Abstract
Ex vivo–expanded mesenchymal stem cells (MSCs) were transduced with a green fluorescent protein (GFP) retroviral construct and subsequently infused into 3 adult baboons following lethal total body irradiation and hematopoietic support or without any prior conditioning. To study the long-term fate of these MSCs, necropsies were performed between 9 and 21 months following MSC infusion, and an average of 16 distinct tissues were recovered from each recipient and evaluated for the presence of the GFP transgene in purified genomic DNA by sensitive real-time polymerase chain reaction (PCR). Two baboons received autologous and one allogeneic GFP-transduced MSCs. Both allogeneic and autologous MSCs appeared to distribute in a similar manner. Gastrointestinal tissues harbored high concentrations of transgene per microgram of DNA. Additional tissues including kidney, lung, liver, thymus, and skin were also found to contain relatively high amounts of DNA equivalents. Estimated levels of engraftment in these tissues ranged from 0.1% to 2.7%. The nonconditioned recipient appeared to have less abundant engraftment. These data suggest that MSCs initially distribute broadly following systemic infusion and later may participate in ongoing cellular turnover and replacement in a wide variety of tissues.
Introduction
Human postnatal bone marrow contains mesenchymal stem cells (MSCs) capable of differentiating into multiple mesenchymal tissues such as bone, adipose, cartilage, and myelosupportive stroma.1-3 In vitro studies have demonstrated the capacity of human MSCs (hMSCs) to secrete numerous hematopoietic cytokines and to support hematopoiesis.4 Studies involving a variety of animal models have shown that MSCs may be useful in the repair or regeneration of damaged or mutated bone, cartilage, or myocardial tissues.1,5-7 MSCs may also provide a useful target for gene therapy strategies involving genes encoding secreted proteins.8 Moreover, both hMSCs and baboon MSCs (bMSCs) possess in vitro immunosuppressive properties that appear not to be major histocompatibility complex (MHC) restricted.5,9,10 A recent study demonstrated the capacity of systemically infused bMSCs to prolong skin allograft survival, confirming that such immunosuppressive effects may be operational in vivo.9 These properties may limit the ability of the recipient immune system to recognize and reject allogeneic or gene-modified MSCs following transplantation.
Several clinical applications of MSCs would theoretically involve the administration of MSCs by the intravenous route.7,11-14However, limited data are available regarding the ultimate fate of systemically infused MSCs. Studies in rodents suggest a broad initial biodistribution followed by a limited capacity for sustained engraftment.15 In fetal sheep, hMSCs undergo a wide tissue distribution and can differentiate into multiple mesenchymal tissues following peritoneal implantation.16 Here, we have used an immunocompetent baboon model to study the fate of systemically infused autologous and allogeneic bMSCs.17 Ex vivo–expanded bMSCs were tagged with a green fluorescent protein (GFP) retroviral construct prior to infusion and then, at various time points after infusion, bone, bone marrow, and nonhematopoietic tissues were analyzed for the presence of transgene by a digoxigenin-based polymerase chain reaction (PCR) methodology. The preliminary results in this model suggested preferential distribution to the bone and bone marrow with persistence of transgene through 454 days following infusion.17 In that initial study, 2 baboons necropsied between 5 and 6 weeks following infusion did not have transgene detectable in any of the nonhematopoietic tissues analyzed. In the present study, we used a sensitive real-time PCR technique to evaluate the long-term fate of systemically infused GFP-transduced bMSCs by assessing their biodistribution in several nonhematopoietic tissues obtained via necropsy from 9 to 21 months after infusion. The data presented herein suggest that MSCs distribute widely to a variety of nonhematopoietic tissues following systemic infusion and may possess the capacity to proliferate within these tissues.
Study design
Animals
Healthy juvenile (age 3 to 4 years) baboons (Papio anubis) of both sexes and weighing between 8 and 12 kg were used. The animals were housed under conditions approved by the Association for the Assessment and Accreditation of Laboratory Animal Care International. The studies were performed under protocols approved by the Animal Care Committee of the University of Illinois at Chicago.
Collection, expansion, and characterization of baboon mesenchymal stem cells (bMSCs)
We have previously described the methods used for the collection and expansion of bMSCs from baboon bone marrow.17 This methodology is similar to that described for obtaining and expanding hMSCs, with minor modifications.3 Baboon MSCs express a nonhematopoietic phenotype (negative for CD34 and CD45) and are positive for markers used to characterize hMSCs (SH-2, SH-3, and SH-4; Osiris Therapeutics, Baltimore, MD).17 The cells isolated for these experiments are termed MSCs based on their extensive proliferative capacity and by their ability to differentiate into bone and adipose tissues in vitro and cartilage in vivo.3 17
Genetic marking of bMSCs
Methods used for transduction of bMSCs with the GFP retroviral vector pOT24 have been previously described.17 Briefly, baboon MSCs were transduced (under centrifugal forces) by exposing the cells to retrovirus at day 6 and at day 8 during primary bMSC culture isolation. Centrifugal transduction was performed by the addition of 10 to 15 mL of transduction cocktail and centrifugation in a Beckman GS-6R centrifuge (Beckman Instruments, Palo Alto, CA) using microtiter plate carriers at 1650g for 1 hour at 32°C. Following centrifugation, 15 mL of fresh medium was added to each flask. The transduction cocktail consisted of retroviral supernatant and polybrene (Sigma, St Louis, MO) at a final concentration of 8 μg/mL. After centrifugation, bMSCs were expanded in culture for a total of 14 days. No drug selection was applied to the cells. To determine transduction efficiency, cells were analyzed by flow cytometry and histochemical staining for enhanced GFP (eGFP). Cells were analyzed by collecting at least 10 000 events on a FACS Vantage instrument using CellQuest software (Becton Dickinson, San Jose, CA). Cells were cryopreserved in 85% PlasmaLyte A (Baxter IV Therapy, Deerfield, IL), 10% dimethyl sulfoxide (DMSO), and 5% autologous baboon serum. In these experiments, transduction efficiencies were low (6% to 11%) and, as a result, most of the MSCs infused were not genetically modified. The mean values for number of integrated copies of eGFP per diploid cell infused are shown in Table1. This value did not vary significantly between the recipients.
Study recipients
| Recipient . | Conditioning . | Hematopoietic support . | MSC source . | MSC dose, per kg . | Transduction efficiency . | Mean copies of eGFP per cell infused . | Timing of necropsy . |
|---|---|---|---|---|---|---|---|
| PA6592 | TBI (1000 cGy) | Auto BM | Auto | 30.3 × 106 | 11% | 0.17 | 21 mo |
| PA6590 | None | None | Auto | 17.4 × 106 | 6% | 0.11 | 19.5 mo |
| PA6243 | TBI (1000 cGy) | Auto MPB | Allo | 18.5 × 106 | 11% | 0.15 | 9 mo |
| Recipient . | Conditioning . | Hematopoietic support . | MSC source . | MSC dose, per kg . | Transduction efficiency . | Mean copies of eGFP per cell infused . | Timing of necropsy . |
|---|---|---|---|---|---|---|---|
| PA6592 | TBI (1000 cGy) | Auto BM | Auto | 30.3 × 106 | 11% | 0.17 | 21 mo |
| PA6590 | None | None | Auto | 17.4 × 106 | 6% | 0.11 | 19.5 mo |
| PA6243 | TBI (1000 cGy) | Auto MPB | Allo | 18.5 × 106 | 11% | 0.15 | 9 mo |
MSC indicates mesenchymal stem cell; eGFP, enhanced green fluorescent protein; TBI, total body irradiation; auto, autologous; BM, bone marrow; MPB, growth factor–mobilized peripheral blood; and llo, allogeneic.
MSC infusion
All 3 baboons received either autologous (PA6590 and PA6592) or allogeneic MHC-mismatched (PA6243) GFP-transduced bMSCs by intravenous infusion on day 0 following lethal total body irradiation (TBI) (1000 cGy) and hematopoietic support (PA6243 and PA6592) or without any prior conditioning (PA6590) as previously described.17 The details of the study are outlined in Table 1.
Procurement of tissues for DNA analysis
Prior to necropsy, baboons were humanely killed by administration of an overdose of sodium pentobarbital (100 mg/kg intravenously). Randomly selected tissue samples (100 to 300 mg) were obtained from the liver, lung, kidney, spleen, bladder, thymus, small and large intestines, blood vessels, stomach, pancreas, heart, brain, gallbladder, and skin and then frozen.
Detection of GFP transgene by real-time PCR
DNA was isolated from baboon tissues with a sodium dodecyl sulfate (SDS)/proteinase K–based technique (DNAeasy, Qiagen, Valencia, CA) and analyzed for purity by A260/A280 ratio. Aliquots of DNA (100 to 125 ng) were analyzed by a real-time PCR assay using fluorescence resonance energy transfer (FRET) probes (primers and probes based on GenBank GFP sequence). Baboon genomic DNA spiked with serial dilutions (known number of copies) of the GFP vector was used to prepare standard curves with the aid of the Lightcycler software (Roche Diagnostic, Indianapolis, IN), and the number of copies of GFP transgene per 100 ng tissue-derived DNA were computed. Nontransduced baboon MSCs were used in each run as a negative control. Percent engraftment was calculated based on the assumption that diploid baboon cells contain 6 pg DNA per cell.
Results and discussion
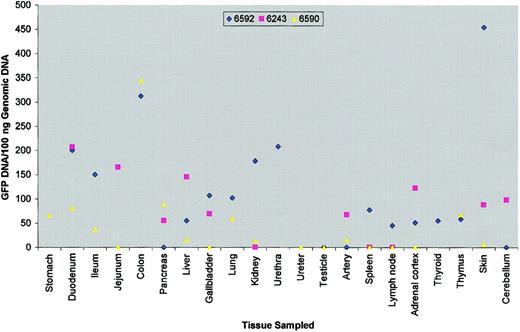
Tissues were obtained by necropsy at 9 (PA6243), 19.5 (PA6590), and 21 (PA6592) months following bMSC infusion (Table 1). An average of 16 different tissues were analyzed per recipient (PA6592: 18; PA6590: 18; PA6243: 12). The results of the analysis for GFP DNA is presented in Figure 1. Both allogeneic and autologous bMSCs appeared to be distributed in a similar manner, although the animal that did not receive conditioning radiation (PA6590) appeared to have lower amounts of GFP-positive DNA equivalents. The mean DNA copy numbers per tissue analyzed for the conditioned animals PA6592 and PA6243 of 114 ± 29 and 84 ± 19, respectively, were higher than the mean copy number in the nonconditioned recipient PA6590 (44 ± 19). Gastrointestinal tissues including colon, duodenum, jejunum, and ileum in general had the highest concentration of engrafted cells per microgram of DNA in each recipient. Additional tissues including kidney, skin, lung, thymus, and liver were found to harbor relatively high amounts of DNA equivalents, with values ranging from 1 × 103 to 2.7 × 104 cell equivalents per microgram of DNA (data not shown). Estimated levels of engraftment ranged from 0.1% to 2.7%, although this may be an underestimate given the low overall transduction efficiency.
Detection of GFP transgene in nonhematopoietic tissues.
Scatter diagram depicts level of green fluorescent protein (GFP)–positive DNA detected per 100 ng baboon genomic DNA from various tissues sampled at necropsy. Symbols were omitted if tissue was not sampled.
Detection of GFP transgene in nonhematopoietic tissues.
Scatter diagram depicts level of green fluorescent protein (GFP)–positive DNA detected per 100 ng baboon genomic DNA from various tissues sampled at necropsy. Symbols were omitted if tissue was not sampled.
These data demonstrate that following systemic infusion of GFP-marked MSCs into an immunocompetent host, these cells can be detected in a wide nonhematopoietic tissue distribution between 9 and 21 months later. In contrast, our previous study failed to detect GFP transgene in several nonhematopoietic organs early (5 to 6 weeks) following bMSC infusion.17 This may be due to the greater sensitivity of the real-time PCR assay used in the present study, proliferation of the GFP-transduced cells over time once engrafted, or possibly redistribution of gene-marked cells from some other tissue reservoir several weeks to months following infusion. Although the number of animals studied here is small, the results suggest that tissue distribution of bMSCs following systemic infusion is not affected by histocompatibility or prior conditioning. The DNA copy numbers were, in general, lower in the nonconditioned recipient. MSCs appear to distribute in a relatively random fashion following intravenous infusion. However, the migration or proliferation of MSCs following infusion may to some degree be influenced by local tissue injury, as may be encountered in the gut following radiation.
Because the baboons were not perfused prior to necropsy, contamination of the tissues sampled by circulating MSCs may have occurred to a limited degree, although we believe this is unlikely. We have consistently failed to identify circulating eGFP-tagged MSCs by any PCR methodology within peripheral blood specimens obtained after days 2 to 3 after transplantation. Bone marrow aspirates, which are highly contaminated by peripheral blood, have been consistently negative for eGFP by PCR after day 100.17 In another recipient of eGFP-tagged MSCs not previously reported (PA6759), we have also failed to detect GFP in peripheral blood or bone marrow aspirates after day 1 after transplantation. Other investigators have failed to identify MSCs in postnatal peripheral blood.18 Taken together, we believe the results obtained here are very unlikely to be the result of contamination by circulating MSCs.
Although we have previously detected GFP-expressing cells in baboon bone marrow following GFP-transduced bMSC infusion, efforts to identify GFP expression in other tissues by immunohistochemical methods have to date failed.17 This is most likely due to gene silencing, although a combination of cell death and immunologic rejection may have contributed. Nevertheless, the GFP vector used here integrates into various sites in the nuclear DNA and becomes a stable part of the genome irrespective of gene silencing events. Further, the method used to detect GFP transgene by real-time PCR in this study allowed for amplification and product analysis in the same reaction vessel, thereby reducing the risk of contamination and false-positive results.
The data obtained in this immunocompetent large-animal model support previous observations in the mouse and the fetal sheep.15 16 In summary, MSCs distribute to a wide variety of tissues following systemic administration and may be capable of participating in ongoing cellular turnover and replacement within an engrafted organ. Immunohistochemical and in situ hybridization techniques, currently being tested by our group, will be required to confirm this hypothesis.
Prepublished online as Blood First Edition Paper, December 12, 2002; DOI 10.1182/blood-2002-06-1830.
Supported by research funding from Osiris Therapeutics, Baltimore, MD (S.M.D., R.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Steven M. Devine, Division of Bone Marrow Transplantation, Leukemia, and Stem Cell Biology, Washington University School of Medicine, 660 S Euclid, Campus Box 8007, St Louis, MO 63110; e-mail: sdevine@im.wustl.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal