The transforming growth factor-β (TGF-β) family of cytokines regulates vascular development and inflammatory responses. We have recently shown that exposure of human umbilical vein endothelial cells (HUVECs) to hypoxia (1% O2) increases gene expression and bioactivation of TGF-β2 and induces its downstream effectors, Smad proteins (Smads), to associate with DNA. In the present study, we show that hypoxia-induced TGF-β2 gene expression is dependent on thrombospondin-1–mediated bioactivation of latent TGF-β. Blocking TGF-β2 but not TGF-β1 in hypoxic endothelial cell cultures inhibited induction of the TGF-β2 gene, indicating that an autocrine mechanism driven by bioactivation of TGF-β2 leads to its gene expression in hypoxic HUVECs. Exposure of HUVECs to hypoxia resulted in phosphorylation and nuclear transportation of Smad2 and Smad3 proteins as well as stimulation of transcriptional activities of Smad3 and the transcription factor hypoxia-inducible factor-1α and culminated in up-regulation of TGF-β2 gene expression. Autocrine regulation of TGF-β2 production in hypoxia may involve cross-talk between Smad3 and HIF-1α signaling pathways, and could be an important mechanism by which endothelial cells respond to hypoxic stress.
Introduction
The transforming growth factor-β (TGF-β) family of growth factors mediates vascular development and regulates endothelial responses to mechanical, inflammatory, and hypoxic stress.1-10 The important role of TGF-β in vascular physiology is indicated by defective vasculogenesis and striking vascular inflammation leading to death in mice null for TGF-βs, their receptors, or their downstream substrates, the Smad proteins.3,7,11,12 We recently have shown that exposure of human umbilical vein endothelial cells (HUVECs) to hypoxia (1% O2) selectively up-regulates transcription and expression of TGF-β2 by as much as 20-fold and induces Smad2, Smad3, and Smad4 to associate with DNA.9
In vascular endothelium, TGF-β2, similar to TGF-β1 and TGF-β3, is produced in a latent form in which the bioactive, 25-kDa TGF-β dimer (mature TGF-β) is noncovalently bound to its propeptide (also known as latency-associated peptide [LAP]) and is unavailable for binding to TGF-β membrane receptors.1 An important physiologic regulator of TGF-β bioactivation is thrombospondin-1 (TSP-1), an extracellular matrix protein that is a member of the TSP family of glycoproteins.13-15 TSP-1, a trimer of disulfide-linked 180-kDa subunits, is secreted from platelet α–granules, endothelial cells, and vascular smooth muscle cells, and is deposited in extracellular matrix.16 Binding of TSP-1 to LAP occurs via amino acid sequence K412RFK415 of TSP-1 and amino acid sequence L54SKL57 of LAP,15,17 and potentially induces a conformational change in LAP that allows interaction of the 25-kDa mature TGF-β peptide with its specific membrane receptors. TSP-1 can activate LAPs associated with both latent TGF-β1 and -β2,15 and similarities reported between TGF-β1–null and TSP-1–null animals17 18 suggest that TSP-1–mediated TGF-β bioactivation is physiologically significant.
Mature TGF-β can bind to its type I, type II, and type III cell membrane receptors, the first 2 of which are serine/threonine kinases.19 Once activated by TGF-β, the type II receptor transphosphorylates the type I receptor, which then phosphorylates Smad2 or Smad3 (receptor-activated Smads [R-Smads]), which in turn heteromerize with Smad4 (Co-Smad) to translocate to the nucleus. Smad complexes accumulate in the nucleus, where they regulate gene transcription by recruiting transcriptional coactivators or inhibitors to DNA.19 This cross-talk created by the interplay between Smads and other signaling pathways is largely responsible for the diverse and context-specific effects of the TGF-β family of proteins.
The Smad signaling pathway was recently shown to interact with the transacting protein complex hypoxia-inducible factor-1 (HIF-1), which is a well-characterized transcription factor complex that regulates hypoxia-driven gene expression.20,21 HIF-1 binds DNA as a heterodimer of 2 basic helix-loop-helix proteins, HIF-1α and the aryl hydrocarbon receptor nuclear translocator (ARNT, or HIF-1β).22,23 Under normoxic conditions, HIF-1α is rapidly degraded by E3 ubiquitin ligase complex,24,25 whereas in hypoxia or in the presence of transition metals and iron chelators, it is stabilized and accumulates in the nucleus.25-30 Recent studies have shown that under normoxic conditions, HIF-1α is targeted for proteasomal degradation by the ubiquitination complex pVHL, the protein of the von Hippel–Lindau (VHL) tumor suppressor gene and a component of an E3 ubiquitin ligase complex. Hydroxylation of HIF-1α's proline residue(s) facilitates binding to VHL, which targets HIF-1α for degradation by E3 ligase.31,32 Under hypoxic conditions, HIF-1α's association with VHL is disrupted, resulting in inhibition of ubiquitination and proteasomal degradation, leading to accumulation of HIF-1α in hypoxic cells.29 30
In the present report, we show that exposure of endothelial cells to hypoxia results in (a) TSP-1–dependent bioactivation of TGF-β2; (b) activation of Smad2 and Smad3; and (c) an additive interplay between transcriptional activities of Smad3 and HIF-1α.
Materials and methods
Cell isolation and culture
Primary HUVECs were isolated from segments of normal-term cords by digestion with collagenase type II, and were pooled and cultured as previously described.33-35 Cells derived from the same primary culture were used for each set of hypoxic or nonhypoxic culture conditions in experiments performed in parallel. Each type of experiment was repeated at least twice, each time with different primary cell isolates unless otherwise indicated. For exposure to hypoxia, confluent HUVECs were placed in an incubator (NAPCO 7301; Precision Scientific, Chicago, IL) at 37°C in humidified 1% O2/5% CO2/94% N2; for exposure to nonhypoxic conditions, identically prepared cells were maintained in standard culture conditions of 37°C in humidified 5% CO2/95% room air as previously described.9
Reagents
Recombinant human TGF-β2, polyclonal rabbit anti–TGF-β2, monoclonal anti–TGF-β1, and isoform-specific controls for rabbit IgG and mouse IgG were purchased from R&D Systems (Minneapolis, MN). Monoclonal antibody 133 against stripped TSP-1; peptide LSKL, representing amino terminus TGF-β1 LAP amino acid residues 54 to 57; and scrambled peptide SLLK were prepared as described.12 14
Assay for bioactive TGF-β
Bioactive TGF-β levels were determined in HUVEC supernatants before and after inhibition of TSP-1–mediated TGF-β bioactivation. For this purpose, confluent HUVECs, cultured without serum, were exposed to normoxic or hypoxic conditions in the presence or absence of 10 μg/mL anti–TSP-1 monoclonal antibody 133, 28 μM LSKL peptide, 28 μM scrambled peptide SLLK, 10 μg/mL anti–TGF-β2 antibody, or 10 μg/mL mouse IgG. After 18 hours, supernatants were collected, and bioactive TGF-β levels were quantitated by the well-established mink lung epithelial cell (MLEC) bioassay36 within 2 hours of collection. TGF-β–responsive MLECs stably transfected with the expression construct p800neoLUC containing a truncated plasminogen activator inhibitor-1 promoter fused to the firefly luciferase reporter gene were kindly supplied by Dr Daniel B. Rifkin (New York University School of Medicine, New York, NY).37 Cells were maintained in Dulbecco modified Eagle medium (BioWhittaker, Walkersville, MD) containing 10% fetal calf serum, 2 mM l-glutamine, and antibiotics (penicillin G, 100 U/mL; streptomycin, 100 μg/mL; and geneticin, 250 μg/mL [Gibco BRL, Grand Island, NY]), and levels of bioactive TGF-β in HUVEC supernatants were determined as previously described.9,36 Briefly, MLECs were detached with trypsin, washed, plated at 1.6 × 105 cells per well in a 2-mL volume in 24-well tissue culture plates (Costar, Cambridge, MA), and allowed to attach for 18 hours at 37°C in 5% CO2. Medium in the wells was replaced by 2 mL of the following, placed in triplicate wells: control medium, control medium containing increasing concentrations of recombinant TGF-β2 to generate a standard curve, and 1:10 dilution of conditioned HUVEC medium to measure bioactive TGF-β. Incubations were continued for 18 hours at 37°C, and MLEC lysates from each well were prepared using reporter lysis buffer (Luciferase Assay System; Promega, Madison, WI). Luciferase activity was measured as relative light units (RLUs; model TD-20/20 luminometer, Promega), which were converted to TGF-β activity (picomoles) using the TGF-β2 standard curve.36Total protein content of supernatants was quantitated using Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA),38 and TGF-β2 levels obtained by the luciferase assay from each well were normalized to protein. Mean RLUs from triplicate wells (which were within 5%-7% of one another) were converted to picomoles of TGF-β. This bioassay is sensitive to all 3 isoforms of TGF-β, and because measurement of bioactive TGF-βs by enzyme-linked immunosorbent assay (ELISA) is not yet reliably accomplished by existing commercial kits, isoform specificity of bioactive TGF-β in our experiments was determined by measuring TGF-β in supernatants from HUVECs cultured in the presence or absence of neutralizing anti–TGF-β2 antibody as indicated.
RNase protection assay
After exposure to the indicated stimulus, cells were gently rinsed twice with ice-cold phosphate-buffered saline X1 (PBS) and scraped quickly into TRIzol (Gibco BRL) for isolation of total RNA followed by RNase protection analysis using the RiboQuant MultiProbe RNase protection assay system (PharMingen, San Diego, CA) as previously described.9 Quantification of mRNA was done from PhosphorImager (Molecular Dynamics, Sunnyvale, CA) analysis of radioactive bands using ImageQuant software (Molecular Dynamics). To control for differences in sample processing, hybridization signals in each sample were divided by the signal for the ribosomal protein mRNA (L32), which by independent analysis was found not to change with exposure to hypoxia (not shown). For determining fold increases in mRNA levels in hypoxic HUVECs compared with control, the TGF-β2/L32 signal ratio from treated HUVECs was divided by the ratio obtained from nonhypoxic HUVECs.
Immunoprecipitation and Western blot analysis
Cultures of endothelial cells were exposed to hypoxic or normoxic conditions for 10 minutes, 30 minutes, 1 hour, or 4 hours, or were exposed to 100 pM TGF-β2 for 30 minutes. At the end of the culture period, HUVECs were washed twice with ice-cold PBS and lysed using a Mammalian Cell Lysis Kit (Sigma, St Louis, MO) in the presence of additional protease inhibitors (Complete Mini Protease Inhibitor Cocktail Tablets; Roche Diagnostics, Indianapolis, IN). From each cell lysate, 1 mL (containing 400-500 μg protein) was subjected to immunoprecipitation with goat anti–Smad2 IgG (10 μg/mL; Santa Cruz Biotechnology, Santa Cruz, CA) or 10 μg/mL goat anti–Smad3 IgG (Santa Cruz Biotechnology) for 16 hours at 4°C with mixing, followed by adsorption to protein G–Sepharose beads (Santa Cruz Biotechnology). Immunoprecipitates were eluted from beads by boiling, separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidenedifluoride membranes (Immobilon-P; Millipore, Bedford, MA). Nonspecific binding was blocked by 5% nonfat milk powder in PBS. Immunoblotting was performed by using 2 μg/mL each of rabbit antiphosphoserine (Zymed Laboratories, South San Francisco, CA) or rabbit anti–phospho-Smad2 (Upstate Biotechnology, Lake Placid, NY), and rabbit anti–Smad2 IgG or rabbit anti–Smad3 IgG (both from Zymed Laboratories). Horseradish peroxidase–linked donkey anti–rabbit IgG F(ab′)2(Amersham Pharmacia Biotech, Piscataway, NJ) was used as secondary antibody at 1:4000 dilution. Immunodetection was carried out by enhanced chemiluminescence (ECL Plus Western blotting detection system; Amersham Pharmacia Biotech) and autoradiography. Autoradiograms were scanned and bands were quantified by ImageQuant software. Fold increases in Smad2 and Smad3 phosphorylation were calculated by dividing the phospho-Smad2 or phosphoserine values by their respective total Smad2 or Smad3 values.
Cellular immunofluorescence imaging
For assessing intracellular localization of Smad2 and Smad3, HUVECs were seeded into gelatin-coated 8-well Permanox chamber slides (Lab-Tek Chamber Slide System; Nalge Nunc International, Naperville, IL) in triplicate and exposed to hypoxic or nonhypoxic conditions for one hour, after which cells were washed with ice-cold PBS, fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 0.4% fetal calf serum. Incubation for 1 hour with primary antibodies was done using 0.2 μg/mL rabbit anti–human Smad2 IgG (Zymed Laboratories) or goat anti–human Smad3 IgG (Santa Cruz Biotechnology) followed by the secondary antibodies, which were, respectively, fluorescein isothiocyanate–conjugated donkey anti–goat IgG (3 μg/mL) or rhodamine-conjugated donkey anti–goat IgG (3 μg/mL; The Jackson Immunoresearch Laboratories, West Grove, PA) as previously described.39 Negative controls included secondary antibody alone or replacement of primary antibody with rabbit IgG or goat IgG. To ensure translocation of Smads from cytoplasm to the nucleus, cells were also cultured in the presence or absence of 100 pM recombinant human TGF-β2 (R&D Systems) for one hour. Immunofluorescence microscopy was performed with a Radiance 2000 confocal laser scanning microscope (Bio-Rad, San Francisco, CA) attached to an Axioskop 2 microscope (Carl Zeiss Microimaging, Thornwood, NY). Quantitation was performed by scoring 300 HUVECs/slide as showing predominantly nuclear or cytoplasmic immunofluorescence. Images of 512 × 512 pixels were obtained and processed using Adobe Photoshop 6.0 (Adobe Systems, Mountain View, CA).
Plasmid constructs
Plasmid construct pB2-77, containing a deletion mutant of the human TGF-β2 gene promoter (−77 to +63 base pair [bp]) linked to a chloramphenicol acetyltransferase (CAT) gene, was generously donated by Dr S.-J. Kim (National Cancer Institute, Bethesda, MD).40Expression vectors for Smad3 and Smad4 that contain the FLAG epitope tag at the amino domain have been described previously41 and were kindly supplied by Drs Rik Derynck and Ying Zhang (University of California, San Francisco, CA). The p3TP-Lux construct (the generous gift of Dr Joan Massagué, Memorial Sloan-Kettering Cancer Center, New York, NY) is a well-described artificial promoter construct that was designed to have maximal responsiveness to TGF-β.42 Expression plasmid encoding human HIF-1α was generously donated by Dr Steven McKnight (University of Texas Southwestern Medical Center, Dallas, TX).43 The HIF-1α–responsive EPO-luciferase construct (EPO-Lux), which has been previously described,24 44 is the hypoxia-responsive enhancer from the human erythropoietin gene (150-bp ApaI/PstI fragment) and was the generous gift of Dr J. Caro (Jefferson Medical College, Philadelphia, PA).
Transfection of recombinant plasmids and reporter gene assays
For transient transfections, plasmids with indicated expression vectors or corresponding empty constructs were introduced by electroporation into HepG2 cells or early-passage (second or third) HUVECs that were 70% confluent, as previously described.9Plasmid cytomegalovirus β-galactosidase (pCMVβ-gal; 0.5 μg; Promega) was included in each transfection as internal control. When multiple plasmids were cotransfected, the total amount of DNA was kept constant by supplementing the samples with empty expression vectors. To ensure that the same amount of protein was analyzed, protein concentration of each sample was determined by Bradford assay, and transfection efficiency was determined by β-gal activity.CAT/β-gal/protein or RLU/β-gal/protein ratios for hypoxic and nonhypoxic transfectants were compared to determine the fold change in response to hypoxia, as previously described.9
Quantitative polymerase chain reaction (PCR)
Fluorogenic 5′ nuclease quantitative RT-PCR was performed as described.45 Total RNA was extracted from A549 and human breast carcinoma cell line MDA-MB-468, and reverse transcription of 2 μg total RNA to cDNA was done with a RETROscript kit (Ambion, Austin, TX). The primer and probe sequences for TGF-β2, designed with Primer Express 2.0 software (Applied Biosystems, Foster City, CA), were as follows: forward, 5′-GACCAACCGGCGGAAGA-3′; reverse, 5′-CAGCAATTATCCTGCACATTTCTAA-3′; probe, 6FAMCGTGCTTTGGATGCGGCCTATTGTAMRA. The primers and probes for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Applied Biosystems. Agarose gel electrophoresis was used to verify that the PCR products corresponded to the size predicted for the amplified fragment. Standard curves of TGF-β2 and GAPDH (not shown) were prepared with first-strand cDNA, and the mean amount of TGF-β2 obtained from triplicate samples was normalized to GAPDH RNA for the same sample.
Statistical analysis
Student t test, 2-tailed, was used to assess differences between data from hypoxic and normoxic culture conditions. Means are presented ± 1 SD. Significance level was set atP ≤ .05.
Results
We previously have shown that exposure of HUVECs to hypoxia stimulates TGF-β2 gene expression at a transcriptional level and causes a parallel induction in TGF-β production and bioactivation in these cells.9 Since the TGF-β family of proteins can regulate their own transcription and synthesis,40 it is possible that hypoxia-driven up-regulation of TGF-β2 gene expression is part of an autocrine pathway triggered by hypoxia-induced bioactivation of the TGF-β ligand. Since transcript and protein levels of TSP-1 have been shown to be increased in hypoxic endothelial cells in a time course parallel to hypoxic induction of the TGF-β2 gene,46 we tested whether hypoxia-induced bioactivation of TGF-β was TSP-1–dependent.
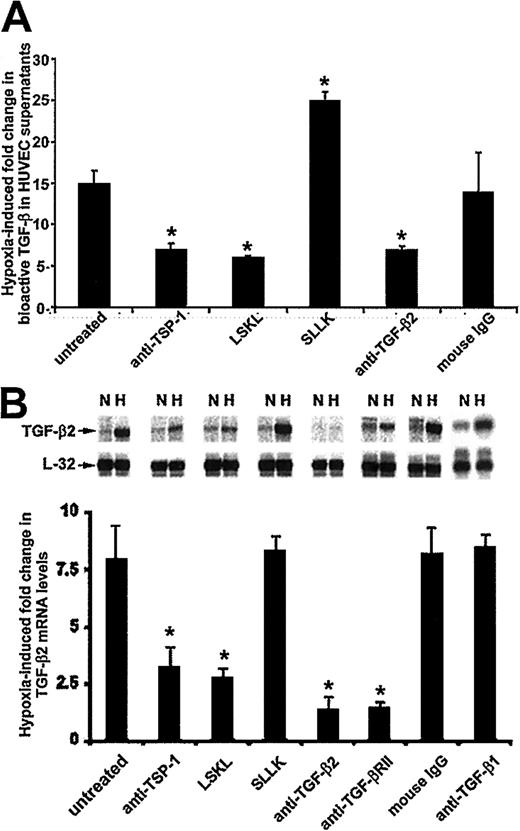
As shown in Figure 1A, the level of bioactive TGF-β was 15.0-fold ± 1.0-fold higher in supernatants from hypoxic HUVECs compared with supernatants from nonhypoxic cells (P < .05). In contrast, exposure of HUVECs to hypoxia in the presence of either anti–TSP-1 antibody or the peptide LSKL, both of which are known inhibitors of TGF-β activation,14reduced bioactive TGF-β levels in supernatants by 54.0% ± 5.8% and 60.7% ± 1.3%, respectively (P < .05 for both conditions), compared with the increase observed in control (untreated) hypoxic HUVECs (leftmost bar). Neutralization of TGF-β2 by exposure of HUVECs to hypoxia in the presence of specific antibody caused an equivalent (54% ± 2.6%; P < .05) inhibition in bioactive TGF-β levels compared with supernatants from control untreated HUVECs, whereas there was no change in supernatants from HUVECs treated with mouse IgG (P < .05 in both conditions; Figure 1A). This latter observation is consistent with the possibility that at least 50% of total bioactive TGF-β in supernatants from hypoxic HUVECs consists of TGF-β2. Interestingly, the increase in bioactive TGF-β levels in the presence of control scrambled peptide SLLK (1.6-fold ± 0.9-fold) was also significant. However, a corresponding increase in TGF-β2 mRNA levels was not observed in these HUVECs. Whether the effect of SLLK is directed specifically to bioactivation of TGF-β isoforms other than TGF-β2 will be determined by measuring bioactive TGF-β in supernatants from HUVECs cultured in the presence of SLLK and neutralizing antibodies to both TGF-β1 and TGF-β3 in hypoxic conditions.
Mechanisms responsible for hypoxia-induced activation of TGF-β2 gene expression in HUVECs.
(A) TSP-1–dependent increases in bioactive TGF-β2 levels in hypoxic HUVEC supernatants. Confluent HUVECs, cultured without serum, were exposed to 20% or 1% O2 in the presence or absence of 10 μg/mL anti–TSP-1 monoclonal antibody 133; 28 μM LSKL peptide, corresponding to TSP-1–binding amino acid residues 54-57 from TGF-β1 LAP; 28 μM scrambled peptide SLLK; 10 μg/mL anti–TGF-β2 antibody; or 10 μg/mL mouse IgG. After 18 hours, supernatants were collected, and bioactive TGF-β levels were quantitated by MLEC bioassay. Results from 3 independent experiments are expressed as fold change (±SD) in bioactive TGF-β levels in hypoxic HUVECs. Values were derived by dividing the levels obtained from hypoxic supernatants by each condition's normoxic counterpart (not shown). Asterisks indicate significant differences (P ≤ .05, Studentt, 2-tailed) from the untreated condition (leftmost bar). (B) TSP-1–dependent increases in TGF-β2 mRNA levels in hypoxic HUVECs. Confluent HUVECs, cultured without serum, were exposed to 20% or 1% O2 in the presence or absence of 10 μg/mL anti–TSP-1 monoclonal antibody 133, 28 μM LSKL (inhibitory) peptide, 28 μM SLLK (scrambled) peptide, 10 μg/mL anti–TGF-β2 antibody, 10 μg/mL anti–TGF-β type II receptor (anti–TGF-β RII) antibody, 10 μg/mL mouse IgG, or 10 μg/mL anti–TGF-β1 antibody for 18 hours and then subjected to total RNA extraction. For RNase protection assays, 10 μg total RNA was hybridized to an antisense RNA probe cocktail containing the templates for TGF-β2 and L-32 (ribosomal protein subunit) genes. Representative results from 1 of 3 independent experiments are shown in the top panel, in which protected fragments of TGF-β2 and L-32 mRNAs are indicated by arrows. The bottom panel presents quantification of results from the 3 experiments. TGF-β2 signal was normalized to that from L32. Each bar represents the increase in mRNA levels from hypoxic HUVECs compared with their own normoxic controls at 18 hours. Asterisks indicate significant differences (P ≤ .05, Student t, 2-tailed) from the untreated condition (leftmost bar). N indicates normoxic conditions; H, hypoxic conditions.
Mechanisms responsible for hypoxia-induced activation of TGF-β2 gene expression in HUVECs.
(A) TSP-1–dependent increases in bioactive TGF-β2 levels in hypoxic HUVEC supernatants. Confluent HUVECs, cultured without serum, were exposed to 20% or 1% O2 in the presence or absence of 10 μg/mL anti–TSP-1 monoclonal antibody 133; 28 μM LSKL peptide, corresponding to TSP-1–binding amino acid residues 54-57 from TGF-β1 LAP; 28 μM scrambled peptide SLLK; 10 μg/mL anti–TGF-β2 antibody; or 10 μg/mL mouse IgG. After 18 hours, supernatants were collected, and bioactive TGF-β levels were quantitated by MLEC bioassay. Results from 3 independent experiments are expressed as fold change (±SD) in bioactive TGF-β levels in hypoxic HUVECs. Values were derived by dividing the levels obtained from hypoxic supernatants by each condition's normoxic counterpart (not shown). Asterisks indicate significant differences (P ≤ .05, Studentt, 2-tailed) from the untreated condition (leftmost bar). (B) TSP-1–dependent increases in TGF-β2 mRNA levels in hypoxic HUVECs. Confluent HUVECs, cultured without serum, were exposed to 20% or 1% O2 in the presence or absence of 10 μg/mL anti–TSP-1 monoclonal antibody 133, 28 μM LSKL (inhibitory) peptide, 28 μM SLLK (scrambled) peptide, 10 μg/mL anti–TGF-β2 antibody, 10 μg/mL anti–TGF-β type II receptor (anti–TGF-β RII) antibody, 10 μg/mL mouse IgG, or 10 μg/mL anti–TGF-β1 antibody for 18 hours and then subjected to total RNA extraction. For RNase protection assays, 10 μg total RNA was hybridized to an antisense RNA probe cocktail containing the templates for TGF-β2 and L-32 (ribosomal protein subunit) genes. Representative results from 1 of 3 independent experiments are shown in the top panel, in which protected fragments of TGF-β2 and L-32 mRNAs are indicated by arrows. The bottom panel presents quantification of results from the 3 experiments. TGF-β2 signal was normalized to that from L32. Each bar represents the increase in mRNA levels from hypoxic HUVECs compared with their own normoxic controls at 18 hours. Asterisks indicate significant differences (P ≤ .05, Student t, 2-tailed) from the untreated condition (leftmost bar). N indicates normoxic conditions; H, hypoxic conditions.
The degree to which hypoxia's effect on bioactivation of LAP might up-regulate TGF-β2 gene expression was examined by inhibiting TSP-1–mediated TGF-β activation in hypoxic HUVECs. As shown in Figure 1B, addition of either monoclonal antibody to TSP-1 or the inhibitory peptide LSKL produced reductions in hypoxia-induced increases in TGF-β2 mRNA levels (by 58.9% ± 9.8% and 65.2% ± 5.4%, respectively; P < .05 for both). Likewise, reducing available receptor ligand by addition of neutralizing antibody to TGF-β2 decreased hypoxia-induced up-regulation of TGF-β2 mRNA levels by 83.0% ± 6.3% (P < .05). Similar decreases (82.1% ± 3.6%;P < .05) were observed when the TGF-β type II receptor was blocked by receptor antibody (Figure 1B).
In contrast, neutralization of TGF-β1 in cultures by specific antibody did not affect hypoxic induction of TGF-β2 mRNA in HUVECs, and in fact yielded identical results as treatment with mouse IgG (Figure 1B), indicating that hypoxia-induced bioactivation of TGF-β2, but not of TGF-β1, is responsible for autocrine up-regulation of TGF-β2 gene expression.
The effect of hypoxia on phosphorylation of Smad2 and Smad3 in HUVECs
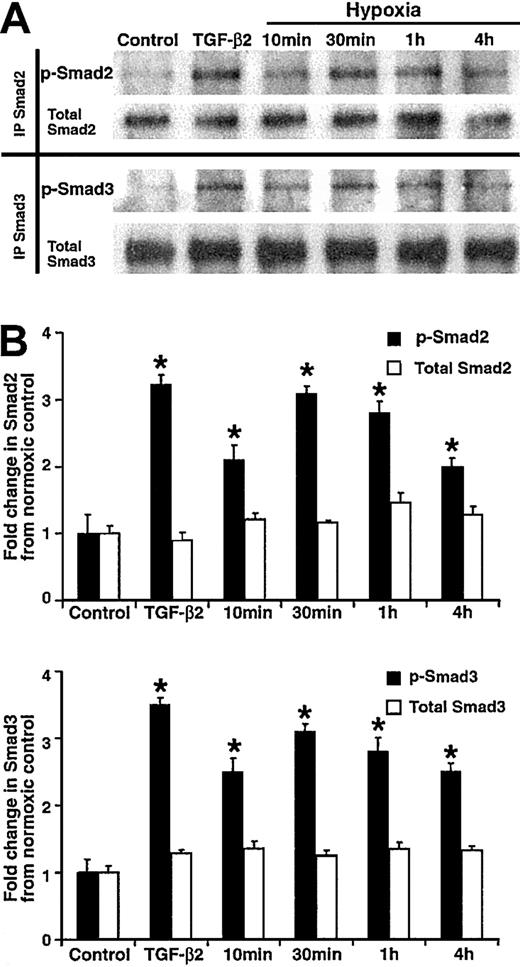
In order to investigate the downstream effects of hypoxia-induced stimulation of TGF-β2 gene expression in HUVECs, the phosphorylation of Smad2 and Smad3 proteins under hypoxic conditions was undertaken. In HUVECs exposed to hypoxia for periods of 10 minutes, 30 minutes, 1 hour, and 4 hours, levels of phosphorylated Smad2 increased by 2.1-fold ± 0.3-fold, 3.1-fold ± 0.1-fold, 2.8-fold ± 0.2-fold, and 2.0-fold ± 0.1-fold, respectively, compared with levels in nonhypoxic HUVECs (P < .05 for all time points; Figure2). Similarly, the level of phosphorylated Smad3 in HUVECs exposed to hypoxia for 10 minutes, 30 minutes, 1 hour, and 4 hours was increased by 2.5-fold ± 0.2-fold, 3.1-fold ± 0.5-fold, 2.8-fold ± 0.2-fold, and 2.5-fold ± 0.1-fold, respectively, compared with nonhypoxic HUVECs (P < .05 for all time points). In contrast, no change was observed in the levels of total Smad2 or Smad3 at any duration of exposure to hypoxia (Figure 2). The effect of hypoxia-induced Smad2 and Smad3 phosphorylation in HUVECs was essentially equivalent to that observed after exposure to 100 pM TGF-β2, a natural activator of Smads (Figure 2B). This finding suggests that TGF-β2 and hypoxia may have comparable effects on Smad activation in vivo as well.
Hypoxia-induced Smad2 and Smad3 phosphorylation in HUVECs.
HUVECs were grown to confluence and were serum starved overnight, followed by exposure to 100 pM TGF-β2 for 30 minutes or to 1% O2 (hypoxia) for 10 minutes, 30 minutes, 1 hour, 2 hours, or 4 hours. As controls, untreated cells were simultaneously analyzed at the same time points (not shown). (A) Lysates were prepared from whole cells and quantified. Half were immunoprecipitated (IP)with goat anti–Smad2 antibody (top panel) and half with goat anti–Smad3 antibody (bottom panel) overnight at 4°C. Immunoprecipitates and molecular weight markers (not shown) were resolved by 10% SDS-polyacrylamide gels. Following electrophoresis and blotting, the membranes were developed by means of rabbit antibodies specific for the phosphorylated form of Smad2 (p-Smad2), total Smad2, phosphoserine (p-Smad3), or total Smad3 followed by chemiluminescence and autoradiography. Chemiluminescent bands were quantified using ImageQuant software. Shown are representative results from 1 of 3 independent experiments. (B) Hypoxia-induced fold increases in Smad phosphorylation or in total Smad protein were calculated by dividing the values for p-Smad2, p-Smad3, Smad2, and Smad3 found in hypoxic HUVECs by their respective normoxic control values. The graph presents mean fold changes (±SD) from 3 independent experiments (*P < .05).
Hypoxia-induced Smad2 and Smad3 phosphorylation in HUVECs.
HUVECs were grown to confluence and were serum starved overnight, followed by exposure to 100 pM TGF-β2 for 30 minutes or to 1% O2 (hypoxia) for 10 minutes, 30 minutes, 1 hour, 2 hours, or 4 hours. As controls, untreated cells were simultaneously analyzed at the same time points (not shown). (A) Lysates were prepared from whole cells and quantified. Half were immunoprecipitated (IP)with goat anti–Smad2 antibody (top panel) and half with goat anti–Smad3 antibody (bottom panel) overnight at 4°C. Immunoprecipitates and molecular weight markers (not shown) were resolved by 10% SDS-polyacrylamide gels. Following electrophoresis and blotting, the membranes were developed by means of rabbit antibodies specific for the phosphorylated form of Smad2 (p-Smad2), total Smad2, phosphoserine (p-Smad3), or total Smad3 followed by chemiluminescence and autoradiography. Chemiluminescent bands were quantified using ImageQuant software. Shown are representative results from 1 of 3 independent experiments. (B) Hypoxia-induced fold increases in Smad phosphorylation or in total Smad protein were calculated by dividing the values for p-Smad2, p-Smad3, Smad2, and Smad3 found in hypoxic HUVECs by their respective normoxic control values. The graph presents mean fold changes (±SD) from 3 independent experiments (*P < .05).
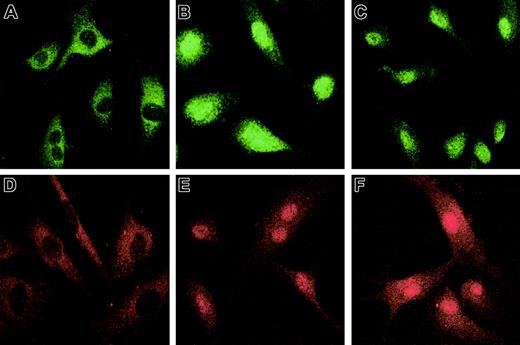
The nuclear translocation of Smad2 and Smad3 (R-Smads) subsequent to their hypoxia-induced phosphorylation was studied by immunofluorescence using specific antisera. Treatment of HUVECs with 100 pM recombinant TGF-β2 under nonhypoxic conditions was used as a positive control. In unstimulated HUVECs cultured under normoxic conditions, Smad2 (Figure3A) and Smad3 (Figure 3D) were localized predominantly in the cytoplasm, with less than 2% of HUVECs showing nuclear staining for either Smad. The intensity of nuclear staining for Smad2 and Smad3 in HUVECs exposed to hypoxia (Figure 3C,F) was similar to that observed in HUVECs exposed to TGF-β2 (Figure3B,E). These data support the notion that the effect of hypoxia on activation of Smad2 and Smad3 is mediated by TGF-β2. This proposition will be tested in future experiments by determination of nuclear translocation of Smad2 and Smad3 in hypoxic HUVECs cultured in the presence of antibody to TGF-β isoforms.
Hypoxia-induced Smad2 and Smad3 nuclear translocation in HUVECs.
HUVECs were grown to confluence on gelatin-coated chamber slides, serum starved overnight, and exposed for 1 hour to 1% O2 or 20% O2 with or without 100 pM recombinant TGF-β2. After cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, immunostaining was performed with rabbit anti–Smad2 (10 μg/mL) followed by fluorescein isothiocyanate–conjugated donkey anti–rabbit IgG (3 μg/mL); or rabbit anti–Smad3 (10 μg/mL), followed by rhodamine-conjugated donkey anti–rabbit IgG (3 μg/mL). Immunofluorescence microscopy was performed with a Bio-Rad Radiance 2000 Confocal laser scanning microscope. Immunostaining for Smad2 is shown (A) in normoxic HUVECs, (B) after treatment with 100 pM TGF-β2, and (C) after exposure to hypoxia; immunostaining for Smad3 is shown (D) in normoxic HUVECs, (E) after treatment with 100 pM TGF-β2, and (F) after exposure to hypoxia. Original magnification, × 60.
Hypoxia-induced Smad2 and Smad3 nuclear translocation in HUVECs.
HUVECs were grown to confluence on gelatin-coated chamber slides, serum starved overnight, and exposed for 1 hour to 1% O2 or 20% O2 with or without 100 pM recombinant TGF-β2. After cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, immunostaining was performed with rabbit anti–Smad2 (10 μg/mL) followed by fluorescein isothiocyanate–conjugated donkey anti–rabbit IgG (3 μg/mL); or rabbit anti–Smad3 (10 μg/mL), followed by rhodamine-conjugated donkey anti–rabbit IgG (3 μg/mL). Immunofluorescence microscopy was performed with a Bio-Rad Radiance 2000 Confocal laser scanning microscope. Immunostaining for Smad2 is shown (A) in normoxic HUVECs, (B) after treatment with 100 pM TGF-β2, and (C) after exposure to hypoxia; immunostaining for Smad3 is shown (D) in normoxic HUVECs, (E) after treatment with 100 pM TGF-β2, and (F) after exposure to hypoxia. Original magnification, × 60.
Downstream effects of Smad3 activation in hypoxia
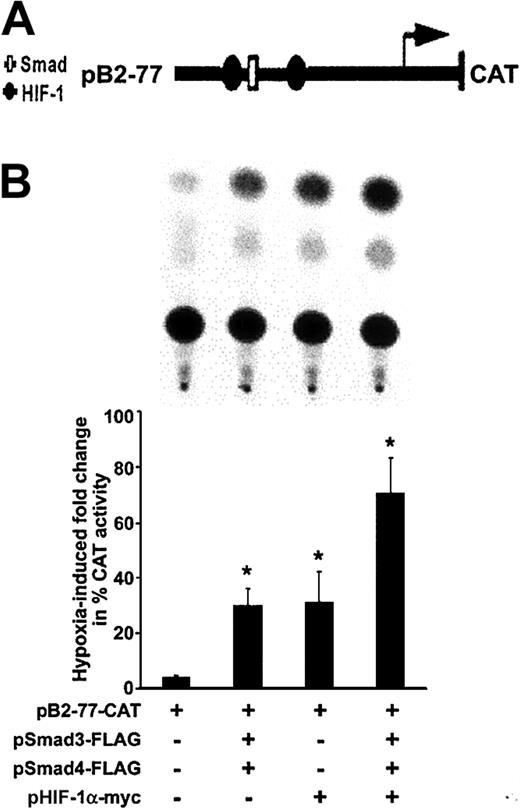
Cooperation between transcriptional mechanisms involving Smad3 and HIF-1α is suggested by their synergistic stimulation of vascular endothelial growth factor transcription.20 We recently have shown9 that the region spanning −77 to −40 bp within the 5′ TGF-β2 promoter, which harbors a Smad binding site and 2 HIF-1α binding sites, is necessary and sufficient to confer hypoxia responsiveness to the 5′ promoter spanning −778 to −40 bp and containing cis-acting elements that regulate transcription of the 5.8-kb TGF-β2 transcript.40 To explore downstream effects of hypoxia-induced phosphorylation and nuclear translocation of Smad3 and its functional relationship to HIF-1α, transient transfection experiments were performed with TGF-β2 promoter CAT construct pB2-77, which harbors the 5′ TGF-β2 promoter spanning −77 to −63 bp (Figure 4A), before and after overexpression of Smad3 and HIF-1α in HUVECs. Since Co-Smad4 is necessary for nuclear translocation of Smad3, a Smad4-overexpressing vector was also used. As shown in Figure 4B, activity of this construct was increased by 4.8-fold ± 0.6-fold in hypoxic conditions compared with that in normoxic conditions (P < .05). As expected, co-overexpression of Smad3 and Smad4 augmented the hypoxia-induced activity of pB2-77 by a further 7.4-fold ± 1.3-fold (P < .05) compared with hypoxic HUVECs, which did not overexpress Smads (Figure 4B). When pB2-77 was cotransfected with an expression vector coding for HIF-1α, there was a 6.3-fold ± 2.2-fold increase in hypoxia-induced activity of this promoter construct. The highest activity of pB2-77 in response to hypoxia was observed after co-overexpression of Smad3, Smad4, and HIF-1α, which resulted in a 14.6-fold ± 2.6-fold increase in activity of pB2-77 compared with its baseline activity in hypoxic HUVECs (P < .05). Thus, both Smad3 and HIF-1α are sufficient to induce hypoxic responsiveness to the TGF-β2 promoter, and, in combination, their effects are additive.
Effect of hypoxia on transcriptional activity of the TGF-β2 promoter.
(A) Schematic structure of recombinant plasmid construct pB2-77 containing the human TGF-β2 gene promoter between −77 and +63 bp, and that is linked to a CAT reporter gene. The transcriptional start site (arrow) driving the CAT gene and selected protein-binding sites (Smad, HIF-1) are indicated. (B) HUVECs were transfected with pB2-77, and cotransfected with indicated Smad and HIF-1α expression vectors and Rous sarcoma virus (RSV)–β-gal by electroporation. Each transfection was divided into 2 plates and exposed to 20% or 1% O2 for 36 hours. Representative results of 1 of 3 independent CAT assays are shown in the panel on the left. CAT activity was determined in whole-cell extracts, and its level, given as percent acetylation, was normalized to protein and to β-gal activity in each experiment. Results are expressed as fold increases (±SD) in CAT activity in indicated hypoxic conditions compared with that found in their normoxic counterparts for each transfection. Fold changes observed in each experimental group are compared with the hypoxia-induced fold change in HUVECs transfected with the pB2-77-CAT alone (*P < .05).
Effect of hypoxia on transcriptional activity of the TGF-β2 promoter.
(A) Schematic structure of recombinant plasmid construct pB2-77 containing the human TGF-β2 gene promoter between −77 and +63 bp, and that is linked to a CAT reporter gene. The transcriptional start site (arrow) driving the CAT gene and selected protein-binding sites (Smad, HIF-1) are indicated. (B) HUVECs were transfected with pB2-77, and cotransfected with indicated Smad and HIF-1α expression vectors and Rous sarcoma virus (RSV)–β-gal by electroporation. Each transfection was divided into 2 plates and exposed to 20% or 1% O2 for 36 hours. Representative results of 1 of 3 independent CAT assays are shown in the panel on the left. CAT activity was determined in whole-cell extracts, and its level, given as percent acetylation, was normalized to protein and to β-gal activity in each experiment. Results are expressed as fold increases (±SD) in CAT activity in indicated hypoxic conditions compared with that found in their normoxic counterparts for each transfection. Fold changes observed in each experimental group are compared with the hypoxia-induced fold change in HUVECs transfected with the pB2-77-CAT alone (*P < .05).
The effect of Smad3 and HIF-1α on hypoxia-induced increases in TGF-β2 mRNA levels
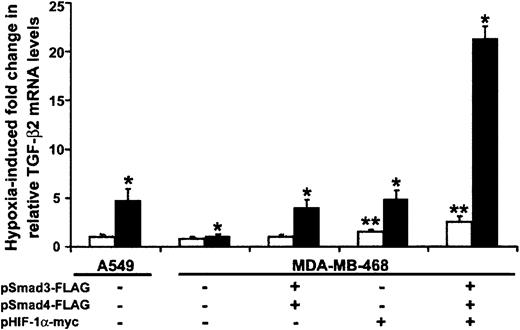
Based on their stimulatory effect on transcription from the TGF-β2 promoter–containing CAT construct, we asked whether either Smads or HIF-1α were necessary and/or sufficient for the occurrence of hypoxia-induced increases in TGF-β2 mRNA levels in HUVECs. Because the absence of Smad4 impairs translocation of Smad3 to the nucleus and impedes its functions,9 we examined the role of Smad3 on hypoxia-induced up-regulation of TGF-β2 mRNA levels in Smad4-deficient and Smad4-replete epithelial cell lines. In real-time quantitative PCR experiments not shown here, we found that overexpression of Smad3 alone did not increase TGF-β2 mRNA levels in hypoxic Smad4-deficient cells. However, co-overexpression of Smad3 and Smad4 caused approximately 15% higher TGF-β2 mRNA levels than were seen in hypoxic cells overexpressing Smad4 alone (P < .05). We then cotransfected Smad3 as well as Smad4 into Smad4-deficient and control cell lines. As seen in Figure5, hypoxia had no effect on TGF-β2 mRNA levels in the Smad4-null epithelial breast carcinoma cell line. However, hypoxia induced a 4.8-fold ± 1.2-fold increase in TGF-β2 mRNA in A549 cells, which do express Smad4. As expected, overexpression of both Smad4 and Smad3 led to a 4.0-fold ± 0.8-fold increase in hypoxia-induced TGF-β2 mRNA levels in MDA-MB-468. Although overlap between any of the functions of HIF-1α and Smad4 has not been described, HIF-1α overexpression resulted in a 3.2-fold ± 0.9-fold increase in TGF-β2 mRNA levels in hypoxic MDA-MB-468 cells compared with untransfected controls (P < .05), indicating that both Smad3/Smad4 and HIF-1α independently stimulate TGF-β2 mRNA levels in response to hypoxia. Co-overexpression of Smad3, Smad4, and HIF-1α resulted in an 8.5-fold ± 1.3-fold induction of TGF-β2 mRNA levels, mirroring the additive effect of Smads and HIF-1α on transcriptional activity of the TGF-β2 promoter construct p2B-77 (Figure 4B), and suggesting that overexpression of HIF-1α provides at least partial rescue for Smad4-null cells in hypoxic conditions. In comparison, under normoxic conditions, TGF-β2 mRNA levels were similar in A549 and MDA-MB-468 cells (Figure 5). When Smad4-deficient cells were transiently transfected with Smad3/Smad4, there was no change in TGF-β mRNA levels in normoxic conditions. While overexpression of HIF-1α resulted in a 1.5-fold increase in TGF-β2 mRNA levels, co-overexpression of Smad3/Smad4 in addition to HIF-1α resulted in a further increase to 2.5-fold in Smad4-deficient cells (P < .05 for both; Figure 5). This modest induction of TGF-β2 mRNA levels in normoxia in MDA-MB-468 cells was equivalent to that seen in control A549 epithelial cells (data not shown). Whether overexpression of HIF-1α potentiates a signaling pathway downstream of Smad4 or whether HIF-1α overexpression provides Co-Smad activity for Smad3 remains to be determined.
Smad3 and HIF-1α are independently capable of inducing TGF-β2 mRNA in response to hypoxia.
Smad4-deficient MDA-MB-468 and Smad4-expressing A549 were transiently transfected with indicated Smad and HIF-1α expression vectors and RSV–β-gal by electroporation. Each transfection was divided into 2 plates and exposed to either 20% or 1% O2for 18 hours. TGF-β2 mRNA levels were determined by real-time RT-PCR analysis, using TGF-β2–specific TaqMan primers and probe. Amount of mRNA in each group is normalized to GAPDH. Results are presented as fold change in TGF-β2 mRNA in normoxic (■) or hypoxic (▪) conditions relative to the levels observed in cell line A549 under basal normoxic conditions. Bars represent mean ±SD from 3 independent experiments (*P < .05 for hypoxic and **P < .05 for normoxic results).
Smad3 and HIF-1α are independently capable of inducing TGF-β2 mRNA in response to hypoxia.
Smad4-deficient MDA-MB-468 and Smad4-expressing A549 were transiently transfected with indicated Smad and HIF-1α expression vectors and RSV–β-gal by electroporation. Each transfection was divided into 2 plates and exposed to either 20% or 1% O2for 18 hours. TGF-β2 mRNA levels were determined by real-time RT-PCR analysis, using TGF-β2–specific TaqMan primers and probe. Amount of mRNA in each group is normalized to GAPDH. Results are presented as fold change in TGF-β2 mRNA in normoxic (■) or hypoxic (▪) conditions relative to the levels observed in cell line A549 under basal normoxic conditions. Bars represent mean ±SD from 3 independent experiments (*P < .05 for hypoxic and **P < .05 for normoxic results).
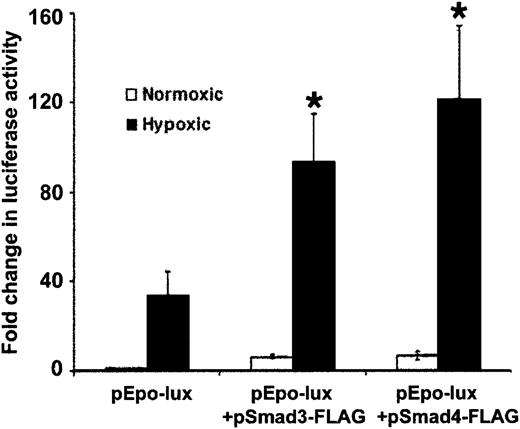
To explore the effect of Smad3 on HIF-1α–mediated transcription in hypoxic cells, we studied the effect of Smad3 overexpression on hypoxia-induced transcription from the luciferase constructEPO-Lux, which contains an erythropoietin enhancer highly responsive to activation by HIF-1α during hypoxia.24,44Similar to other reports,24,44 activity ofEPO-Lux increased 34.4-fold ± 10.1-fold in hypoxic cells compared with normoxic controls (Figure6). Overexpression of Smad3 and Smad4 caused further 2.7-fold ± 0.6-fold and 3.6-fold ± 0.9-fold increases in EPO-Lux activity, respectively, compared with hypoxic baseline activity of this plasmid construct (P < .05 for both). The results are consistent with the notion that between Smad3 and HIF-1α signaling pathways, there is a bidirectional stimulatory effect that possibly increases promoter sensitivity to hypoxia. Whether this effect is due to the interaction of Smad3 and HIF-1α, as suggested by a recent report20showing their coprecipitation in hypoxic cells, or is via independent interactions of the proteins with DNA or other transcription factors remains to be proved. In addition, transcriptional activity ofEPO-Lux was induced by 5-fold after Smad3 and Smad4 overexpression in normoxic conditions (Figure 6). Whether Smads play a role in the activity of HIF-1α in nonhypoxic conditions remains to be determined.
Hypoxia-induced increases in transcription fromEPO-Lux are mediated by Smad proteins and HIF-1α.
Confluent HepG2 cells were transfected with EPO-Lux, and cotransfected with indicated Smad and HIF-1α expression vectors, or empty vector and RSV–β-gal by electroporation. Each transfection was divided into 2 plates and exposed to either 20% or 1% O2for 24 hours. Luciferase activity was determined and results normalized to RSV–β-gal activity and protein content of the extracts. Results are expressed as fold increase in luciferase activity in hypoxic conditions compared with that found in normoxic conditions for each transfection. The figure presents the mean (±SD) of 3 independent experiments (*P < .05).
Hypoxia-induced increases in transcription fromEPO-Lux are mediated by Smad proteins and HIF-1α.
Confluent HepG2 cells were transfected with EPO-Lux, and cotransfected with indicated Smad and HIF-1α expression vectors, or empty vector and RSV–β-gal by electroporation. Each transfection was divided into 2 plates and exposed to either 20% or 1% O2for 24 hours. Luciferase activity was determined and results normalized to RSV–β-gal activity and protein content of the extracts. Results are expressed as fold increase in luciferase activity in hypoxic conditions compared with that found in normoxic conditions for each transfection. The figure presents the mean (±SD) of 3 independent experiments (*P < .05).
Discussion
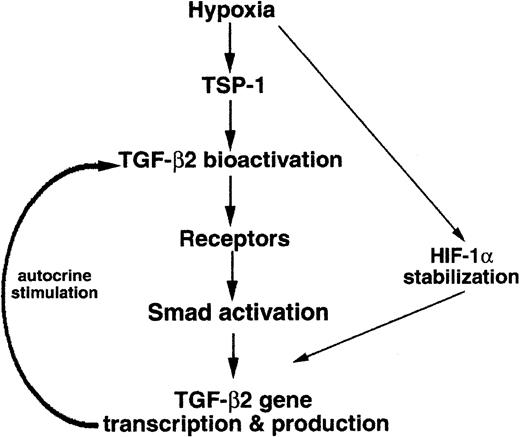
In HUVECs exposed to hypoxia, significant up-regulation of TGF-β2 gene expression was found to occur via an autocrine loop dependent upon TSP-1–mediated TGF-β2 bioactivation, presented schematically in Figure7. Furthermore, both Smad3 and HIF-1α were shown to contribute to hypoxia-induced transcriptional up-regulation of TGF-β2 in endothelial cells. The effects of proteolysis by plasmin, exposure to reactive oxygen species, and binding to integrins,47 48 all of which bioactivate TGF-β, on hypoxia-induced elevation of TGF-β2 mRNA levels remain to be elucidated.
Schematic drawing of the hypoxia-induced signaling pathway leading to TGF-β2 gene expression.
Schematic drawing of the hypoxia-induced signaling pathway leading to TGF-β2 gene expression.
The blocking of TSP-1–mediated bioactivation of latent TGF-β2 during exposure to hypoxia inhibited up-regulation of TGF-β2 mRNA in HUVECs. It thus is likely that TGF-β2 mediates at least some actions of TSP-1 on hypoxic endothelial cells. Exposure of endothelial cells to hypoxia results in up-regulation of gene expression of TSP-1, thus inhibiting neovascularization, in part by its dampening effect on endothelial response to proangiogenic stimuli,16 and also by its proapoptotic effects on endothelial cells via downstream signaling from membrane receptor CD36.49 Inhibition of retinal angiogenesis by interruption of TSP-1's activation of TGF-β50 suggests that bioactivation of TGF-β may be one mechanism responsible for TSP-1–induced antiangiogenesis. An antiangiogenic function in vivo was suggested for TGF-β2 based on its inhibitory effect on hepatocyte growth factor–mediated angiogenesis,51 whereas experimental evidence suggests that TGF-β1 has proangiogenic52 and proapoptotic effects.53,54 Since TGF-β2 is only minimally expressed in vascular endothelium under normoxic conditions,55,56 the degree to which hypoxic induction of TGF-β2, as distinct from TGF-β1 or TGF-β3, is associated with regulation of apoptosis and angiogenesis in endothelium has remained unclear. The low level of expression of TGF-β2 seen in normoxic conditions in vascular endothelium56 suggests that TGF-β2 gene expression and function may be more relevant under hypoxic conditions. Deletion of the TGF-β2 gene did not result in the severe vascular developmental abnormalities seen in deletions of the genes encoding for Smad5,12 type I TGF-β receptor,10 endoglin,57-59 or HIF-1α60 but resulted in perinatal death with cyanosis and cardiac and pulmonary abnormalities.7 Similar to our findings of hypoxia-induced Smad activation, the phosphorylation and subsequent nuclear translocation of Smad proteins were reported to occur after exposure of HUVECs to laminar shear,61indicating that the Smad signaling pathway is an important mechanism by which endothelial cells respond to stress.
Activity of the 5′ TGF-β2 promoter spanning −778 and +63 bp, which regulates transcription of the 5.8-kb TGF-β2 transcript, is significantly increased by hypoxia, and within this region, sequences spanning −77 to −40 bp are necessary and sufficient to mediate the hypoxia response.9 Our current results show that activity of the latter hypoxia-responsive promoter region is also increased in an additive fashion by HIF-1α and Smad3/Smad4, indicating that HIF-1α and Smad proteins have a potentiating effect on hypoxic expression of TGF-β2. The additive effect of Smad3/Smad4 and HIF-1α is likely to be a result of their independent binding sites within the TGF-β2 promoter, since in our previous studies,9 we found that a HIF-1–binding oligonucleotide did not compete with binding of hypoxic nuclear extracts to DNA containing 2 palindromic Smad-binding sequences. In addition to having an additive effect on TGF-β2 promoter activity, our results suggest the possibility of a stimulatory effect of Smad3/Smad4 on HIF-1α function, indicated by the increase in activation of the HIF-1α–responsive EPO promoter construct after Smad3/Smad4 overexpression. These findings are in agreement with existing reports that indicate cooperation between Smad and HIF-1α signaling pathways, as, for example, in the observed reduction of TGF-β1 gene expression in fetal skin by HIF-1α,62in regulation of TGF-β3 gene expression by HIF-1α in trophoblasts,62,63 and, particularly, in the recent demonstration of a synergistic stimulatory effect of Smad3 and HIF-1α on hypoxia-induced transcription of vascular endothelial growth factor gene expression.20 Furthermore, nuclear transcriptional coactivator p300/cyclic AMP response element binding protein (CREB)–binding protein (CBP) can associate with both Smads64,65 and HIF-1α66,67 and thus can play a role in their transcriptional interactions. Lastly, hypoxia could also increase stabilization of Smad proteins and enhance their accumulation in the nucleus. Similar to results observed with HIF-1α Smad3 is also degraded by E3 ubiquitin ligase complex68; thus, it is conceivable that Smad3 could also be subject to posttranslational modifications during hypoxia that interfere with Smad3 ubiquitination and subsequent degradation. The role of proline hydroxylation or pVHL in facilitating Smad degradation is not known. The role, if any, of hypoxia on constitutive phosphorylation of Smad4 is also unknown at this time.69 Our findings also suggest that the supportive bidirectional effects between Smad and HIF-1α signaling pathways in modulating transcription may not be restricted to hypoxia, but may also occur to a lesser extent in normoxic conditions as well by an as-yet-unknown mechanism.
Taken together, these results suggest that Smads and HIF-1α can affect each other's transcriptional activities via different mechanisms, and their interaction may contribute to the isoform- and context-specific actions of TGF-βs. A further understanding of hypoxia-driven bioactivation of TGF-β2 and its downstream effects on signaling by Smads as well as by HIF-1α may help elucidate cell-specific actions of TGF-β isoforms in the vascular system, and may clarify the role of this family of cytokines in disease.70
We are grateful to Dr Jaime Caro (Cardeza Foundation for Hematologic Research, Jefferson Medical College, Thomas Jefferson University, Philadelphia, PA) for his helpful advice on this project.
Prepublished online as Blood First Edition Paper, October 31, 2002; DOI 10.1182/blood-2002-02-0629.
Supported by a Multiple Myeloma Research Foundation Senior Research Award (O.A.B.) and by grants from the American Lung Association Brooklyn Chapter (O.A.B.), the National Institute of Mental Health (MH599990, E.L.P.S.), and the National Heart, Lung, and Blood Institute (HL50061, J.E.M.-U.; HL53573, M.A.Q.S.).
H.Z. and H.O.A. contributed equally to this paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Olcay Ayanlar Batuman, Division of Hematology/Oncology, Department of Medicine, SUNY Downstate Medical Center, Box 20, 450 Clarkson Ave, Brooklyn, NY 11203; e-mail:obatuman@downstate.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal