Inappropriate expression of the multidrug resistance(MDR1) gene encoding the P-glycoprotein (Pgp) has been frequently implicated in resistance to different chemotherapeutic drugs. We have previously generated chronic myeloid leukemia (CML) cell lines resistant to the tyrosine kinase inhibitor imatinib mesylate (STI571), and one line (LAMA84-r) showed overexpression not only of the Bcr-Abl protein but also of Pgp. In the present study, we investigated this phenomenon in other cell lines overexpressing exclusively Pgp. Thus, cells from the K562/DOX line, described as resistant to doxorubicin due to MDR1 gene overexpression, grew continuously in the presence of 1 μM imatinib, but died in 4 to 5 days if the Pgp pump modulators verapamil or PSC833 were added to the imatinib-treated culture. Analysis of cell proliferation by the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay confirmed the differential sensitivity of K562/DOX to imatinib, which was also reversed by verapamil or PSC833. Flow cytometric analysis of the total phosphotyrosine content by intracytoplasmic staining after a 2-hour incubation with escalating doses of imatinib showed that the inhibitory concentrations of 50% (IC50) for inhibition of cellular protein tyrosine phosphorylation were 15, 10, and 5 μM for K562/DOX, K562/DOX plus verapamil, and K562, respectively. Retroviral-mediated transfection of theBCR-ABL+ AR230 cell line with theMDR1 gene decreased its sensitivity to imatinib, an effect that was also reversed by verapamil. The possible role of MDR overexpression in clinical resistance to imatinib remains to be defined. We therefore confirm that imatinib should be added to the extensive list of drugs that can be affected by the MDR phenomenon.
Introduction
Multidrug resistance (MDR) is a well-known phenomenon of cross-resistance of mammalian cells to a number of anticancer agents following exposure to one such drug.1 A diverse range of agents involved in MDR include alkaloid compounds and bacterial and fungal antibiotics, such as anthracyclines and etoposide. An accepted mechanism of MDR is a reduced cellular accumulation and an altered subcellular distribution of cytotoxic drugs. In many cases, this is mediated by increased expression at the cell surface of theMDR1 gene product, P-glycoprotein (Pgp), a 170-kDa energy-dependent efflux pump.2 3 MDR mediated by Pgp is thought to be primarily due to reduced intracellular concentrations and failure of the drug to reach its target.
The causative event in the initiation of chronic myeloid leukemia (CML) is the formation of the BCR-ABL oncogene, which codes for a constitutively active Bcr-Abl tyrosine kinase, on the Philadelphia (Ph) chromosome (22q−), which is itself the result of the t(9;22)(q34;q11) reciprocal translocation. This Ph chromosome and BCR-ABL oncoprotein are also found in about 25% of adults and 5% of children with acute lymphoblastic leukemia (ALL).4
STI571 or imatinib mesylate, a 2-phenylaminopyrimidine derivative, inhibits the Bcr-Abl tyrosine kinase with high selectivity.5 Preliminary results from 2 phase 1/2 clinical trials were recently reported.6,7 Of the 54 patients treated with at least 300 mg in chronic phase (CP), 98% showed hematologic and 53% cytogenetic responses, which have so far been largely durable. When given in advanced phase of CML or in Ph+ ALL, imatinib was able to induce hematologic responses, but these were nearly invariably followed by relapse, suggesting development of de novo resistance to the drug.7
In anticipation of drug resistance, we and other groups have isolatedBCR-ABL+ cell lines with differential sensitivity to imatinib.8-10 Our initial studies showed that in some cell lines amplification of the BCR-ABL gene plays a major role in the development of imatinib resistance. Furthermore, one resistant line showed overexpression not only of the Bcr-Abl protein but also of the MDR Pgp, indicating that at least 2 mechanisms of resistance operate in this cell line.10
In the current study, we have investigated further the role of Pgp in the resistance to imatinib. We show here that the K562/DOX cell line, which displays resistance to several drugs and overexpresses Pgp, also exhibits a reduced sensitivity to imatinib as compared to the parental K562 cells, and that this resistance can be modulated by blockers of the Pgp pump. We also show that retroviral-mediated introduction and overexpression of the MDR1 gene in AR230, another CML cell line, leads to resistance to imatinib, also reversible by Pgp inhibitors.
Materials and methods
Cell lines
All cell lines were grown in RPMI 1640 medium (Gibco, Paisley, United Kingdom) supplemented with penicillin, streptomycin,l-glutamine (Life Technologies, Grand Island, NY), 10% fetal bovine serum (FBS), herein referred to as RF-10. K562, HL60, and AR230 lines were purchased from cell repository banks or kindly donated by the originators.11 K562/DOX (a gift from J. P. Marie, INSERM, E9912, University of Paris 6, France) was obtained by in vitro passaging of K562 in progressively increasing doses of daunorubicin.12
Cell proliferation assay
Proliferation was assessed by the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) tetrazolium assay (Cell Titer96 Aqueous, Promega, Madison, WI), which measures the number of viable cells. Between 2 × 103 and 104 cells were washed twice in RF-10 and plated in quadruplicate into microtiter-plate wells in 100 μL RF-10 plus various doses of imatinib. Controls using the same concentrations of imatinib without cells were set up in parallel. MTS (20 μL) was added to the wells at daily intervals. Two hours after adding MTS the plates were read in a microplate autoreader (Dynex Technologies, Billingshurst, United Kingdom) at 490 nm wavelength. The results were expressed as the mean optical density (OD) of the 4-well set for each imatinib (Novartis, Basel, Switzerland) dose. All the experiments were repeated at least 3 times.
Cell viability assay
Cells were plated at a density of 2 to 2.5 × 105cells/mL in RF-10 with or without the inhibitor and, in some experiments, with or without 5 μg/mL verapamil (Isoptine, Laboratoires Knoll, Levallois-Perret, France) or 1 μM PSC833 (Novartis Pharma, Basel, Switzerland). Aliquots were taken out at 24-hour intervals for assessment of cell viability by trypan blue dye exclusion.
Apoptosis assay
For determination of apoptotic death the level of caspase-3 activation was assessed by measurement of its capacity to cleave an Ac-Asp-Glu-Val-Asp-amino-4-methyl-coumarine (Ac-DEVD) substrate, as modified from a previously published method.10 Aliquots of 7.5 × 104 cells were cultured in triplicate in RF-10 into 96-well plates in the presence or absence of 1 μM imatinib. Triplicate wells with RF-10 only were used as background control. After 3 days, the plate was centrifuged at 1500 rpm for 5 minutes, the culture supernatant was removed, and the cells were resuspended in 50 μL of a buffer containing 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 5 mM dithiothreitol (DTT), 2 mM EDTA (ethylenediaminetetraacetic acid), 0.02% saponin (Sigma Aldrich, St Quentin Fallavier, France), 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL pepstatin A, 10 μg/mL leupeptin, and 1.4 μM fluorogenic Ac-DEVD substrate (UBI Euromedex, Souffelweyerssheim, France). Development of the reaction was read on an automatic spectrofluorometer (Victor 2 Multilabel Counter, Wallac & Perkin Elmer, Akron, OH), using λexc = 380 nm and λem = 480 nm. After this reading, 200 μL of a 15-μg/mL propidium iodide solution was added to each well and a new reading was taken at λexc = 360 nm and λem = 600 nm, to evaluate DNA content in the wells. A calibration curve was performed using increasing amounts of cells per well and the cell number in the wells was calculated from this curve. The caspase-3 activity was calculated for 105 cells.
Western blot analysis
Protein lysates were prepared according to Kabarowski et al.13 Protein concentrations were determined by the Bradford method (DC Protein Assay, Bio-Rad, Hercules, CA). Approximately 250 μg protein was resolved on 7% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels, blotted onto polyvinylidene (PVDF) membranes (Immobilon-P, Millipore, Bedford, MA) by semidry electrophoretic transfer, probed with individual antibodies, and visualized by the enhanced chemiluminescence (ECL) system (Amersham, Little Chalfont, United Kingdom). The following antibodies were used: PY-99 antiphosphotyrosine (Santa Cruz Biotechnology, Santa Cruz, CA), Ab-3 anti-Abl (Calbiochem-Novabiochem, Nottingham, United Kingdom), antiphosphorylated STAT5 (Santa Cruz), and A-2066 antiactin (Sigma Chemical, St Louis, MO). The secondary antibodies were horseradish peroxidase (HRP)–conjugated rabbit antimouse IgG and swine antirabbit IgG (Dako, Glostrup, Denmark).
Flow cytometric analysis
A phycoerythrin (PE)–conjugated monoclonal antibody (MoAb) Fab′ fragment of a murine antihuman Pgp (clone UIC2; Immunotech, Marseille, France) was used to determine the expression of the MDR1gene product on the different cell lines. For detection of the tyrosine phosphorylated proteins, cells were fixed in 1% paraformaldehyde/phosphate-buffered saline (PBS) for 10 minutes at room temperature, permeabilized with 0.3% saponin, and stained with the PY-99 antiphosphotyrosine antibody followed by a fluorescein isothiocyanate (FITC)–conjugated goat antimouse IgG (Becton Dickinson, San Jose, CA) as the secondary reagent. Appropriate isotypic controls were used in all experiments. Stained cells were analyzed with an XL Coulter instrument (Coultronics, Margency, France).
Transduction of BCR-ABL+ cell line
The proviral plasmid pSF-MDR (kindly donated by Dr Christopher Baum, Heinrich-Pette-Institut, Hamburg, Germany) was transfected by calcium phosphate precipitation into the ecotropic packaging cell line ψCRE as described.14 Filtered supernatants from the ecotropic packaging line were then added to the amphotropic cell line ψ-CRIP in the presence of 4 μg/mL protamine sulfate. ψ-CRIP clones were selected in the presence of colchicine (20 ng/mL; Sigma), and titered using NIH3T3 fibroblasts as targets. The highest titer clone (105 virus particles/mL) was used in these experiments. Viral supernatant was harvested, filtered, and frozen in ready-to-use aliquots. The supernatant was tested for the presence of replication competent virus and confirmed to be free of helper virus. A 105 cell aliquot from the AR230 CML cell line in log-phase growth was exposed to viral supernatant at a multiplicity of infection of 5 in the presence of 4 μg/mL protamine sulfate. Nontransduced control cells were kept for the same length of time in RF-10 medium and protamine sulfate. Cells were selected and propagated in the presence of 10 ng/mL colchicine.
CFU assay of human imatinib-resistant cells
Peripheral blood or bone marrow cells from CML patients treated with imatinib in the Novartis protocols 0102 or 0115 (blast crisis) were obtained at the time of the relapse. None of these patients exhibited amplification of the BCR-ABL gene or point mutations in its tyrosine kinase domain that could account for the imatinib-resistant phenotype. Mononuclear cells (MNCs) were separated on Lymphoprep (Nycomed, Oslo, Norway) and plated at 5 × 104 cells/mL in Iscove methylcellulose medium (Methocult H4230 or H4330; Stem Cell Technologies, Meylan, France) supplemented with 20 ng/mL recombinant human interleukin-3 (rhIL-3), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-6, (Amgen, Thousand Oaks, CA), and 100 ng/mL Flt3 ligand (R & D Systems, Abingdon, Oxon, United Kingdom). All clonogenic assays were carried out in quadruplicate. Imatinib (1 μM) or PSC833 (1 μM) or both was added directly to the methylcellulose. Granulocyte-macrophage colony-forming units (CFU-GMs) or CFU-blast colonies were evaluated after 14 days of culture.
Results
The K562/DOX cell line overexpresses Pgp and is resistant to imatinib
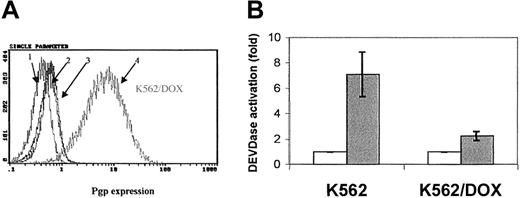
Fluorocytometric analysis confirmed that K562/DOX exhibited a significant overexpression of Pgp; more than 80% of cells showed a strong reaction with the anti-Pgp antibody, in contrast to only 2% of the parental K562 line (Figure 1A). There was no Bcr-Abl overexpression in K562/DOX as compared to the parental clone (data not shown).
Expression of the Pgp
MDR-1 gene product in K562 and K562/DOX. (A) Flow cytometric analysis of Pgp expression in K562 and K562/DOX: (1) isotypic control for K562/DOX; (2) isotypic control for K562; (3) Pgp for K562; (4) Pgp for K562/DOX. (B) Caspase-3 activity in 105 cells after 24 hours in culture without (■, control) or with (1 μM) imatinib (░). Results represent the mean ± SD of triplicate cultures. The difference between the 2 treated cultures is statistically significant (P = .004; Student t test).
Expression of the Pgp
MDR-1 gene product in K562 and K562/DOX. (A) Flow cytometric analysis of Pgp expression in K562 and K562/DOX: (1) isotypic control for K562/DOX; (2) isotypic control for K562; (3) Pgp for K562; (4) Pgp for K562/DOX. (B) Caspase-3 activity in 105 cells after 24 hours in culture without (■, control) or with (1 μM) imatinib (░). Results represent the mean ± SD of triplicate cultures. The difference between the 2 treated cultures is statistically significant (P = .004; Student t test).
A 24-hour exposure of the 2 cell lines to imatinib resulted in increased caspase-3 activity in K562 but not in K562/DOX (Figure 1B). This difference was also illustrated by the fact that less than 10% of the parental K562 cells survived a 4-day treatment with 1 μM imatinib, whereas the viability of the K562/DOX line was above 80% at the end of the same period of exposure to the inhibitor (Figure2A).
Growth characteristics of K562/DOX.
(A) Cell viability assessed by trypan blue exclusion of K562/DOX and the parental K562 cell line cultured in the presence or absence of 1 μM imatinib (STI) or 5 μg/mL verapamil (VP) or both. (B) Cell viability of K562/DOX and K562 in the presence or absence of 1 μM imatinib (STI) or 1 μM PSC833 or both.
Growth characteristics of K562/DOX.
(A) Cell viability assessed by trypan blue exclusion of K562/DOX and the parental K562 cell line cultured in the presence or absence of 1 μM imatinib (STI) or 5 μg/mL verapamil (VP) or both. (B) Cell viability of K562/DOX and K562 in the presence or absence of 1 μM imatinib (STI) or 1 μM PSC833 or both.
Resistance of K562/DOX to imatinib is reversed by verapamil or PSC833
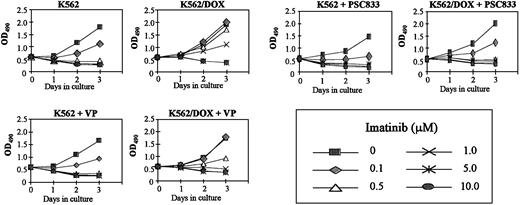
K562/DOX cells were incubated with verapamil or PSC833, 2 known inhibitors of the Pgp pump, and assessed for viability on exposure to imatinib. A clear enhancement of sensitivity to 1 μM imatinib was observed when either verapamil (5 μg/mL) or PSC833 (1 μM) was added to the culture (Figure 2). MTS cell proliferation assays were performed on cells exposed to 0.1, 0.5, 1, 5, and 10 μM imatinib (Figure3). On day 3 we evaluated the dose of imatinib necessary to induce a 50% decrease in the uptake of MTS (IP50), as measured by the OD index. Thus, the IP50 for K562 was 0.1 μM, whereas that for K562/DOX was 1 μM. This indicated that the concentration of imatinib needed for a 50% reduction in the number of viable cells after 3-day exposure to the compound was on average 10 times higher in the resistant as compared to the sensitive cells (Figure 3). This was, however, still lower than the respective concentration for 2 other lines resistant to imatinib, LAMA84-r (IP50 = 2 μM) and K562-r (IP50 = 3 μM).10 The resistance of K562/DOX to imatinib was partially reversed by simultaneous incubation with verapamil (IP50 = 0.5 μM), and completely reversed by PSC833 (IP50 = 0.1 μM).
Effect of Pgp inhibitors on cell proliferation.
Cell proliferation of K562/DOX cells, under the effect of various concentrations of imatinib with or without 5 μg/mL verapamil (VP) or 1 μM PSC833, as assessed by MTS uptake. Results are expressed as the mean OD490 of quadruplicate cultures, which is directly proportional to the number of viable cells.
Effect of Pgp inhibitors on cell proliferation.
Cell proliferation of K562/DOX cells, under the effect of various concentrations of imatinib with or without 5 μg/mL verapamil (VP) or 1 μM PSC833, as assessed by MTS uptake. Results are expressed as the mean OD490 of quadruplicate cultures, which is directly proportional to the number of viable cells.
Reduced inhibition of cellular tyrosine phosphorylation of K562/DOX by imatinib
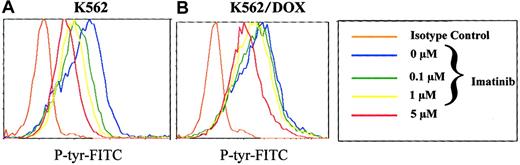
To analyze the effect of imatinib on the phosphorylation pattern of K562 and K562/DOX cells, we established a new assay using flow cytometry on permeabilized cells stained with a phosphotyrosine antibody. We therefore washed K652 and K562/DOX with RF-10, incubated the individual cell lines in graded concentrations of 0 to 10 mM imatinib for 2 hours, and analyzed their phosphotyrosine content by fluorocytometry (Figure 4). Phosphorylation of various proteins decreased in a dose-dependent manner in the sensitive and resistant clones (Figure 4). However, the doses of imatinib necessary to inhibit overall tyrosine phosphorylation were higher for K562/DOX than for K562. Estimation of the IC50 of the compound for inhibition of 50% of total cellular tyrosine phosphorylation from the fluorocytometric data showed that it was 5 μM for K562 and 15 μM for K562/DOX. When the latter cells were cotreated with verapamil, the IC50 was reduced to 10 μM, whereas no change was detected in the parental line on treatment with verapamil (data not shown).
Total phosphotyrosine content.
Total cellular phosphotyrosine content as detected by staining with the Py 99 antibody analyzed by flow cytometry after incubation of K562 (A) and K562/DOX (B) during 2 hours with different doses of imatinib (0, 0.1, 1, 5 μM).
Total phosphotyrosine content.
Total cellular phosphotyrosine content as detected by staining with the Py 99 antibody analyzed by flow cytometry after incubation of K562 (A) and K562/DOX (B) during 2 hours with different doses of imatinib (0, 0.1, 1, 5 μM).
K562/DOX requires higher imatinib concentrations for inhibition of STAT5 phosphorylation
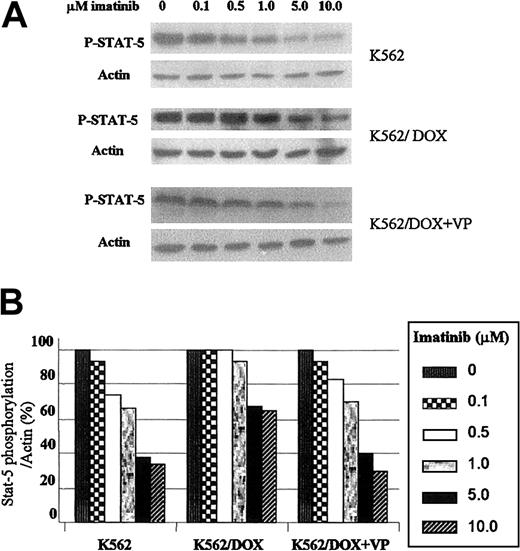
Because the transcription factor STAT5 is constitutively tyrosine phosphorylated and activated after transformation of hematopoietic cells by p210Bcr/Abl, we have studied the effect of imatinib on this target protein.15 The 2 K562 cell lines were incubated with graded concentrations of imatinib during 2 hours and immunoblotted with an antibody that recognizes the phosphorylated isoform of STAT5. As shown in Figure 5A, the concentration of imatinib necessary to inhibit STAT5 phosphorylation was higher in K562/DOX as compared to K562. Thus, when exposed to 5 μM imatinib the level of STAT5 phosphorylation in K562 was 38% of that observed in the control (nonexposed) cells; in contrast, under the same conditions, the amount of phospho-STAT5 in K562/DOX cells was 67% of the control and decreased to 40% after the addition of verapamil (Figure 5B).
Effect of imatinib on STAT5 phosphorylation in K562 and K562/DOX.
(A) Cells were incubated during 2 hours with escalating doses of imatinib with or without verapamil (VP) and total protein lysates were immunoblotted with an antibody against phosphorylated STAT5 (P-STAT-5). (B) Percentage of STAT5 phosphorylation in the presence of different concentrations of imatinib relative to the control (0 μM) cultures, as calculated by the phopho-STAT5/actin densitometric ratio of the respective bands.
Effect of imatinib on STAT5 phosphorylation in K562 and K562/DOX.
(A) Cells were incubated during 2 hours with escalating doses of imatinib with or without verapamil (VP) and total protein lysates were immunoblotted with an antibody against phosphorylated STAT5 (P-STAT-5). (B) Percentage of STAT5 phosphorylation in the presence of different concentrations of imatinib relative to the control (0 μM) cultures, as calculated by the phopho-STAT5/actin densitometric ratio of the respective bands.
Ectopic overexpression of Pgp in a CML cell line leads to resistance to imatinib
To demonstrate a direct causal relationship between Pgp overexpression and resistance to imatinib, we infected AR230, aBCR-ABL+ CML cell line with a retrovirus carrying the MDR1 gene. Transduced cells were selected with colchicine and tested by flow cytometry for the expression of Bcr-Abl and Pgp, in parallel with control, nontransfected cells (Figure6A). The MDR1-transduced cells showed significant overexpression of Pgp in comparison with the negligible, baseline level of expression in the parental, nontransfected line. As expected, expression of the endogenous Bcr-Abl protein was unaffected by the transduction procedure (data not shown).
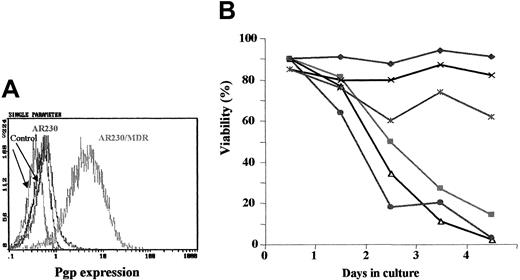
Effect of imatinib on
MDR1-transduced AR230 cell line. (A) Flow cytometric analysis of Pgp expression on AR230 and AR230/MDR cells labeled with a PE-conjugated Pgp antibody or a PE-conjugated isotype control. (B) Cell viability of AR230 and AR230/MDR in the presence or absence of 1 μM imatinib and/or 5 μg/mL verapamil (VP): AR230 (♦); AR230/MDR (×); AR230/MDR + imatinib (∗); AR230 + imatinib (▪); AR230/MDR + imatinib + VP (●); AR230 + imatinib + VP (▵).
Effect of imatinib on
MDR1-transduced AR230 cell line. (A) Flow cytometric analysis of Pgp expression on AR230 and AR230/MDR cells labeled with a PE-conjugated Pgp antibody or a PE-conjugated isotype control. (B) Cell viability of AR230 and AR230/MDR in the presence or absence of 1 μM imatinib and/or 5 μg/mL verapamil (VP): AR230 (♦); AR230/MDR (×); AR230/MDR + imatinib (∗); AR230 + imatinib (▪); AR230/MDR + imatinib + VP (●); AR230 + imatinib + VP (▵).
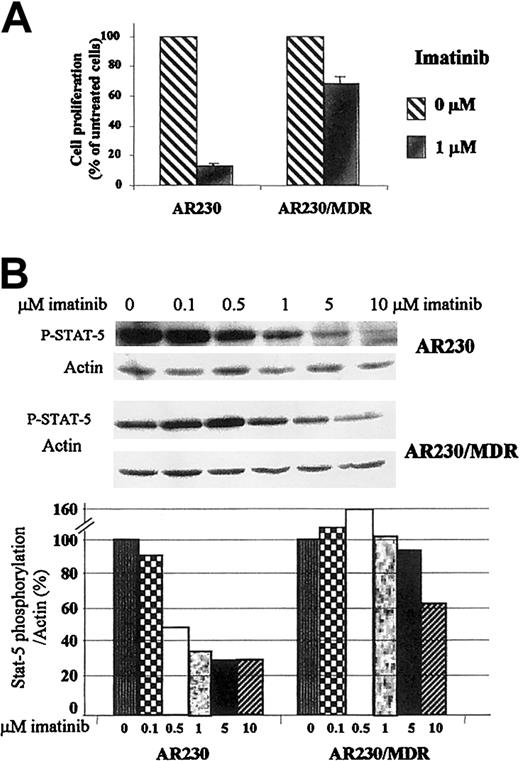
The effect of imatinib on the viability of theMDR1-transduced AR230 cell line was compared to that of the parental AR230 cells. Whereas the latter died within 4 days of exposure to 0.5 μM imatinib, the MDR1+ cells were still 65% viable at the end of the same period (Figure 6B). This longer survival was, however, abrogated when verapamil was also included in the imatinib culture. The viability of these cell lines on exposure to DOX was also examined, with similar results: 70% of the AR230/MDR cells were alive after a 3-day exposure to 100 ng/mL DOX but none of the parental AR230 cells survived this treatment. Analysis of proliferation after 3 days in 1 μM imatinib showed that the growth of the nontransfected AR230 cells was significantly reduced to 13% of the control (untreated cells); in contrast, proliferation of theMDR1-transduced line was reduced to only 70% of the control (Figure 7A). Finally the concentration of imatinib necessary to inhibit STAT5 phosphorylation was significantly higher in AR230/MDR as compared to the parental AR230 cells (Figure7B).
Effect of imatinib on the proliferation and phosphorylation of
MDR1-transduced AR230 cell line. (A) Proliferation of AR230 and AR230/MDR cells after 3 days of culture in the presence or absence of 1 μM imatinib, as assessed by MTS uptake. (B) STAT5 phosphorylation assessed by Western blot after a 2-hour incubation with imatinib. The bottom panel shows the percentage of STAT5 phosphorylation in the presence of different concentrations of imatinib relative to the control (0 μM) cultures, as calculated by the phospho-STAT5/actin densitometric ratio of the respective bands.
Effect of imatinib on the proliferation and phosphorylation of
MDR1-transduced AR230 cell line. (A) Proliferation of AR230 and AR230/MDR cells after 3 days of culture in the presence or absence of 1 μM imatinib, as assessed by MTS uptake. (B) STAT5 phosphorylation assessed by Western blot after a 2-hour incubation with imatinib. The bottom panel shows the percentage of STAT5 phosphorylation in the presence of different concentrations of imatinib relative to the control (0 μM) cultures, as calculated by the phospho-STAT5/actin densitometric ratio of the respective bands.
An inhibitor of the Pgp pump reduces imatinib resistance of primary CML cells
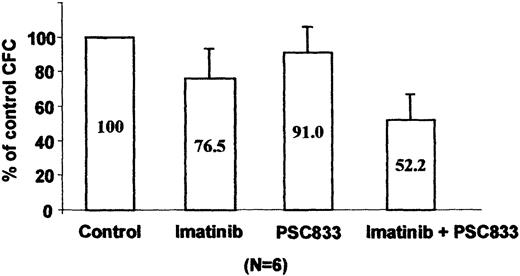
We tested blast cells from 6 patients with CML in blast crisis clinically resistant to imatinib for Pgp expression by cytometry analysis. No overexpression of MDR-1 was detected using UIC2 antibody staining. Because an association between early progenitors and functional MDR-1 has been reported,16 we tested the combination of imatinib and the PSC833 pump inhibitor in a CFU assay using total MNCs from these imatinib-resistant patients. As shown on Figure 8, 1 μM imatinib induced a weak inhibition of CFU formation (77%) as compared to the clonogenicity of control untreated cells. However, addition of 1 μM PSC833 to the imatinib-treated cultures led to a significant decrease in colony formation (52%, P = .001; Student t test), whereas PSC833 alone had no effect on the CFU growth (91%). Taken together, these functional results suggest that Pgp overexpression could contribute to acquired imatinib resistance in the primary leukemic cells, even though increased levels of the protein are not demonstrable by conventional immunostaining methods.
Effect of imatinib and PSC833 on imatinib-resistant primary CML cells.
Progenitor cell colony formation after 14 days in quadruplicate methylcellulose cultures in the presence or absence of 1 μM imatinib or 1 μM PSC833 or both. Results represent the mean ± SE of the average colony counts in samples from 6 patients with CML and are expressed as a percentage of the control, untreated cultures.
Effect of imatinib and PSC833 on imatinib-resistant primary CML cells.
Progenitor cell colony formation after 14 days in quadruplicate methylcellulose cultures in the presence or absence of 1 μM imatinib or 1 μM PSC833 or both. Results represent the mean ± SE of the average colony counts in samples from 6 patients with CML and are expressed as a percentage of the control, untreated cultures.
Discussion
The results on the first clinical trials with imatinib have suggested that this drug has the potential for long-term if not indefinite control of chronic phase CML.17 The same cannot be said for more advanced stages of the disease, where patients nearly always experience an initial clinical response to the inhibitor but subsequently lose it relatively early.7,18 Studies on imatinib-resistant CML cell lines suggested that BCR-ABLamplification and overexpression might be the most common mechanism by which the leukemic cells evade the action of imatinib.9,10This together with specific mutations in the adenosine triphosphate (ATP)–binding region of the Bcr-Abl tyrosine kinase are, indeed, being progressively identified in a variable proportion of CML patients who become refractory to imatinib treatment.19-23
It appears, however, that other single or multiple mechanisms must operate in a substantial proportion of patients. Among these, MDR is probably one of the most likely candidates, because it has already been shown to participate in the phenomenon of imatinib resistance of at least one cell line, that is, LAMA84-r.10 In this line, however, the contribution of excess Pgp to the overall resistant phenotype was difficult to evaluate because the cells also overexpress Bcr-Abl. In the present study we therefore analyzed the effect of theMDR1 gene in cell lines where its overexpression is independent of previous exposure to imatinib. In one of the lines (K562/DOX), Pgp-overproducing cells were selected in response to anthracycline exposure, whereas in the other (AR230/MDR), ectopic overexpression of the pump protein was obtained by retroviral-mediatedMDR1 gene transfer. Despite the difference in the origin of their MDR1 overexpression, both lines were markedly resistant to imatinib, as shown by their sustained viability and proliferation, reduced caspase-3 activity, unaltered tyrosine phosphorylation profile, and elevated STAT5 activity when exposed to the inhibitor. Furthermore, the imatinib-resistant phenotype could be significantly alleviated by Pgp pump blockers, in particular PSC833, previously shown to be more potent than verapamil.24 25
It is interesting to observe the variable degree of resistance to imatinib in CML cell lines with different mechanisms of resistance. Thus, by using the imatinib IP50 as defined, we demonstrated that K562/DOX, whose only mechanism of resistance is MDR1 overexpression, is less resistant to imatinib than LAMA84-r, which overexpresses both MDR1 and BCR-ABL and this, in turn, is less resistant than K562-r whose mechanism of resistance is still unknown.10
The possible role of Pgp overexpression in the acquired in vivo resistance of primary CML cells to the kinase inhibitor is less well defined. In none of the 6 patients who lost their initial response to the drug could MDR1 up-regulation be detected on the relapse specimens. However, addition of a pump blocker to imatinib was able to significantly increase its capacity to inhibit CFU-GM or CFU-blast (or both) proliferation, suggesting though not proving, that a pump-mediated drug resistance effect might be operating in these progenitors. In none of these patients did we observe amplification of the BCR-ABL gene or a point mutation in the tyrosine kinase domain.19 The apparent paradox may be explained by the known difficulty in accurately measuring relatively low levels of Pgp in clinical samples, as largely documented, for instance, in acute myeloid leukemia.26 Despite this problem, various workshops devoted to this phenomenon have concluded that even a weak Pgp activity may lead to an MDR phenotype.27-29 In this context, it should be noted that whereas development of imatinib resistance is the rule rather than the exception in blast crisis of CML, in chronic phase it has been only rarely documented so far. This may be related, at least in part, to the relatively increased basal level of MDR1 gene expression in blast crisis as compared to chronic phase cells, as reported by some groups.30,31 It is also possible that the apparent pump-mediated imatinib resistance in CML blasts is due to overexpression of an MDR gene other than MDR1.32
The practical implications of our study are manifold. In the first instance, it is evident that BCR-ABL+ leukemic cells that develop MDR-1 up-regulation by previous induction with another drug (eg, anthracyclines) are likely to be cross-resistant to imatinib. In clinical terms, documentation of Pgp overexpression in a patient with CML in blast crisis might preclude prescription of imatinib as an alternative to the previous chemotherapy, because the tyrosine kinase inhibitor should also be excluded from the cell by the pump protein. Another concern is based on the fact that Pgp is physiologically expressed in normal hematopoietic stem cells for their own protection against accumulation of toxic metabolites.33 Being a stem cell disorder, CML leukemic progenitors are likely to express similar if not higher levels of Pgp and this could adversely affect their total elimination by imatinib. This has, in fact, been postulated as a possible mechanism to explain the inherent resistance to imatinib of very primitive, quiescent CML stem cells.34 Finally, a similar argument may be applicable to the possible refractoriness or early relapse while on imatinib therapy of solid tumors, many of which overexpressMDR1.35-38
We are very grateful to Dr J. P. Marie (INSERM, E9912, University Paris 6, France) for providing us the K562/DOX cell line and Dr S. Mizutani (National Children's Medical Research Center, Tokyo, Japan) for AR230; to Dr Elisabeth Buchdunger (Novartis, Basel, Switzerland) for imatinib and PSC833; and to Dr Christopher Baum (Heinrich-Pette-Institut, Hamburg, Germany) for the proviral plasmid pSF-MDR. We thank Dr Patrick Berthaud and Novartis Pharma (Rueil-Malmaison, France) for supporting this work.
Supported by grants from the Association pour la recherche sur le cancer, Fondation de la recherche medicale, and Novartis Pharma France (F.-X.M., J.R. and V.L.) and the Leukaemia Research Fund, United Kingdom (J.M.G. and J.V.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
François-Xavier Mahon, Laboratoire Greffe de Moelle UMR CNRS 5540, Université Victor Ségalen Bordeaux 2, 146 rue Léo Saignat, 33076 Bordeaux, France; e-mail:francois-xavier.mahon@umr5540.u-bordeaux2.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal