Previous studies have revealed that that approximately 10% to 15% of multiple myelomas (MMs) are characterized by a reciprocal t(4;14)(p16;q32) translocation that activates expression ofFGFR3 and creates an IGH/MMSET fusion transcript. Current data suggest that activation of FGFR3is the oncogenic consequence of this rearrangement. Using a combination of microarray profiling, reverse transcriptase–polymerase chain reaction (RT-PCR), and interphase fluorescence in situ hybridization (FISH), we show that 32 (18%) of 178 newly diagnosed cases of MM harbor the t(4;14)(p16;q32). Importantly, 32% of these cases lack expression of FGFR3, yet express MMSET and have an IGH/MMSET fusion transcript. Interphase FISH showed that whereas the IGH/MMSET fusion was present in more than 80% of the clonotypic plasma cells in these novel cases, there was typically a complete loss of one copy of FGFR3. These data indicate that the t(4;14)(p16;q32) and loss of FGFR3occurred at a very early stage and suggest that activation ofMMSET, not FGFR3, may be the critical transforming event of this recurrent translocation.
Introduction
Like many tumors of the B-cell lineage, multiple myeloma (MM) shows recurrent rearrangements of the immunoglobulin heavy-chain (IGH) locus at 14q32 and molecular studies have identified FGFR3, CCND1, CCND3, and MAF genes as targets of primary translocations in this malignancy.1 The multiple myeloma (MM) specific t(4;14)(p16;q32) not only results in the activation of FGFR3 but also the creation of a chimeric fusion transcript between IGH and MMSET. Given the transcription-activating nature of 14q32 translocations, microarray profiling of global gene expression has emerged as a powerful method for identifying IGH–associated rearrangements in B-cell malignancies. We have previously used this strategy to identify a novel 14q32 translocation involving the cyclin D3 gene (CCND3)2 as well as all known 14q32 translocations in MM.3 Using a combination of gene expression profiling reverse transcriptase–polymerase chain reaction (RT-PCR) and interphase fluorescence in situ hybridization (FISH) we present evidence that nearly 20% of newly diagnosed cases of MM harbor the t(4;14)(p16;q32), and approximately 32% of these cases, while expressing the IGH/MMSET fusion transcript, lackFGFR3 expression.
Study design
Cell procurement, cell purification, and RNA isolation
Bone marrow (BM) aspirates from 178 subjects with newly diagnosed MM were taken after obtaining written informed consent in keeping with institutional policies. Plasma cell (PC) isolation from the mononuclear cell fraction was performed by immunomagnetic bead selection with monoclonal mouse anti–human CD138 antibodies as described.3 PC purity of more than 90% was confirmed by 2-color flow cytometry using CD138+/CD45− and CD38+/CD45− antibodies (Becton Dickinson, San Jose, CA), immunocytochemistry for cytoplasmic light chain immunoglobulin, and morphology by Wright-Giemsa staining on all samples.
Conversion of total RNA to labeled cRNA and hybridization to microarray
Total RNA isolation, preparation of labeled cRNA, and hybridizations to U95Av2 microarrays were performed as described.3 Data were analyzed and signal derived using Analysis Suite 5.01 (Affymetrix, Santa Clara, CA).
Interphase FISH analysis of t(4;14)(p16;q32)
Disassociation of the MMSET and FGFR3 andVH and CH signals and fusion of the VHand MMSET and CH and FGFR3 signals were performed essentially as described.3,4 TheVH and CH IGH probes have been described.2 The MMSET (RPCI11-262P20) andFGFR3 (RPCI11-20I20) probes were obtained by hybridization screening (BAC/PAC Resources, Oakland, CA) and mapped to 4p16 using normal human bone marrow lymphocyte metaphases. All probes were labeled by nick translation in the presence of directly conjugated Spectrum-red dUTP (Vysis, Downers Grove, IL) in the case of RPCI11-262P20 and VH or Spectrum-green dUTP (Vysis) in the case of RPCI11-20I20 andCH. A total of 100 clonotypic PCs, identified by positive staining with AMCA (7-amino-4-methylcoumarin-3-acetic acid)–conjugated anti-kappa or anti-lambda immunoglobulin light chain antibody (Vector Laboratories, Burlingame, CA), were counted for abnormal signal patterns using a BX-60 epifluoresence microscope and images captured using a Vysis Genetics Workstation (Vysis). Probe disassociation or fusion in more than 10% of clonotypic PCs was evidence of rearrangement. FISH on 22 cases with both FGFR3and MMSET spikes showed VH/MMSET and FGFR3/CH fusions in more than 80% of clonotypic cells in all cases (data not shown).
RT-PCR
Reverse transcriptase–polymerase chain reactions (RT-PCRs) were performed as described3 using primers IGJH2 (5′-CAATGGTCACCGTCTCTTCA-3′) or JH1 (5′-CCCTGGTCACCGTCTCCTCA-3′) and MMSET (5′-CCTCAATTTCCTGAAATTGGTT-3′).
Results and discussion
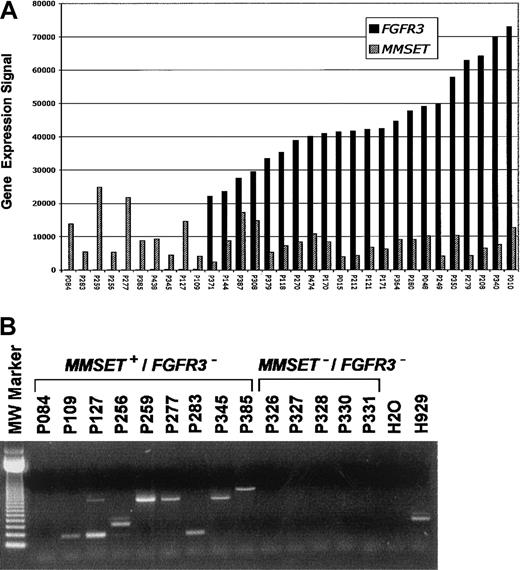
Expression of both MMSET and FGFR3 is low or undetectable in normal bone marrow PCs and a majority of MMs3. A total of 22 of 178 (12.3%) newly diagnosed cases of MM had spiked expression of FGFR3 and MMSETand an additional 10 of 178 (5.6%) had elevated expression of onlyMMSET (Figure 1A). TheIGH-MMSET fusion transcript was present in all cases with spikes for both MMSET and FGFR3 (data not shown) and 8 of 9 cases with only MMSET spikes (Figure 1B). Five cases lacking spikes of either gene also lacked the fusion transcript (Figure 1B).
FGFR3 and/or MMSET activation in a subset of MM is associated with the presence of the t(4;14)(p16;q32).
(A) A bar graph of normalized mRNA expression levels forFGFR3 (▪) and/or MMSET (▨) from CD138-enriched PCs from 32 MM cases. The gene expression signal (a quantitative value of gene expression derived from fluorescence intensity of cRNA hybridization derived from Affymetrix microarray hybridization) is on the vertical axis and samples are on the horizontal axis. The samples are ordered from left to right based on increasing level of expression of FGFR3. (B) A total of 14 MM samples, 9 with MMSET expression but lackingFGFR3 (MMSET+/FGFR3−) and 5 lacking expression of both, (MMSET−/FGFR3−) as observed in microarray experiments, were analyzed for the presence of the t(4;14)(p16;q32) translocation–specific IGHJH2/FMMSET fusion transcript. There were 8 MMSET+/FGFR3− samples positive for the fusion transcript. The size of the PCR product is dependent on the breakpoint position. One MMSET+/FGFR3− case, (P084), negative for both JH1 and JH2 fusion transcripts, was confirmed to have the t(4;14)(p16;q32) by FISH. All 5 MMSET−/FGFR3−cases were negative for the fusion transcript. Previous studies have shown that FGFR3 spike cases exhibit fusion transcripts.3 A negative control (H2O) and positive control (human MM cell line H929) are located to the right. The 123–base pair DNA molecular-weight size standard is to the left.
FGFR3 and/or MMSET activation in a subset of MM is associated with the presence of the t(4;14)(p16;q32).
(A) A bar graph of normalized mRNA expression levels forFGFR3 (▪) and/or MMSET (▨) from CD138-enriched PCs from 32 MM cases. The gene expression signal (a quantitative value of gene expression derived from fluorescence intensity of cRNA hybridization derived from Affymetrix microarray hybridization) is on the vertical axis and samples are on the horizontal axis. The samples are ordered from left to right based on increasing level of expression of FGFR3. (B) A total of 14 MM samples, 9 with MMSET expression but lackingFGFR3 (MMSET+/FGFR3−) and 5 lacking expression of both, (MMSET−/FGFR3−) as observed in microarray experiments, were analyzed for the presence of the t(4;14)(p16;q32) translocation–specific IGHJH2/FMMSET fusion transcript. There were 8 MMSET+/FGFR3− samples positive for the fusion transcript. The size of the PCR product is dependent on the breakpoint position. One MMSET+/FGFR3− case, (P084), negative for both JH1 and JH2 fusion transcripts, was confirmed to have the t(4;14)(p16;q32) by FISH. All 5 MMSET−/FGFR3−cases were negative for the fusion transcript. Previous studies have shown that FGFR3 spike cases exhibit fusion transcripts.3 A negative control (H2O) and positive control (human MM cell line H929) are located to the right. The 123–base pair DNA molecular-weight size standard is to the left.
To investigate the molecular cytogenetics of the 10 MMSET+/FGFR3− cases, we performed triple-color interphase FISH using 4 different probe combinations:MMSET/FGFR3, VH/CH, MMSET/VH, andFGFR3/CH (Table 1). Disassociation of the MMSET from FGFR3 and theVH from CH probes was observed in more than 65% of the clonotypic PCs (cPCs) of all 10MMSET+/FGFR3− samples (Table 1). Importantly, FGFR3 was deleted in more than 90% of cPCs in 5 and more than 80% in 7 of 10 patients. MMSET andVH colocalized in more than 90% of cPCs in all 10 cases. However, colocalization of FGFR3 and CH was seen in only 7 of 10 patients. Three samples with 0%, 4%, and 7% of cPCs harboring the FGFR3/CH fusions were deemed to lack the der14 chromosome. There were 2 samples with 10% and one with 14% that were considered boarderline positive, but also having clear evidence of loss of the der14 chromosome. In 4 cases, 25%, 41%, 71%, and 87% of the cPCs had fusion signals.
Expression and FISH data from MM with MMSET,but not FGFR3, activation
| PT . | FGFR3 signal . | MMSETsignal . | FGFR3/MMSET split . | VH/CH split . | MMSET/VH fusion . | FGFR3/CH fusion . | FGFR3 DEL . | CH DEL . |
|---|---|---|---|---|---|---|---|---|
| P345 | 117 | 4 420 | 92 | 85 | 88 | 87 | 17 | 7 |
| P109 | 211 | 4 098 | 97 | 91 | 100 | 25 | 81 | 95 |
| P385 | 91 | 8 830 | 100 | 95 | 89 | 41 | 14 | 0 |
| P277 | 68 | 21 686 | 92 | 100 | 93 | 4 | 84 | 67 |
| P127 | 148 | 14 564 | 100 | 98 | 100 | 7 | 100 | 0 |
| P084 | 28 | 13 846 | 96 | 66 | 100 | 10 | 90 | 80 |
| P256 | 61 | 5 338 | 100 | 79 | 86 | 10 | 91 | 96 |
| P438 | 101 | 9 266 | 91 | 87 | 86 | 71 | 6 | 12 |
| P259 | 53 | 24 885 | 100 | 98 | 96 | 0 | 94 | 4 |
| P283 | 38 | 5 372 | 100 | 97 | 77 | 14 | 92 | 11 |
| PT . | FGFR3 signal . | MMSETsignal . | FGFR3/MMSET split . | VH/CH split . | MMSET/VH fusion . | FGFR3/CH fusion . | FGFR3 DEL . | CH DEL . |
|---|---|---|---|---|---|---|---|---|
| P345 | 117 | 4 420 | 92 | 85 | 88 | 87 | 17 | 7 |
| P109 | 211 | 4 098 | 97 | 91 | 100 | 25 | 81 | 95 |
| P385 | 91 | 8 830 | 100 | 95 | 89 | 41 | 14 | 0 |
| P277 | 68 | 21 686 | 92 | 100 | 93 | 4 | 84 | 67 |
| P127 | 148 | 14 564 | 100 | 98 | 100 | 7 | 100 | 0 |
| P084 | 28 | 13 846 | 96 | 66 | 100 | 10 | 90 | 80 |
| P256 | 61 | 5 338 | 100 | 79 | 86 | 10 | 91 | 96 |
| P438 | 101 | 9 266 | 91 | 87 | 86 | 71 | 6 | 12 |
| P259 | 53 | 24 885 | 100 | 98 | 96 | 0 | 94 | 4 |
| P283 | 38 | 5 372 | 100 | 97 | 77 | 14 | 92 | 11 |
Signal values for FGFR3 (mean 208, range 39-536) andMMSET (mean 615, range 120-2673) in 33 normal bone marrow-derived PCs and FGFR3 (mean 163, range 18-3789) andMMSET (mean 691, range 298-1179) in the 156 MM cases lacking the t(4;14)(p16;q32) are provided for reference. Columns 4 through 7 represent the percentage of clonotypic plasma cells with either a “split” or “fusion” of the indicated probes in triple-color interphase FISH. Columns 8 and 9 represent a mean percentage of clonotypic plasma cells showing deletion of the FGFR3 or CH probe in the 2 FISH experiments depicted in columns 4 and 7 (FGFR3) and columns 5 and 7 (CH).
PT indicates patient identifier; FGFR3 and MMSETsignal, gene expression level; signal is a quantitative value of gene expression derived from Analysis Suite 5.01.
Data presented here suggest that the t(4;14)(p16;q32) translocation is present in 18% of newly diagnosed cases of MM, with 32% of these cases lacking expression of FGFR3. While all cases retained the der4 chromosome, loss of FGFR3 expression was linked with deletion of the der14 chromosome in virtually all cPCs in 4 cases. There were 3 cases that had deletion of FGFR3 in more than 92% of cPCs with no evidence of CH deletion, and 3 cases that showed little evidence of deletion of either FGFR3 or CH. Although a deletion mechanism could explain FGFR3 loss in most cases, unrecognized cryptic mechanisms are likely at work in cases with no gross abnormalities. Thus, although spikedFGFR3 expression in 13% of newly diagnosed cases of MM is consistent with the incidence of the t(4;14)(p16;q32) as detected by FISH,5,6 our data suggest that this number underestimates the true percentage of patients with this translocation. Thus, the poor survival attributed to the t(4;14)(p16;q32) by Moreau and colleagues would be more pronounced if this novel class of t(4;14)(p16;q32), lacking FGFR3 expression, were included.7 This possibility is strengthened in light of recent data showing that the t(4;14)(p16;q32) is an adverse prognostic factor irrespective ofFGFR3 expression.8
Given that a vast majority of the clonal PCs in the 10 MM cases with a t(4;14)(p16;q32) but lacking expression of FGFR3 had FISH evidence of disassociation of MMSET and FGFR3 and fusion of MMSET and VH suggests that the translocation was an early, if not an initiating, event. In addition, the deletion of FGFR3 in more than 90% of the clonal PCs in 5 cases suggests that FGFR3 loss also occurred at a very early stage of tumorigenesis. Since the t(4;14)(p16;q32) translocation has been observed in monoclonal gammopathy of undetermined significance (MGUS),5 9 it is possible that these novel cases represent MM cases that have converted from an MGUS, in which FGFR3was present and important for initiation and clonal expansion, but dispensable for progression. This hypothesis might hold more weight if the patients in this study had been previously treated and theFGFR3-positive cells selectively purged by chemotherapy, leaving only the FGFR3-negative cells. However, given that all the patients in this study were newly diagnosed and that virtually all the clonal PCs harbored FGFR3 deletion, this scenario seems unlikely. In conclusion, activation of MMSET, notFGFR3, may be the critical transforming event in myelomas harboring the t(4;14)(p16;q32) and consistent retention of the der4 chromosome suggests that MMSET may also be critical to maintaining the malignant phenotype.
Prepublished online as Blood First Edition Paper, November 14, 2002; DOI 10.1182/blood-2002-09-2801.
Supported through private funding and by grants CA55819 (B.B. and J.S.) and CA97513 (J.S.) from the National Cancer Institute, Bethesda, MD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John Shaughnessy Jr, Donna and Donald Lambert Laboratory of Myeloma Genetics, University of Arkansas for Medical Sciences, 4301 W Markham St, Slot 776, Little Rock, AR 72205; e-mail: shaughnessyjohn@uams.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal