A combination of magnetic cell sorting (MACS) and fluorescent in situ hybridization (FISH) techniques was used to detect clonal cytogenetic markers in different myeloid and lymphoid cell types of the peripheral blood from 4 patients with myelofibrosis with myeloid metaplasia (MMM) that was associated with either a 13q− or a 20q− karyotypic abnormality. Interphase cytogenetics studies demonstrated abnormal clonal FISH signal patterns in neutrophil, myeloid, erythroid, megakaryocyte, and B- and T-cell preparations in 3 of the 4 patients. In one patient, FISH results were within normal limits in T cells and slightly abnormal in B cells. In general, the percentage of abnormal nuclei was variable in both lymphocyte populations but always higher in B lymphocytes compared with T lymphocytes. The current study provides direct evidence for the clonal involvement of both B and T lymphocytes in MMM. A larger study is needed to clarify the relevance of the observed interpatient heterogeneity in clonal constitution.
Introduction
In 1951, Dameshek1 classified myelofibrosis with myeloid metaplasia (MMM) as a chronic myeloproliferative disorder (CMPD) along with essential thrombocythemia (ET), polycythemia vera (PV), and chronic myeloid leukemia (CML) because of similarities in both clinical and laboratory features. Between 1967 and 1981, Fialkow and colleauges,2-6 using glucose-6-phosphate dehydrogenase isoenzyme analysis, showed that CMPDs are also biologically interrelated on the basis of being clonal stem cell disorders with involvement of all myeloid cell types (granulocytes, erythrocytes, platelets, monocytes). In the latter part of the same period and using similar clonality assays, evidence emerged that B lymphocytes but not T lymphocytes were clonally involved in CML, PV, and ET.7-9 More recent studies, based primarily on X-linked DNA analysis, have supported these early observations.10-13 Other studies, using different clonal assays, have suggested monoclonality of T lymphocytes in both MMM (based on Ras mutational analysis)14 and CML (based on fluorescent in situ hybridization [FISH] analysis).15 In the current study, we evaluated 4 patients with MMM who had clonal chromosome abnormalities that could be identified by FISH in interphase nuclei in highly purified cell fractions.
Study design
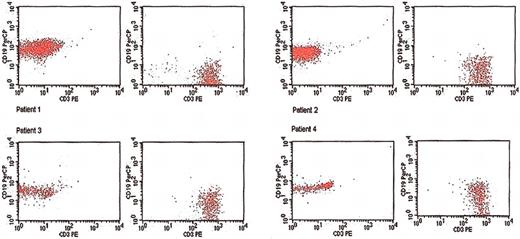
After approval by the Mayo Clinic institutional review board, peripheral blood (30 mL) was collected from 4 patients with MMM whose bone marrow karyotype analysis revealed a deletion of the long arm of either chromosome 13 (13q−) or chromosome 20 (20q−). Hypaque density gradient centrifugation was used to separate out the granulocyte and mononuclear cell layers (Sigma Diagnostics, St Louis, MO) from each of the 4 samples. Each mononuclear cell layer was then further cell fractionated by magnetic cell sorting (MACS; Miltenyi Biotech, Auburn, CA) by using antibodies that are specific to myeloid (CD34+), megakaryocyte (CD61+), and erythroid (CD71+) precursor cells as well as T (CD3+) and B (CD19+) lymphocytes. The particular procedure was performed according to the manufacturer's recommendation. Sample purity in regard to B- and T-cell preparations was confirmed by flow cytometry (Figure 1). In addition, polymerase chain reaction (PCR)–based T-cell–receptor (TCR) and immunoglobulin gene rearrangement studies were performed in lymphocyte-rich, peripheral blood mononuclear cells from all 4 study patients.
Flow cytometric confirmation of sample purity of B (CD19) and T (CD3) lymphocyte fractions that are sorted by antibody-linked immunomagnetic beads from 4 patients with myelofibrosis with myeloid metaplasia.
Flow cytometric confirmation of sample purity of B (CD19) and T (CD3) lymphocyte fractions that are sorted by antibody-linked immunomagnetic beads from 4 patients with myelofibrosis with myeloid metaplasia.
Standard cytospin preparations were made from each of the described cell preparations and submitted to FISH studies. Each slide was fixed in methanol/glacial acetic acid (3:1) and air-dried. Slides were pretreated in 2 × standard saline citrate (SSC) at 37°C for 60 minutes and dehydrated in a series of three 2-minute solutions of 70%, 85%, and 100% ethanol and air-dried. Commercial probes from Vysis (Downers Grove, IL) for red light scattering index (LSI) D13S319 and green CEP9 were used together to detect 13q− anomalies. D13S319 hybridizes to 13q14 and CEP9 hybridizes to the alpha Satellite DNA in the centromere region (9p11-q11) of chromosome 9. Commercial probes from Vysis for LSI D20S108 (red) and CEP9 (green) were used together to detect 20q− anomalies. D20S108 hybridizes to 20q12. The cutoff for false-positive nuclei was less than 9.5% for 13q− and less than 11.5% for 20q−. Any specimens with percentages of nuclei with less than the cutoff were classified as “normal.” Slides were then cover-slipped, sealed with rubber cement, denatured at 75°C for 3 minutes, and allowed to hybridize for 18 to 22 hours at 37°C.
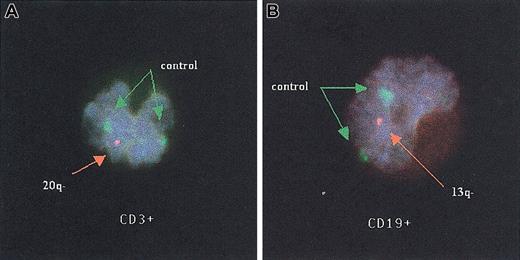
After hybridization, coverslips were removed, slides were washed in 0.4% SSC at 70°C for 2 minutes, and then transferred to 1 × phosphate-buffered detergent at room temperature for 1 minute. The slides were counterstained with a mixture of 5 μL 4′-6′-diamidine-2-phenylindole dihydrochloride (DAPI) and Vectashield antifade (Vector Labs, Burlingame, CA) at a ratio of 1:10. Cells were viewed with a fluorescent microscope equipped with a triple-band pass filter for fluoroisothiocyanate, Texas red, and DAPI (Chromatechnology, Brattleboro, VT). Each specimen was studied by 2 microscopists independently. Each microscopist scored 100 consecutive interphase nuclei, exhibiting 2 clear control signals: the percentages of abnormal nuclei for the 2 microscopists were averaged to produce a single percentage of abnormal nuclei for each specimen. Representative B and T cells were photographed with a computer-based imaging system (Figure2).
Fluorescent in situ hybridization studies of T and B lymphocytes.
T (A) and B (B) lymphocytes from 2 patients with myelofibrosis with myeloid metaplasia that exhibit a single orange signal revealing a deletion of the long arm of either chromosomes 20 (20q−) or 13 (13q−), respectively, by fluorescent in situ hybridization studies. Both green signals of the control probe (CEP9) are visible in each figure. Original magnification, × 100.
Fluorescent in situ hybridization studies of T and B lymphocytes.
T (A) and B (B) lymphocytes from 2 patients with myelofibrosis with myeloid metaplasia that exhibit a single orange signal revealing a deletion of the long arm of either chromosomes 20 (20q−) or 13 (13q−), respectively, by fluorescent in situ hybridization studies. Both green signals of the control probe (CEP9) are visible in each figure. Original magnification, × 100.
Results and discussion
The 4 study patients (3 men and 1 woman) with MMM had bone marrow chromosomal abnormalities including either 13q− [del(13)(q12q14); del(13)(q12q21.2)] seen in 2 patients or 20q− [del(20)(q13.1)] seen in the other 2 patients. Three of the 4 patients had either 13q− or 20q− in all metaphases (Table 1). In the fourth patient, 20q− was carried by 16 of 21 metaphases. In 3 of the 4 patients, FISH analysis revealed an abnormal signal pattern in neutrophils, erythroid (CD71+), megakaryocyte (CD61+), and myeloid (CD34+) cells as well as B (CD19+) and T (CD3+) lymphocytes (Table 1). The results in the 4th patient were similar, except that the T-cell population FISH results were within normal limits (Table 1). We observed substantial interpatient heterogeneity in the percentage of abnormal nuclei in both B (13%-96%) and T lymphocytes (6%-57%) as well as CD34+ cells (36%-87%) (Table 1). Among the 3 patients with only abnormal metaphases, the highest percentage of abnormal nuclei was demonstrated by neutrophils (82%-98%), megakaryocyte (CD61+) (75%-94%), and erythroid (CD71+) (78%-88%) cell types.
Percentage of abnormal nuclei in each cell lineage of 4 patients with myelofibrosis with myeloid metaplasia studied with fluorescent in situ hybridization using probes to detect 13q− and 20q−
| Patient . | CD3+ . | CD19+ . | PMN . | CD34+ . | CD61+ . | CD71+ . |
|---|---|---|---|---|---|---|
| 1 del(13)(q12q21.2) in 20 of 20 metaphases | 42 | 61 | 82 | 58 | 75 | 78 |
| 2 del(13)(q12q14) in 20 of 20 metaphases | 6 | 13 | 90 | 51 | 94 | 88 |
| 3 del(20)(q13.1) in 20 of 20 metaphases | 57 | 96 | 98 | 87 | 77 | 81 |
| 4 del(20)(q13.1) in 16 of 21 metaphases | 21 | 30 | 86 | 36 | 67 | 57 |
| Patient . | CD3+ . | CD19+ . | PMN . | CD34+ . | CD61+ . | CD71+ . |
|---|---|---|---|---|---|---|
| 1 del(13)(q12q21.2) in 20 of 20 metaphases | 42 | 61 | 82 | 58 | 75 | 78 |
| 2 del(13)(q12q14) in 20 of 20 metaphases | 6 | 13 | 90 | 51 | 94 | 88 |
| 3 del(20)(q13.1) in 20 of 20 metaphases | 57 | 96 | 98 | 87 | 77 | 81 |
| 4 del(20)(q13.1) in 16 of 21 metaphases | 21 | 30 | 86 | 36 | 67 | 57 |
Each column represents T cells (CD3+), B cells (CD19+), neutrophils (PMN), myeloid progenitor cells (CD34+), megakaryocyte precursor cells (CD61+), and erythroid cells (CD71+). Normal ranges are < 9.5% for 13q− and < 11.5% for 20q−.
Information from X chromosome–based clonality assays may not be totally adequate because of the occurrence of both excessive Lyonization and acquired skewing patterns of hematopoiesis in normal elderly women.16-18 Therefore, the application of alternative methods is essential for further clarification of clonal constitution in myeloid disorders. In this regard, karyotype,19,20 FISH,15,21 or Ras mutational analysis14 have been applied to either fractionated cell populations or in vitro progenitor colonies. In most, but not all,14 15 instances, the results have been consistent with those derived from X-linked clonality assays.
The current clonality study was FISH based and clearly indicates that both B and T lymphocytes may be clonally involved in MMM. The absence of clonal immunoglobulin and TCR gene rearrangements, in lymphocyte-rich cell fractions from our patients, is consistent with the current assumption that the disease-initiating mutation in MMM and related disorders occurs before the earliest biologic events that are associated with lymphocyte commitment. In addition, the study suggests substantial interpatient heterogeneity in clonal constitution of myeloid progenitor cells (CD34+) as well as both B and T lymphocytes. However, this variation in the expression of a FISH-detected “clonal” marker in a selected cell population may reflect dynamic differences in clonal evolution rather than a difference in the distribution of the original disease clone.22 In other words, a FISH-detected lesion may represent a cytogenetic subclone, and the absence of such a marker does not necessarily imply nonclonality. Similarly, the observed heterogeneity in the degree of clonal signal distribution may apply only to cytogenetic subclones and not to the original disease clone. Nevertheless, the current study raises questions about the validity of using T cells as controls in biologic studies of myeloid disorders.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-07-2341.
Supported by Mayo Clinic Hematology Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ayalew Tefferi, Division of Hematology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail:tefferi.ayalew@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal