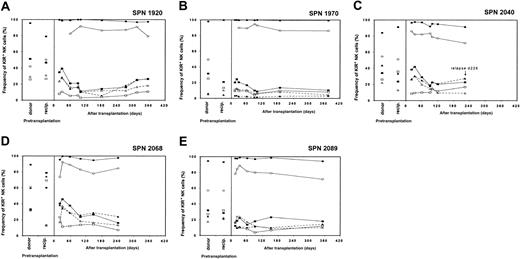
Reconstitution of NK cell receptor expression in patients from group 2 following transplantation
The recovery of the NK cell receptor repertoire for each of the 5 patients of this group is illustrated in Figure 6A-E. For each patient, the frequency of KIR-positive cells was low throughout the first year following transplantation, whereas the frequency of CD94:NKG2A-positive NK cells remained high. Despite lower KIR expression frequencies, the hierarchy of KIR expression within the KIR-positive population for each patient resembled that of the donor, indicating that KIR expression was generally depressed in these patients during the first year, but not selectively perturbed for any particular KIR.
KIR repertoire comparisons for patients in group 2 during the first year after transplantation
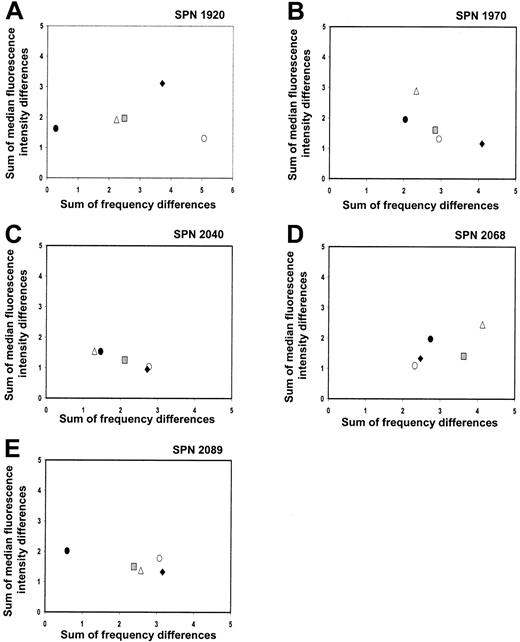
Summed frequency and MFI differences for each KIR-specific antibody were calculated and plotted for each donor-recipient pair prior to and following transplantation (Figure 7A-E). In contrast to the patients in group 1, the KIR repertoire of the patients in group 2 did not change to resemble that of the donor during the first year after transplantation. One donor-recipient pair in this group was KIRmatched (SPN 2089) and, as a result, had very similar pretransplantation KIR repertoires (Figure 7E; solid circle). A second transplant pair also had similar pretransplantation repertoires (Figure7A; solid circle), although this donor and recipient were KIRdisparate (SPN 1920). Despite having similar repertoires, donor-recipient comparisons at 1 year after transplantation differed (gray squares) for both transplant pairs, showing that the posttransplantation KIR repertoire in patients of this group is distinct from that of the donor and recipient prior to transplantation. The clustering of all data points for the different time points under comparison for the remaining 3 patients of this group (SPN 1970, 2040, 2068) further indicates that the KIR repertoire of expression does not change dramatically during the first year following transplantation. The measurement of differences in KIR expression level in the patients from group 2 is less accurate than for patients in group 1, because of the low frequency of KIR-expressing NK cells and the limited number of NK cells available for analysis. Accordingly, more weight should be given to the differences in frequency of expression than levels of expression.
NK cell receptor expression in patients from group 2 following hematopoietic cell transplantation.
Posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for each of the 5 patients in group 2 are compared with donor and recipient pretransplantation expression (separate panel per patient). Patients in group 2 are characterized by depressed KIR expression and high CD94:NKG2A expression during the first year following hematopoietic cell transplantation. Symbols representing receptor-specific antibodies are the same as in Figure3.
NK cell receptor expression in patients from group 2 following hematopoietic cell transplantation.
Posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for each of the 5 patients in group 2 are compared with donor and recipient pretransplantation expression (separate panel per patient). Patients in group 2 are characterized by depressed KIR expression and high CD94:NKG2A expression during the first year following hematopoietic cell transplantation. Symbols representing receptor-specific antibodies are the same as in Figure3.
Changes in KIR repertoire in each patient from group 2 over a 1-year time period following hematopoietic cell transplantation.
Each panel shows comparisons between the KIR repertoires of the donor and patient at different time points prior to and following hematopoietic cell transplantation. Each panel represents a different patient, and each symbol represents a different time point under comparison. Symbols representing each time point under comparison are the same as in Figure 4.
Changes in KIR repertoire in each patient from group 2 over a 1-year time period following hematopoietic cell transplantation.
Each panel shows comparisons between the KIR repertoires of the donor and patient at different time points prior to and following hematopoietic cell transplantation. Each panel represents a different patient, and each symbol represents a different time point under comparison. Symbols representing each time point under comparison are the same as in Figure 4.
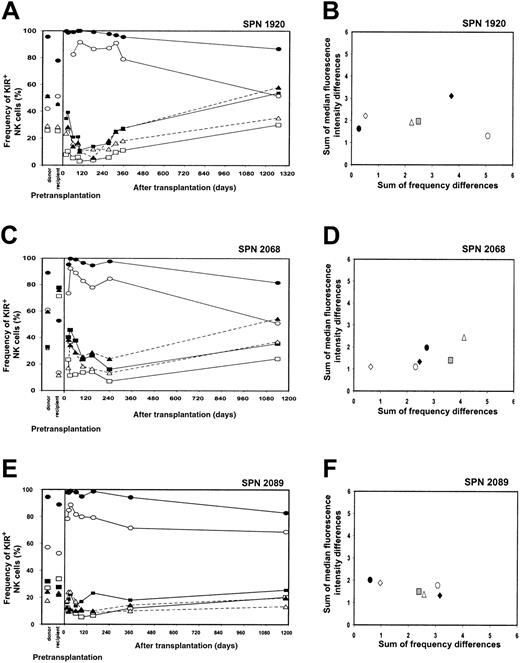
KIR expression patterns 3 years after transplantation (SPN 1920, 2068, 2089)
Peripheral blood was obtained from the 3 living patients belonging to group 2 (SPN 1920, 2068, 2089) at 42.5, 37, and 36.5 months after transplantation, respectively. The NK cell receptor reconstitution and repertoire comparisons for each patient are illustrated in Figure 8A-F. In each patient, the frequencies of KIR-positive NK cells at the 3-year time point were at levels comparable to those of the donor (Figure 8A,C,E). As shown in Figure8B, D, and F, repertoire comparisons clearly demonstrate that at 3 years after transplantation, the KIR repertoire of expression is more similar to that of the donor (open diamonds) and distinct from that reconstituted at 1 year after transplantation (gray squares). These comparisons indicate that the low levels of KIR expression observed in these patients during the first year following transplantation do not persist, there being an eventual increase in the frequencies of KIR expression that leads to reconstitution of a donorlike KIR phenotype. Further investigation is needed to determine the recovery kinetics of KIR expression in patients from group 2 during the period from 1 to 3 years after transplantation. In conclusion, however, the important difference between the patients in group 1 and group 2 appears to be in the kinetics and time frame of recovery of a donorlike repertoire of KIR expression by NK cells, not in the potential for such recovery.
NK cell repertoire of expression for 3 group 2 patients 3 years after transplantation.
(A,C,E) These panels show posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for patients in group 2 (SPN 1920, 2068, and 2089, respectively) followed for a 3-year time period. Posttransplantation expression frequencies are compared with donor and recipient pretransplantation expression. Symbols representing receptor-specific antibodies are the same as in Figure 3. (B,D,F) These panels show comparisons between the NK cell repertoires of the donors and patients at different time points following transplantation. Symbols representing each time point under comparison are the same as in Figure 4, with the addition of the following: donor verus patient 3 years after transplantation (⋄).
NK cell repertoire of expression for 3 group 2 patients 3 years after transplantation.
(A,C,E) These panels show posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for patients in group 2 (SPN 1920, 2068, and 2089, respectively) followed for a 3-year time period. Posttransplantation expression frequencies are compared with donor and recipient pretransplantation expression. Symbols representing receptor-specific antibodies are the same as in Figure 3. (B,D,F) These panels show comparisons between the NK cell repertoires of the donors and patients at different time points following transplantation. Symbols representing each time point under comparison are the same as in Figure 4, with the addition of the following: donor verus patient 3 years after transplantation (⋄).
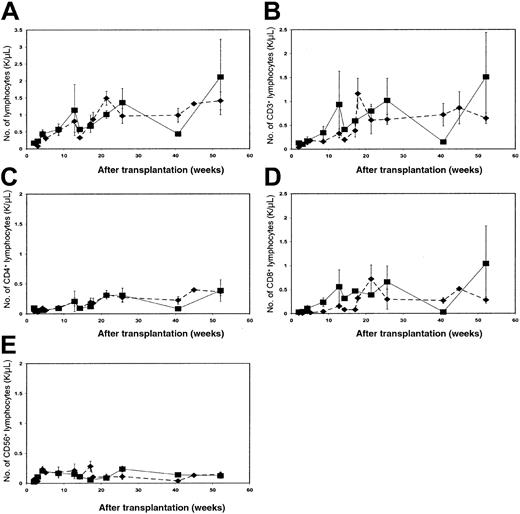
Lymphocyte reconstitution of patients in groups 1 and 2
To assess whether NK cell receptor reconstitution patterns reflected underlying differences in hematopoietic recovery, rates of lymphocyte reconstitution, evaluated by total lymphocyte count, total T-cell count, CD4+ and CD8+ T-cell subsets, and total NK cell count, were compared. All patients demonstrated a rapid recovery of NK cells. As expected, recovery of total lymphocyte numbers and T-cell counts (bulk T cells and CD4+ subsets) was faster in PBPC than BM recipients (data not shown). There were no significant differences in reconstitution rates between patients receiving BM from related versus unrelated donors, nor did reconstitution differ between CML and AML patients (data not shown). Likewise, there were no significant differences in the kinetics of lymphocyte reconstitution between patient groups 1 and 2 (Figure 9A-E), indicating that any skewing of reconstitution rate between these patient groups was more subtle than variation because of different hematopoietic cell sources.
NK cell populations were further subdivided into CD56brightand CD56dim. These phenotypes may reflect different stages in NK development, or distinct regulatory and cytolytic NK subpopulations, with CD56dim cells being the major population of KIR-expressing cells.25-27 The proportion of CD56dim NK cells was similar in patients in group 1 and 2 early after transplantation, but significantly different after 1 year (Table 2). Despite this difference, reduced KIR expression in the bulk NK cell population in patients from group 2 (data not shown) could not be attributed to an altered ratio of CD56dim and CD56bright cells, as lack of KIR expression was seen in both the CD56bright and CD56dim cells.
Reconstitution of the CD56+ lymphocyte subsets in BM versus PBPC recipients, and patients in group 1 versus group 2
| . | BMT (n = 14) . | PBPC (n = 4) . | Group 1 (n = 8) . | Group 2 (n = 5) . |
|---|---|---|---|---|
| CD56+, K/μL | ||||
| Day 30 | 0.22 ± 0.03 | 0.26 ± 0.04 | 0.26 ± 0.04 | 0.18 ± 0.04 |
| Day 60 | 0.17 ± 0.03 | 0.16 ± 0.06 | 0.17 ± 0.04 | 0.17 ± 0.05 |
| Day 90/100 | 0.17 ± 0.03 | 0.19 ± 0.06 | 0.18 ± 0.06 | 0.17 ± 0.04 |
| Day 180 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.11 ± 0.02 | 0.24 ± 0.07 |
| Day 365 | 0.14 ± 0.03 | 0.07 ± 0.03 | 0.11 ± 0.04 | 0.13 ± 0.05 |
| CD56dim, % of CD56+ | ||||
| Day 30 | 51.9 ± 6.7 | 66.6 ± 7.2 | 55.5 ± 10.2 | 62.7 ± 7.8 |
| Day 60 | 54.3 ± 5.4 | 72.3 ± 7.6 | 59.1 ± 7.8 | 57.1 ± 11.5 |
| Day 90/100 | 55.6 ± 6.6 | 67.2 ± 5.0 | 53.3 ± 10.0 | 51.9 ± 10.2 |
| Day 180 | 76.4 ± 5.5 | 79.2 ± 12.5 | 81.5 ± 2.4 | 61.8 ± 13.8 |
| Day 365 | 79.3 ± 4.7 | 84.5 ± 6.3 | 86.1 ± 2.1* | 58.1 ± 6.2* |
| . | BMT (n = 14) . | PBPC (n = 4) . | Group 1 (n = 8) . | Group 2 (n = 5) . |
|---|---|---|---|---|
| CD56+, K/μL | ||||
| Day 30 | 0.22 ± 0.03 | 0.26 ± 0.04 | 0.26 ± 0.04 | 0.18 ± 0.04 |
| Day 60 | 0.17 ± 0.03 | 0.16 ± 0.06 | 0.17 ± 0.04 | 0.17 ± 0.05 |
| Day 90/100 | 0.17 ± 0.03 | 0.19 ± 0.06 | 0.18 ± 0.06 | 0.17 ± 0.04 |
| Day 180 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.11 ± 0.02 | 0.24 ± 0.07 |
| Day 365 | 0.14 ± 0.03 | 0.07 ± 0.03 | 0.11 ± 0.04 | 0.13 ± 0.05 |
| CD56dim, % of CD56+ | ||||
| Day 30 | 51.9 ± 6.7 | 66.6 ± 7.2 | 55.5 ± 10.2 | 62.7 ± 7.8 |
| Day 60 | 54.3 ± 5.4 | 72.3 ± 7.6 | 59.1 ± 7.8 | 57.1 ± 11.5 |
| Day 90/100 | 55.6 ± 6.6 | 67.2 ± 5.0 | 53.3 ± 10.0 | 51.9 ± 10.2 |
| Day 180 | 76.4 ± 5.5 | 79.2 ± 12.5 | 81.5 ± 2.4 | 61.8 ± 13.8 |
| Day 365 | 79.3 ± 4.7 | 84.5 ± 6.3 | 86.1 ± 2.1* | 58.1 ± 6.2* |
Means are shown ± SEM. Differences between patient groups tested by Student t-test.
Significant difference (P < .05).
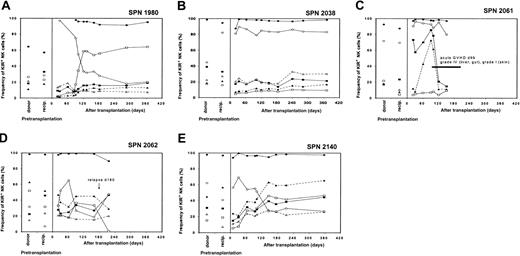
Remaining, nongroup 1 or 2, patients (SPN 1980, 2038, 2061, 2062, 2140)
The remaining 5 patients (3 CML, 2 AML) examined are grouped together not on the basis of any similarity in NK cell receptor expression but because their outcomes were idiosyncratic and different from groups 1 and 2. One donor-recipient pair comprised MUD individuals and 4 were MRDs, of which 2 pairs were also KIR identical. These patients reconstituted KIR and CD94:NKG2A expression patterns in ways that were distinct from either the pretransplantation patient or donor repertoires (Figure 10A-E). As a group, they were distinguished from patients in groups 1 and 2 in that all suffered serious clinical complications within the first year after transplantation, including clinically extensive chronic GVHD (SPN 2038, 2061, 2140), grade IV acute GVHD (SPN 2061), and CML relapse (SPN 1980, 2062). Three patients were alive and in continuous complete remission (SPN 1980, 2038, 2140), one patient underwent a second transplantation (SPN 2062), and one patient had died (SPN 2061) at the time of last follow-up (Table 1).
Reconstitution of NK cell receptor expression in the remaining patients following transplantation
The recovery of NK cell receptor expression following transplantation for the nongroup 1 or 2 patients is illustrated in Figure 10A-E. Each patient had a unique reconstitution pattern. One patient (SPN 1980) expressed high levels of KIR2DL1, whereas expression of the other NK cell receptors was low (Figure 10A). Patient 2038 expressed intermediate levels of KIR, but the overall expression pattern differed from that of the donor and recipient prior to transplantation (Figure 10B). Patient 2061 was characterized by high expression of KIR2DL3/2DS2 and 3DL2 immediately after engraftment, followed by low total KIR expression day 100 after transplantation (Figure 10C). Patients 2062 and 2140 expressed novel KIR reconstitution patterns, with low or intermediate NKG2A expression levels (Figure 10D-E, respectively). The unique features of the NK cell reconstitution patterns seen in these 5 patients cannot readily be explained in light of current knowledge and likely involve both genetic and environmental factors. The former could include the KIRand HLA genes and their differences between transplant patient and transplant donor; the latter could include the clinical history of the patient, the procedures and treatments used in transplantation, and the clinical complications following transplantation.
Reconstitution of lymphocyte subsets in patients from group 1 versus 2 over time.
Frequencies of CD3+, CD3+CD4−CD8+, CD3+CD4+CD8−, and CD3−CD56+ lymphocytes were calculated by flow cytometry using specific monoclonal antibodies. Values were used to back-calculate absolute numbers (× 103 cells per microliter) for each lymphocyte subset, from the absolute number of lymphocytes obtained from complete blood counts. Each panel compares the mean reconstitution values between the 8 patients from group 1 (▪, solid line) and 5 patients from group 2 (♦, broken line) for a different lymphocyte subset over a 1-year time period. Standard error bars for each patient group are shown. (A) Compares total lymphocyte reconstitution between patients from groups 1 and 2, (B) compares the number of CD3+ T lymphocytes, (C) compares the number of CD4+ T cells, (D) compares the number of CD8+ T cells, and (E) compares the number of CD56+ NK cells.
Reconstitution of lymphocyte subsets in patients from group 1 versus 2 over time.
Frequencies of CD3+, CD3+CD4−CD8+, CD3+CD4+CD8−, and CD3−CD56+ lymphocytes were calculated by flow cytometry using specific monoclonal antibodies. Values were used to back-calculate absolute numbers (× 103 cells per microliter) for each lymphocyte subset, from the absolute number of lymphocytes obtained from complete blood counts. Each panel compares the mean reconstitution values between the 8 patients from group 1 (▪, solid line) and 5 patients from group 2 (♦, broken line) for a different lymphocyte subset over a 1-year time period. Standard error bars for each patient group are shown. (A) Compares total lymphocyte reconstitution between patients from groups 1 and 2, (B) compares the number of CD3+ T lymphocytes, (C) compares the number of CD4+ T cells, (D) compares the number of CD8+ T cells, and (E) compares the number of CD56+ NK cells.
NK cell receptor expression in the nongroup 1 and 2 patients following hematopoietic cell transplantation.
Posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for each of the 5 remaining nongroup 1 and 2 patients are compared with donor and recipient pretransplantation expression. Each of these patients has idiosyncratic patterns of KIR reconstitution that differ from the donor and recipient following hematopoietic cell transplantation. Symbols representing each receptor-specific antibody are the same as in Figure 3.
NK cell receptor expression in the nongroup 1 and 2 patients following hematopoietic cell transplantation.
Posttransplantation KIR and CD94:NKG2A NK cell expression frequencies for each of the 5 remaining nongroup 1 and 2 patients are compared with donor and recipient pretransplantation expression. Each of these patients has idiosyncratic patterns of KIR reconstitution that differ from the donor and recipient following hematopoietic cell transplantation. Symbols representing each receptor-specific antibody are the same as in Figure 3.
Discussion
We show here that clinical hematopoietic cell transplantation provides a good system for examining the reconstitution of NK cell repertoires under conditions of HLA and/or KIRdifference. In the allogeneic HLA-matched,KIR-mismatched transplants studied here, most of the recipients reconstituted NK cell populations with a donorlike pattern of KIR expression distinct from that of the recipient prior to transplantation. Thus, regeneration of KIR repertoire was governed by the genotype of the NK cells, not the genotype of the new “host” environment. This finding demonstrates the importance of KIRgenotype in controlling KIR expression, as first revealed in comparisons of healthy sibling pairs.17
The patients studied here formed 3 groups, discriminated by posttransplantation patterns of NK cell receptor expression during the first year after transplantation. In the first group, which included equal numbers of MUD and MRD transplant recipients, the patterns of KIR expression became like that of the donor within the first year after transplantation and remained that way when assessed 2 years later. Immediately following engraftment, the frequency of KIR-positive NK cells for each patient in group 1 was low, whereas most NK cells expressed CD94:NKG2A. The frequency of KIR-positive NK cells then increased over time, reaching donor levels within the first year. This pattern of reconstitution is consistent with a previous report showing that the first NK cells appearing in patients following MUD transplantation had an immature phenotype and primarily expressed CD94:NKG2A, with relatively low expression of KIR.28
In the second group of patients, all MRD transplant recipients, the relative expression of the different KIR genes after transplantation was similar to the donor, but the overall percentage of KIR-expressing NK cells was substantially reduced during the first year after transplantation. When a subset of the patients in group 2 was examined 3 years after transplantation, they had recovered a donor-type pattern of KIR expression. Thus, the reconstitution of KIR expression in patients from group 2 was delayed compared with patients in group 1. The cause of this difference is unknown, but it could involve genetic factors as well as aspects of the transplantation protocol. That patients receiving autologous transplants exhibited depressed KIR expression after transplantation17 suggests that some degree of genetic incompatibility between transplant donor and recipient may speed recovery of KIR expression. Further support for this possibility is that xenogeneic mouse feeder cells were needed to induce KIR expression by human NK cell precursors in vitro.29 Another potential factor was the more common use of FTBI in the conditioning regimens for patients in group 1 than in group 2. Irradiation may be more efficient at ablating recipient hematopoietic cells than chemical agents, thereby minimizing low-level alloreactions that could influence NK cell development. Or, FTBI and chemical ablation may have different effects on the bone marrow stromal environment, which is critical for NK development.
The third group of patients included those suffering major clinical complications following transplantation. Whether their idiosyncratic patterns of NK cell receptor expression reflects influences from GVHD, early relapse, and other complications or clinical interventions remains to be determined. Intriguingly, a previous study shows lower frequencies of KIR and CD94:NKG2A expression in patients with chronic GVHD compared with those without, although the patient and donorKIR types were not taken into account.30 31
Unexpected were the kinetics of NK cell receptor repertoire reconstitution. Although NK cell recovery was rapid and comparable for all patients, the expression of KIR and CD94:NKG2A by the NK cells emerging immediately after engraftment was usually different from both donor and recipient. Establishment of a donorlike KIR repertoire was gradual, taking place over the course of 6 to 9 months for patients in group 1 and longer than 1 year for patients in group 2. In a study ofHLA haplotype-mismatched hematopoietic cell transplants, alloreactive NK cell clones were observed soon after engraftment, but by 4 months after transplantation they were no longer detectable.32 This time frame coincides with the development of donor-type KIR expression in the patients from group 1 studied here, suggesting that NK cell receptor repertoire reconstitution is a genetically programmed process, overlaid by HLA-mediated selection or education. Furthermore, this observation indicates that transplantation should prove a tractable system for assessing the influence of HLA environment on KIR repertoire development.
For all patients, CD94:NKG2A and KIR frequencies were inversely correlated during reconstitution, having a relationship that was similar to, but more dramatic than, the inverse correlation observed for healthy sibling pairs.17 Taken together, these data provide direct evidence for the 2 types of receptors sharing common purpose and for some degree of coordination in their differential expression during NK cell development. It will be of interest to examine posttransplantation outcomes in a larger group of transplant recipients to determine whether the patients having delayed KIR reconstitution patterns, and retaining CD94:NKG2A as their main inhibitory HLA class I receptor, are more susceptible to viral infections or other transplantation complications.
Several studies have correlated mismatching of KIRs and their HLA class I ligands (KIR epitopes) with clinical outcome following transplantation. In KIR-epitope incompatible, haploidentical transplants, donor-derived alloreactive NK cells protect patients with AML against relapse, GVHD, and graft failure.33 In contrast, De Santis et al34 observed higher engraftment and survival rates when haploidentical transplant pairs were matched for the KIR epitopes carried by HLA-C. However, this group did not evaluate their results according to the type of leukemia, an important variable as seen from the observation of Ruggeri et al33that alloreactive NK cells offer no protection from ALL following haploidentical transplantation. KIR-epitope matching was also examined in unrelated transplants with some degree of HLA mismatch. A retrospective analysis found no advantage for KIR epitope-incompatible patients in terms of survival, suggesting that the beneficial alloreactivities observed in haploidentical transplantation may not extend to other transplantation protocols.35 However, Giebel et al36 report better outcome for recipients following unrelated transplantation when antithymocyte globulin was used as part of the preparative regimen. Discrepancies in the results reported between groups likely reflect differences in the transplantation settings used at different treatment centers, such as conditioning regimens, stem cell and T-cell content, and the different forms of GVHD prophylaxis. Clearly, further study will be required before these results can be fully reconciled.
Recent reports have also examined the relationship between KIRtype and clinical outcome following transplantation. Gagne et al37 report an increase in GVHD in unrelated transplants when the donor has more activating KIR than the recipient. Analogous results were observed in HLA-identical sibling transplant and in haploidentical transplant recipients.34Similarly, Chida et al39 noted a slight increase in graft rejection in a panel of 87 HLA-matched transplants when the donor and recipient were KIR mismatched, although levels did not reach significance. Taken together, these studies support a role for NK cell activity in affecting clinical outcome.
In conclusion, this study has shown that most HLA-matched hematopoietic cell transplants involve varying degrees of KIRmismatch. In addition to providing valuable markers for assessing the reconstitution of NK receptor repertoire in transplant recipients,KIR polymorphisms and KIR compatibility are potential factors to influence transplantation outcome. As predicted from the analysis of healthy sibling pairs,17reconstitution of NK cell populations in transplant recipients leads to a pattern of KIR expression like that of the donor. Two patterns of reconstitution with different kinetics were observed, the first in which recovery was completed during the first year following transplantation and the second in which expression levels were depressed throughout the first year and only recovered after longer times. The cause of this difference and its effect on NK cell function following transplantation are important questions that should be addressed by extending the analysis to larger cohorts of transplant patients, experiencing similar disease and treatment, and correlating parameters of clinical outcome with those of NK cell function and receptor reconstitution. This type of study would also assess whether the idiosyncratic changes in NK cell receptor expression observed in patients experiencing clinical complications are in any way associated with those complications.
We thank Kathryn Tierney, RN, Drs Neil Young and Karl Blume for their help in organizing and facilitating the collection of patient samples, and Dr Byron Brown for advice regarding statistical methods. We would like to acknowledge the staff of the clinical BMT Laboratory at the Stanford Medical Center for their assistance in patient sample preparation and Gary E. Peacock for technical support. We also thank Dr Karl Blume for his encouragement to begin this project and for his continuing support throughout its progress.
Prepublished online as Blood First Edition Paper, January 2, 2003; DOI 10.1182/ blood-2002-08-2568.
Supported by National Institutes of Health Grant CA49605, a translational grant from the Leukemia and Lymphoma Society, National Institutes of Health Training Grant AI07290 (H.G.S.), and a Cancer Research Institute Fellowship (K.L.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Karina McQueen, Department of Structural Biology, Fairchild Building, D-155, Stanford University School of Medicine, Stanford, CA 94305-5126; e-mail:kmcqueen@stanford.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal