Acute promyelocytic leukemia (APL) is associated with a reciprocal and balanced translocation involving the retinoic acid receptor-α(RARα). All-trans retinoic acid (ATRA) is used to treat APL and is a potent morphogen that regulatesHOX gene expression in embryogenesis and organogenesis.HOX genes are also involved in hematopoiesis and leukemogenesis. Thirty-nine mammalian HOX genes have been identified and classified into 13 paralogous groups clustered on 4 chromosomes. They encode a complex network of transcription regulatory proteins whose precise targets remain poorly understood. The overall function of the network appears to be dictated by gene dosage. To investigate the mechanisms involved in HOX gene regulation in hematopoiesis and leukemogenesis by precise measurement of individual HOX genes, a small-array real-timeHOX (SMART-HOX) quantitative polymerase chain reaction (PCR) platform was designed and validated. Application of SMART-HOX to 16 APL bone marrow samples revealed a global down-regulation of 26 HOX genes compared with normal controls. HOX gene expression was also altered during differentiation induced by ATRA in thePML-RARα+ NB4 cell line. PML-RARα fusion proteins have been reported to act as part of a repressor complex during myeloid cell differentiation, and a model linkingHOX gene expression to this PML-RARα repressor complex is now proposed.
Introduction
Acute promyelocytic leukemia (APL) is a subgroup of acute myelogenous leukemia (AML) typified by the t(15;17) leading to fusion of the putative growth suppressor PML to the retinoic acid receptor α (RARα), which has been implicated in normal myelopoiesis.1 Recent studies suggest that PML-RARα, which retains the ligand (RA)–binding domain and functions as an aberrant retinoid receptor at physiological levels of RA, leads to repression of downstream target genes (reviewed by Lin et al2). This is due to recruitment of nuclear corepressor/histone deacetylase (HDAC) complexes and DNA methyltransferases,3 contributing to the differentiation block that characterizes the disease. However, the block can be overcome by pharmacologic levels of ligand in the form of all-trans retinoic acid (ATRA). The result is the induction of a conformational change in the fusion protein accompanied by binding of coactivators and release of corepressors, converting it from a repressor to a transcriptional activator. ATRA-induced degradation of PML-RARα provides a further mechanism contributing to alleviation of the block in differentiation.2 The precise molecular consequences of the presence of the PML-RARα fusion and the changes that follow ATRA therapy are still far from clear. However, retinoids are potent morphogens and regulators in embryogenesis and organogenesis and have been shown to modulate HOX gene expression. Furthermore, the RARα molecule has been hypothesized to act upstream of HOX genes.4 5
HOX genes encode a subset of homeodomain-containing proteins, thought to bind DNA, which act as major regulators of embryogenesis and organogenesis.6HOX genes contain a highly conserved 183–base pair (bp) region termed the homeobox.7-9 The genomes of all animals analyzed to date exhibit a distinct clustering of the homeobox genes, which are assumed to have arisen by duplication and divergence from a primordialHOX gene. In nonvertebrates one cluster exists—that is, HOM-C of insects and nematodes—whereas 4 clusters (HOX-A, B, C, and D) are present in mammals. The complex organization of the mammalian HOX gene cluster is based on 13 identifiable genes termed paralogs 1-13. Paralogs are distinguished by homeobox sequence similarity and position within the cluster. A total of 39 human HOX genes have been assigned to clusters A through D located at 4 different loci: 7p15, 17p21, 12q13, and 2q31, respectively.10 During body patterning the expression of a particular HOM-C or HOX gene is related to its position within the chromosome—the colinearity rule. Recent work involving replacement transgenics suggests that the roles attributed to paralogous genes are the result of “quantitative modulations in gene expression” or gene dosage,11 but the temporal order of expression of individual HOX genes throughout hematopoiesis remains to be elucidated.
HOX genes play a significant role in both normal and dysregulated hematopoiesis.12-15 In rare cases of AML involving a t(7;11)(p15;p15) or t(2;11)(q31;p15) translocation, theHOXA9 or HOXD13 genes, respectively, are fused with the NUP98 nucleoporin gene, underlying the disease state.16-20 Furthermore, HOXA9 showed strong correlation with AML and treatment failure when an oligo expression array containing 6817 genes was used to segregate AML and acute lymphoblastic leukemia (ALL) patients.21 The product of the mixed lineage leukemia gene (MLL) is a positive regulator for the maintenance of cell-specific HOXexpression via chromatin remodeling.22,23MLLhas been implicated in fusions with more than 25 other genes in both ALL (eg, MLL-AF4) and AML (eg,MLL-AF9).24,25 Thus, alterations inMLL may also disrupt HOX gene expression patterns in hematopoietic progenitor cells and contribute to a leukemic state.26
We have developed and validated a small-arrayreal-time quantitative polymerase chain reaction (PCR) approach for the simultaneous and rapid measurement of expression of 27 individual HOX genes (SMART-HOX). TaqMan probe–based chemistry (ABI Prism 7700; Applied Biosystems, Foster City, CA) was used to attain high levels of specificity, and application of SMART-HOX to 5 leukemic cell lines revealed novel patterns of gene expression. The aim of this study was to identify interactive HOX gene pathways in the well-defined APL subgroup.
Patients, materials, and methods
Leukemic cell lines and patient samples
Five human leukemic cell lines—KG1a, HL60, NB4, U937, and HEL—were studied. All cell lines were cultured as previously described.27 KG1a, HL60, U937, and HEL were all purchased from European Collection of Animal Cells and Cultures (ECACC; Porton Down, United Kingdom). NB4 cells were kindly provided by Dr M. Lanotte (Hôpital St Louis, Paris, France). Total RNA samples isolated from the bone marrow of 20 APL patients were obtained from the United Kingdom Medical Research Council tissue bank at University College Hospital, London. Presence of the PML-RARα fusion was confirmed in all cases by nested reverse-transcriptase polymerase chain reaction (RT-PCR), as previously described.28 Four of the 20 APL samples were excluded from SMART-HOX analysis: One showed significant RNA degradation as determined by gel electrophoresis, while 3 others had insufficient cDNA. Normal bone marrow samples were obtained from donors following informed consent. Mononuclear cells were isolated by Ficoll-Hypaque (Pharmacia Biotech, Uppsala, Sweden) density centrifugation and washed in 1 × phosphate-buffered saline (PBS) prior to cryopreservation at −80°C in 90% fetal bovine serum (FBS) 10% dimethyl sulfoxide (DMSO) (BDH Laboratories, Poole, United Kingdom). Cryopreserved cells were rapidly thawed in a 37°C water bath, diluted in warm RPMI 1640 media containing 10% FBS, washed (× 2) with RPMI 1640, and then pelleted for RNA isolation.
RNA isolation, DNase treatment, and cDNA preparation
Total RNA was isolated by the guanidinium thiocyanate/acid phenol method29 using Trizol reagent (Life Technologies, Paisley, United Kingdom) in accordance with the manufacturer's standard method. RNA pellets were washed with 70% ethanol containing diethyl polycarbonate (DEPC), air dried, and redissolved in 0.01% DEPC by heating at 55°C. The integrity and purity of the RNA was assessed by agarose gel electrophoresis (data not shown). Following the quality assessment of the RNA, aliquots were quantitated by absorbance at 260 and 280 nm. Aliquots of each patient and cell line sample (10 to 20 μg) were subjected to DNase treatment using Ampligrade enzymes (Life Technologies) for 15 to 30 minutes at 22°C to degrade genomic DNA. The DNase activity was neutralized by 25 mM EDTA (ethylenediaminetetraacetic acid) (Life Technologies) and heat inactivated by incubation at 65°C for 5 minutes. Samples were divided into 2 groups for preparation of cDNA or cDNA controls, termed RT+ and RT−, respectively. Reverse transcription of the RNA pools was performed using murine Maloney leukemia virus native reverse transcriptase (MMLV-RT) at a concentration of 105 U/mL in the presence of 0.5 mM deoxyribonucleoside triphosphate (dNTP), 5 × 103U/mL RNase inhibitor, and 50 μM random hexamer primers (all from Life Technologies) at 42°C for 2 hours (RT+). The RT− samples contained equimolar quantities of all the reactants except the MMLV-RT enzyme and were used to evaluate potential genomic DNA contamination, which was not detected.
Real-time quantitative PCR (RQ-PCR)
RQ-PCR was carried out using TaqMan probe–based chemistry (Applied Biosystems). This chemistry provides for a high level of specificity through the design of complementary oligonucleotide primers and 5′-reporter/3′ quencher fluorogenic probes. The 5′-reporters for the HOX genes and endogenous controls (18SrRNAand GAPDH) were 6-carboxyfluorescein (FAM) and VIC (Applied Biosystems), respectively. In all cases the 3′ quencher was 6-carboxy-tetramethylrhodamine (TAMRA). During the normal PCR process the fluorogenic probe is cleaved by the native 5′-exonuclease activity of Taq polymerase30 that releases the reporter from the quencher, resulting in the generation of a sequence-specific signal. Each additional cycle results in the release of reporter molecules from the respective probes. The fluorescence intensity is related to the initial number of RNA copies, which can be assessed by determining the threshold cycle (CT).31 AllHOX-specific primers and probes were designed against GenBank-published sequences in association with Primer Express (Applied Biosystems). Endogenous controls were purchased as RNA-specific Pre Developed Assay Reagents (PDARs; Applied Biosystems). The universality of the Taqman system arising from the intrinsic constraints of Primer Express was exploited to measure simultaneously the expression of 27 HOX genes in singleplex mode. Control reactions lacking cDNA template, included to assess specificity, showed no appreciable amplification (CT more than 40; data not shown). The amplification reactions (12.5 μL) contained 50 ng cDNA equivalents (or control), 1 × Taqman universal PCR master mix, final concentrations of 5 mM MgCl2, 0.2 mM deoxyadenosine deoxycytosine deoxyguanosine triphosphate (dA/dC/dGTP), 0.4 mM deoxyuridine triphosphate (dUTP), 0.125 U AmpliTaq Gold, 2 μM primers (forward and reverse), and 200 nM TaqMan probe.
Amplifications were performed following an initial 2-minute incubation at 50°C to allow uracil-N-glycosylase (UNG) to destroy any contaminating RNA, followed by treatment at 95°C for 10 minutes to inactivate the UNG enzyme and activate the AmpliTaq Gold DNA polymerase. This was followed by 40 to 45 cycles of denaturing at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute. An ABI Prism 7700 Sequence Detection System equipped with a 96-well thermal cycler was used for the amplifications. Data were collected and analyzed with Sequence Detector v1.6.3 software (Applied Biosystems).
To reduce the possibility of altered reaction kinetics contributing to the difference in the PCR profile and hence the CT values, the starting template concentration must be similar for each patient and normal sample. For this reason, samples that gave an 18S CT value outside the range of 10 ± 3 were excluded from further analysis. This range of template concentration is well within the guidelines of 5 orders of magnitude as set out in Applied Biosystems' user bulletin no. 2. Relative quantitative (Q–PCR) data based on the ΔΔCT method (reviewed by Ginzinger32) was calculated using the following formulas: If CT18S > 10, then normalized CT(HOX) = actual CT (HOX) − (CT 18S−10); and if CT18S < 10, then normalized CT (HOX) = actual CT (HOX) + (10−CT 18S).
Cloning, sequencing, and validation of RQ-PCR amplicons
Oligonucleotides were designed and developed in association with Applied Biosystems. Due to the high degree of sequence homology between and among HOX paralog groups, it was essential to validate that each amplicon generated during the RQ-PCR process was the desired target by sequencing the product. PCR reactions using the gene-specific forward and reverse primers in the absence of the fluorogenic oligonucleotide probe were performed on the ABI Prism 7700 under exactly the same conditions as the RQ-PCR reactions. cDNA from several cell lines, a fetal brain library, and normal bone marrow were “pooled” to provide a source of template that would contain all potential HOX targets. Amplicons were identified by electrophoresis through a 2.5% agarose gel, excised, and purified using a CONCERT Rapid Gel Extraction System (Life Technologies) following the manufacturer's protocol. Purified amplicons were then directly cloned into the TOPO TA Cloning kit (InVitrogen, Paisley, United Kingdom) and transformed into TOP10F cells (InVitrogen). Plasmid DNA was extracted from ampicillin-resistant Luria-Bertani (LB) broth cultures by the CONCERT Rapid Plasmid Miniprep System (Life Technologies) and sequenced using the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems) along with M13F (17 mer) or M13R (21 mer) primers with an ABI 310 sequence detection system (Applied Biosystems). At least 5 individual colonies were sequenced for each cloning reaction. All sequences were searched against GenBank and Swiss Protein Database (SWISSPROT) using Basic Logic Alignment Search Tool (BLAST)33 and fastall protein or nucleotide comparison search tool (FASTA)34 and verified as being specific for the original target gene.
Differentiation of NB4 cells with ATRA treatment assays
NB4 cells were seeded at a concentration of 2 × 104/mL in standard T-75 tissue culture flasks. Cells were either untreated (control) or treated with 1 μM ATRA or an equivalent volume of vehicle (EtOH) for the indicated times. Cell proliferation was monitored by the direct count method of viable cells using standard Trypan Blue exclusion assays. NB4 cell differentiation was monitored using a direct immunofluorescence assay measuring the myeloid cell surface molecule CD11b as a marker. The phycoerythrin (PE)–conjugated monoclonal anti-CD11b clone 2LPM19c (Dako A/S Denmark) antibody was used in all experiments with appropriate isotype controls (IgG1). The percentage of CD11b+ cells was calculated by the EXPO32 analyzer software (Applied Cytometry Systems, Sheffield, United Kingdom). Morphologic assays were carried out on cytospun specimens using May-Grünwald Giemsa staining and light microscopy.
Results
Design, development, and validation of the RQ-PCR oligonucleotides used
The primers and probes used in the reactions (Table1) were designed and developed using the latest available GenBank sequence information and Primer Express. Direct DNA sequencing of at least 5 clones per target (data not shown) validated the specificity of the amplicons. The amplicon lengths ranged from 56 bp to 107 bp (Table 1). Twenty-seven HOX targets were successfully detected by the designed assays. Plasmid DNA from clones that were sequenced showed either the original target or self-ligation of the cloning vector.
Oligonucleotides used in the SMART-HOX system
| HOX . | Amplicon size, bp . | Accession no. . | Forward primer . | Reverse primer . | 5′FAM/3′TAMRA probe . |
|---|---|---|---|---|---|
| A1 | 63 | NM_005522 | CGGACCATAGGATTACAACTTTCC | GTCGCCGCCGCAACT | TGCGCGGTCAGCGCCAA |
| A4 | 70 | NM_002141 | TGGTGTACCCCTGGATGAAGA | CTTAGGCTCCCCTCCGTTATAAC | TCCATGTCAGCGCCGTTAACCCC |
| A5 | 92 | XM_004918 | TCCCATCGCTTCCCTACCT | GCTTTGGAACAGCCTACAGCTT | ACCCATTGCAAAGTTCAGGGCATAAGGT |
| A7 | 79 | NM_006896 | AGCTTGGAAATTCTGCTCACTTCT | TCTGATGTCATGGCCAAATTTG | CTCTTGCTTGCTTCTCTGGTGGGCTTC |
| A9 | 84 | U82759 | GCCGGCCTTATGGCATTAA | CAGGGACAAAGTGTGAGTGTCAA | TGAACCGCTGTCGGCCAGAAGG |
| A10 | 70 | AF040714 | CAAGAGGCAAAGAAAAGCGTTAA | GTAGATGCTTGCAGAAGGAAAGG | TCAAGATGTACAGCCTGCCCTCCCA |
| A11 | 87 | NM_005523 | TTGAGCATGCGGGACAGTT | GTACCAGATCCGAGAGCTGGAA | AGGCGCTTCTCTTTGTTAATGTAGACGCTGAA |
| B1 | 65 | NM_002144 | GCCATATCCTCCGCAGCAT | CATAGGCCGGTGCAAAGCT | CCCCTTATGGGAACGAGCAGACCG |
| B3 | 58 | NM_002146 | TCACCCAGCGATGCAGAA | CGAGGAATAGCCTCCGAAGA | CCACCTACTACGACAACGCCGCG |
| B5 | 71 | NM_002147 | TCCTTCCATGCTCCCAACTC | CCACAGACACAAACATTCAGAAACA | CTTCTGCTTGTCCCAAATCCGCCA |
| B6 | 56 | NM_018952 | AGCATCTCCACTCTGCGTAACA | AGCGTCTGGTAACGTGTGTATGTCT | CCCAGCGGCCGGCGAG |
| B7 | 61 | NM_004502 | TCGAGCCGAGTTCCTTCAAC | CACACCCCGGAGAGGTTCT | TGCACTGCGCGCCCTTTGAG |
| B8 | 64 | NM_024016 | AAGCGGCGAATCGAGGTA | GGAACCAGATTTTGACCTGTCTCT | CGCACGCCCTGGGACTGACA |
| B9 | 77 | X16172 | AACCCCTCCGCCAACTG | CCAGCGTCTGGTATTTGGTGTA | CACGCTCGCTCTTCCCGGAAAAA |
| B13 | 65 | NM_006361 | GCTGATGCCTGCTGTCAACTAT | ATTGCTTTGGCGGCTCC | CCTTGGATCTGCCAGGCTCGGC |
| C4 | 68 | NM_014620 | GCTGCCGGGTATAAGCTGTACT | CAGTTAGCAGGTGAACCCCAAT | AGCGCTTGGGTTCCCCTCCGTTA |
| C5 | 107 | NM_018953 | TCTGGCATAGGTACCACATAAGGA | GACACGGACAGAGCCAGACA | CACAGGGAAACAGAGCAGTCCTCTAGACCA |
| C6 | 69 | NM_004503 | TCAAACGTGGACCTGAAAGTCA | GGCGCGTGGAGAAAGAGA | TCTGGACCCCCTCCCTCACCG |
| C9 | 66 | AK000445 | TCATCCTTCGATTCTGAAACCA | GGTGGCCCGGGTTCTC | ATTTTGACCTGCCGCTCGGTGAGAT |
| C11 | 75 | XM_012215 | GCGGCCGACGAGCTT | GCCTTCGTTTTTCATGAGGATCT | TGCACCGGGAGTGCCTGCC |
| C12 | 69 | AF328963S2 | GCGAGTTTCTGGTCAACGAGTT | AAGATTCAAGCGGTCTGAGAGTTC | CACACGCCAGCGCCGGAG |
| C13 | 78 | AF263466 | GGCTAGCAAGTTCATCACCAAA | GGTTACCTGGCGCTCAGAGA | CGCATCTCCGCCACCACGAA |
| D3 | 77 | Y09980 | CCACGTGATTGGCGAAATAA | CCTTTGCGTGGCAGATTAATG | AATTCAGCACGTCCCTTAAGAAACACGGAGT |
| D4 | 77 | NM_014621 | GAGAGCAGTCTTTGGATGTACCATT | TTTTAGGAAAGCAGCTTAGCAACTT | AAGGAGCCGCTATCCTTAGGCAAGTTGG |
| D8 | 74 | AY014304 | CTGACCAGGAAAAGAAGAATCGA | TGGAACCAGATTTTTACCTGTCTCT | TTCCCACGCCCTAGCCCTCACC |
| D9 | 75 | NM_014213 | CCCCGAAGCGAACTGGAT | GCTCAAGCGTCTGGTATTTGG | CCCGGAAAAAGCGCTGTCCCTACA |
| D10 | 102 | NM_002148 | CAGCAGCGCCAGCATGT | TTCACTTCTCTTTTGGCCAGAGA | CATGCCACCACCTAGCGCAGACA |
| D13 | 69 | NM_000523 | CACTTCGGCAACGGCTACTAC | CTTGAGCGCATTCTGCTGTAAG | CCGTATGTCGCACGGCGTGG |
| HOX . | Amplicon size, bp . | Accession no. . | Forward primer . | Reverse primer . | 5′FAM/3′TAMRA probe . |
|---|---|---|---|---|---|
| A1 | 63 | NM_005522 | CGGACCATAGGATTACAACTTTCC | GTCGCCGCCGCAACT | TGCGCGGTCAGCGCCAA |
| A4 | 70 | NM_002141 | TGGTGTACCCCTGGATGAAGA | CTTAGGCTCCCCTCCGTTATAAC | TCCATGTCAGCGCCGTTAACCCC |
| A5 | 92 | XM_004918 | TCCCATCGCTTCCCTACCT | GCTTTGGAACAGCCTACAGCTT | ACCCATTGCAAAGTTCAGGGCATAAGGT |
| A7 | 79 | NM_006896 | AGCTTGGAAATTCTGCTCACTTCT | TCTGATGTCATGGCCAAATTTG | CTCTTGCTTGCTTCTCTGGTGGGCTTC |
| A9 | 84 | U82759 | GCCGGCCTTATGGCATTAA | CAGGGACAAAGTGTGAGTGTCAA | TGAACCGCTGTCGGCCAGAAGG |
| A10 | 70 | AF040714 | CAAGAGGCAAAGAAAAGCGTTAA | GTAGATGCTTGCAGAAGGAAAGG | TCAAGATGTACAGCCTGCCCTCCCA |
| A11 | 87 | NM_005523 | TTGAGCATGCGGGACAGTT | GTACCAGATCCGAGAGCTGGAA | AGGCGCTTCTCTTTGTTAATGTAGACGCTGAA |
| B1 | 65 | NM_002144 | GCCATATCCTCCGCAGCAT | CATAGGCCGGTGCAAAGCT | CCCCTTATGGGAACGAGCAGACCG |
| B3 | 58 | NM_002146 | TCACCCAGCGATGCAGAA | CGAGGAATAGCCTCCGAAGA | CCACCTACTACGACAACGCCGCG |
| B5 | 71 | NM_002147 | TCCTTCCATGCTCCCAACTC | CCACAGACACAAACATTCAGAAACA | CTTCTGCTTGTCCCAAATCCGCCA |
| B6 | 56 | NM_018952 | AGCATCTCCACTCTGCGTAACA | AGCGTCTGGTAACGTGTGTATGTCT | CCCAGCGGCCGGCGAG |
| B7 | 61 | NM_004502 | TCGAGCCGAGTTCCTTCAAC | CACACCCCGGAGAGGTTCT | TGCACTGCGCGCCCTTTGAG |
| B8 | 64 | NM_024016 | AAGCGGCGAATCGAGGTA | GGAACCAGATTTTGACCTGTCTCT | CGCACGCCCTGGGACTGACA |
| B9 | 77 | X16172 | AACCCCTCCGCCAACTG | CCAGCGTCTGGTATTTGGTGTA | CACGCTCGCTCTTCCCGGAAAAA |
| B13 | 65 | NM_006361 | GCTGATGCCTGCTGTCAACTAT | ATTGCTTTGGCGGCTCC | CCTTGGATCTGCCAGGCTCGGC |
| C4 | 68 | NM_014620 | GCTGCCGGGTATAAGCTGTACT | CAGTTAGCAGGTGAACCCCAAT | AGCGCTTGGGTTCCCCTCCGTTA |
| C5 | 107 | NM_018953 | TCTGGCATAGGTACCACATAAGGA | GACACGGACAGAGCCAGACA | CACAGGGAAACAGAGCAGTCCTCTAGACCA |
| C6 | 69 | NM_004503 | TCAAACGTGGACCTGAAAGTCA | GGCGCGTGGAGAAAGAGA | TCTGGACCCCCTCCCTCACCG |
| C9 | 66 | AK000445 | TCATCCTTCGATTCTGAAACCA | GGTGGCCCGGGTTCTC | ATTTTGACCTGCCGCTCGGTGAGAT |
| C11 | 75 | XM_012215 | GCGGCCGACGAGCTT | GCCTTCGTTTTTCATGAGGATCT | TGCACCGGGAGTGCCTGCC |
| C12 | 69 | AF328963S2 | GCGAGTTTCTGGTCAACGAGTT | AAGATTCAAGCGGTCTGAGAGTTC | CACACGCCAGCGCCGGAG |
| C13 | 78 | AF263466 | GGCTAGCAAGTTCATCACCAAA | GGTTACCTGGCGCTCAGAGA | CGCATCTCCGCCACCACGAA |
| D3 | 77 | Y09980 | CCACGTGATTGGCGAAATAA | CCTTTGCGTGGCAGATTAATG | AATTCAGCACGTCCCTTAAGAAACACGGAGT |
| D4 | 77 | NM_014621 | GAGAGCAGTCTTTGGATGTACCATT | TTTTAGGAAAGCAGCTTAGCAACTT | AAGGAGCCGCTATCCTTAGGCAAGTTGG |
| D8 | 74 | AY014304 | CTGACCAGGAAAAGAAGAATCGA | TGGAACCAGATTTTTACCTGTCTCT | TTCCCACGCCCTAGCCCTCACC |
| D9 | 75 | NM_014213 | CCCCGAAGCGAACTGGAT | GCTCAAGCGTCTGGTATTTGG | CCCGGAAAAAGCGCTGTCCCTACA |
| D10 | 102 | NM_002148 | CAGCAGCGCCAGCATGT | TTCACTTCTCTTTTGGCCAGAGA | CATGCCACCACCTAGCGCAGACA |
| D13 | 69 | NM_000523 | CACTTCGGCAACGGCTACTAC | CTTGAGCGCATTCTGCTGTAAG | CCGTATGTCGCACGGCGTGG |
Amplicons were verified and validated by cloning and sequencing. Amplicon sequences were interrogated by BLAST33 analysis and aligned with the specific accession number.
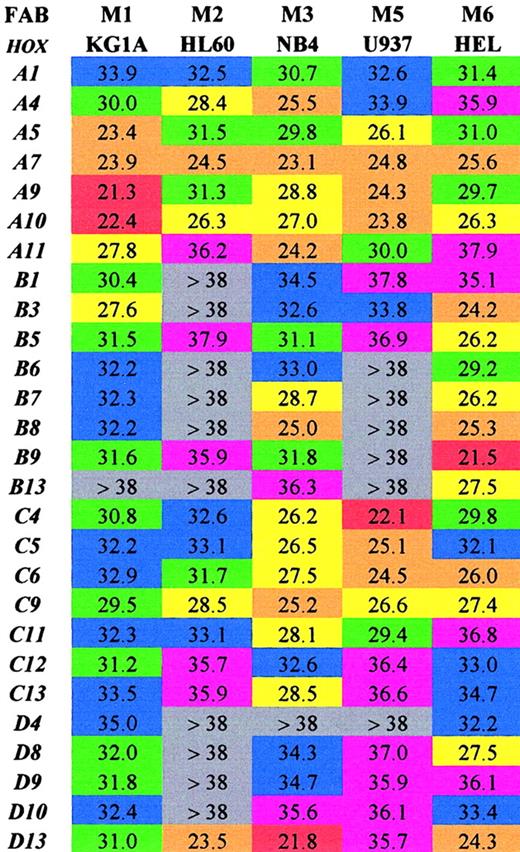
SMART-HOX profiling in leukemic cell lines
Initially the SMART-HOX technology was applied to several well-characterized leukemic cell lines (Figure1). The objective was to assess the expression of 27 HOX genes in various leukemic cell lines in a comprehensive and quantitative manner. Previous studies on leukemic cells have involved the use of standard PCR, Northern blot, or RNase protection assays, which have limitations either due to the amount of template required or the degree of quantitation obtained.15,35-39 SMART-HOX has evolved the measurement of relative gene dosage differences using specific, validated TaqMan reagents. Figure 1 depicts a 2-dimensional readout of the CT values obtained for each cell line (columns) for each particular HOX gene (rows) corrected for starting template concentration by the 18S endogenous control. The values obtained were also validated using GAPDHcontrols, which duplicated the findings in each of the cell lines (data not shown). The CT value for GAPDH in the KG1a cell line was markedly higher than the other cell lines (data not shown), while the 18SrRNA values were consistent among the samples. The CT value reflects the initial concentration of the target cDNA in that it is inversely proportional to the target concentration. Because the CT value is obtained during the exponential phase of the PCR reaction, a decrease of 1 CTvalue between comparative samples is equivalent to a doubling of the initial target concentration—that is, gene expression = 2(ΔCT). The lower the CT value, the higher the expression of that particular HOX gene. A color-coded system is used to highlight differences in HOXgene expression among the leukemic cell lines tested. The highest expressed HOX genes (lowest CT values 20 to 23) are colored red, while very low/absent expression (CTvalues more than 38) are colored gray. The results (Figure 1) were consistent with and expanded on previous studies. There was a preponderance of HOXB expression in the erythroid cell line HEL,27,39 whereas the more myeloid-type cell lines (eg, U937) tended to express the HOXA genes.40 The general high level expression of some 66% of the subset ofHOX genes studied was noted in the NB4 cell line. CertainHOX genes, particularly A7, A10, andC9, appear to be highly expressed ubiquitously throughout the cell lines studied. These HOX genes may play a critical role in basal cellular processes independent of the lineage subtype or differentiation state. In some cases the level of expression of particular HOX genes, A9 in KG1a cells andD13 in NB4 cells, approached the level observed forGAPDH (CT 18 to 20) for the same cells. This was a surprising result because these transcription factors are usually expressed at low levels—hence the use of PCR and RNase protection assays in the past to observe their occurrence. Additional novel findings were in the relatively low expression of most of theHOX genes studied in HL60 cells (only 30% scored as moderately expressed or higher) and in the similarity of profile between HL60 and U937 cells. The high expression of HOXD13in the HL60, NB4, and HEL cell lines was unexpected, becauseD13 was originally considered to be a gene involved in limb development,41,42 although it was recently identified as aNUP98 fusion partner in some AML cases.16,19 20The significance of some of these findings and the link betweenHOX gene expression and potential upstream regulators show the strength of investigating an array of HOX genes at the same time in a quantitative manner. This has previously been difficult and time-consuming. Insights into the role of certain HOXgenes in leukemogenesis are suggested from this approach.
Array of comparative analysis of the CTvalues obtained for 27 HOX genes from 5 well-characterized leukemic cell lines.
Values shown are the mean of 3 experiments corrected for18S. Lower CT values indicate higher expression for the particular gene. Red indicates extremely high (20-23); orange, very high (23-26); yellow, high (26-29); green, moderate (29-32); blue, low (32-35); purple, very low (35-38); and gray, extremely low/not expressed (> 38).
Array of comparative analysis of the CTvalues obtained for 27 HOX genes from 5 well-characterized leukemic cell lines.
Values shown are the mean of 3 experiments corrected for18S. Lower CT values indicate higher expression for the particular gene. Red indicates extremely high (20-23); orange, very high (23-26); yellow, high (26-29); green, moderate (29-32); blue, low (32-35); purple, very low (35-38); and gray, extremely low/not expressed (> 38).
Altered HOX gene expression in differentiating NB4 cells
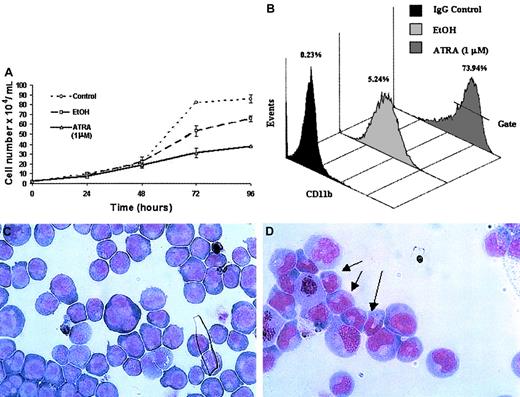
ATRA treatment of NB4 cells over a 96-hour period resulted in a significant decrease in the rate of cell proliferation (Figure2A) as well as the induction of a differentiation response, observed as an increased expression of the cell surface marker CD11b (Figure 2B). Morphologic changes, in particular the presence of band neutrophils, were observed following 96-hour ATRA treatment of NB4 cells but were not seen in vehicle-treated cells (Figure 2C-D). Morphologic changes occurred at the same stage as an increase in apoptosis was observed (data not shown). Taken together these results confirmed the ability of the NB4 cells used to differentiate in the presence of ATRA. To gain functional insights into this differentiation process, the expression of 27 HOX genes was monitored at 2 different time points, one following short-term (4 hours) exposure to ATRA (Figure3A), the other (48 hours) following the establishment of the differentiation response (Figure3B). The levels of HOX gene expression following terminal differentiation were not investigated due to the high level of apoptosis in these cells. HOX gene expression (as actual CT values corrected for 18S) was compared with vehicle. Most HOX genes showed no significant difference in expression 4 hours following ATRA treatment (Figure 3A and data not shown). However, there was a moderate increase in the expression of several HOXA cluster genes (decrease in CT values) and a moderate decrease (increase in CT values) in the expression of other genes (eg,HOXB9). Following differentiation (48-hour time point) there appeared to be a moderate down-regulation (increased CTvalues) for many of the HOX genes studied. The fold difference in HOX gene expression was calculated from the following formula: fold difference = 2ΔCT, where ΔCT = CT (EtOH) − CT(ATRA). The expression of HOXC12 increased 20-fold within 4 hours and was maintained or slightly enhanced (23-fold) following longer-term (48 hours) ATRA treatment (Figure 3B-C). The constitutively highly expressed HOXD13 was down-regulated 5-fold within 4 hours of ATRA treatment that was also maintained (6.5-fold) during the differentiation process. These significant changes occurred in the presence of more moderate differences in several other genes, including HOXA1, HOXA11, and HOXB9, suggesting a degree of specificity in the response to ATRA rather than a global effect on HOX gene expression.
Differentiation of NB4 cells following exposure to ATRA.
NB4 cells seeded at a density of 2 × 104/mL in RPMI 1640 media containing 10% FBS were left untreated (control) or treated with either all-trans retinoic acid (ATRA) at a concentration of 1 μM or vehicle (EtOH) for the indicated times. Differentiation was indicated as a decrease in the rate of proliferation, compared with controls, as measured by the direct counting method of viable cells (A); each data point represents the mean of 3 independent experiments with standard errors. NB4 differentiation was monitored by (B) a direct immunofluorescence assay using a phycoerythrin-conjugated monoclonal anti-CD11b antibody or isotype control. The percentage values of gated CD11b+ cells identified following 48 hours of ATRA or vehicle treatment were evaluated on the FL2 channel. This assay was carried out on all NB4 cells used in subsequent analyses; a representative output of 3 experiments is shown. Terminal differentiation of NB4 cells treated with EtOH (C) or ATRA (D) for 96 hours was analyzed by microscopic examination (original magnification × 1000) of glass slides following May-Grünwald Giemsa staining. Nuclear segmentation (arrow) and granule formation were observed in the ATRA-treated cells. A representative of 3 independent experiments is shown.
Differentiation of NB4 cells following exposure to ATRA.
NB4 cells seeded at a density of 2 × 104/mL in RPMI 1640 media containing 10% FBS were left untreated (control) or treated with either all-trans retinoic acid (ATRA) at a concentration of 1 μM or vehicle (EtOH) for the indicated times. Differentiation was indicated as a decrease in the rate of proliferation, compared with controls, as measured by the direct counting method of viable cells (A); each data point represents the mean of 3 independent experiments with standard errors. NB4 differentiation was monitored by (B) a direct immunofluorescence assay using a phycoerythrin-conjugated monoclonal anti-CD11b antibody or isotype control. The percentage values of gated CD11b+ cells identified following 48 hours of ATRA or vehicle treatment were evaluated on the FL2 channel. This assay was carried out on all NB4 cells used in subsequent analyses; a representative output of 3 experiments is shown. Terminal differentiation of NB4 cells treated with EtOH (C) or ATRA (D) for 96 hours was analyzed by microscopic examination (original magnification × 1000) of glass slides following May-Grünwald Giemsa staining. Nuclear segmentation (arrow) and granule formation were observed in the ATRA-treated cells. A representative of 3 independent experiments is shown.
Real-time quantitative PCR analysis of HOXgene expression in NB4 cells treated with ATRA.
Complementary DNAs obtained from NB4 cells were analyzed forHOX gene expression using the SMART-HOX platform. The data graphically presented are the cycle threshold (CT) values of the particular HOX gene normalized to the endogenous 18SrRNA control. All data points are the mean of 3 independent experiments plus standard errors for the particularHOX genes following (A) short-term ATRA exposure (░, 4-hour EtOH; ▪, 4-hour ATRA) or (B) the onset of differentiation after ATRA exposure (░, 48-hour EtOH; ▪, 48-hour ATRA). The CT value is inversely proportional to the gene expression. The most significant changes in HOX gene expression following ATRA treatment are plotted (C) as relative fold differences in the means of 3 experiments over the appropriate vehicle control, where fold difference = 2ΔCT and ΔCT= CT (EtOH) − CT (ATRA). In the cases where ΔCT was a negative integer, the fold difference was calculated as − 2ΔCT. ░ indicates 4-hour ATRA; and ▪, 48-hour ATRA.
Real-time quantitative PCR analysis of HOXgene expression in NB4 cells treated with ATRA.
Complementary DNAs obtained from NB4 cells were analyzed forHOX gene expression using the SMART-HOX platform. The data graphically presented are the cycle threshold (CT) values of the particular HOX gene normalized to the endogenous 18SrRNA control. All data points are the mean of 3 independent experiments plus standard errors for the particularHOX genes following (A) short-term ATRA exposure (░, 4-hour EtOH; ▪, 4-hour ATRA) or (B) the onset of differentiation after ATRA exposure (░, 48-hour EtOH; ▪, 48-hour ATRA). The CT value is inversely proportional to the gene expression. The most significant changes in HOX gene expression following ATRA treatment are plotted (C) as relative fold differences in the means of 3 experiments over the appropriate vehicle control, where fold difference = 2ΔCT and ΔCT= CT (EtOH) − CT (ATRA). In the cases where ΔCT was a negative integer, the fold difference was calculated as − 2ΔCT. ░ indicates 4-hour ATRA; and ▪, 48-hour ATRA.
SMART-HOX profiling in APL patient samples
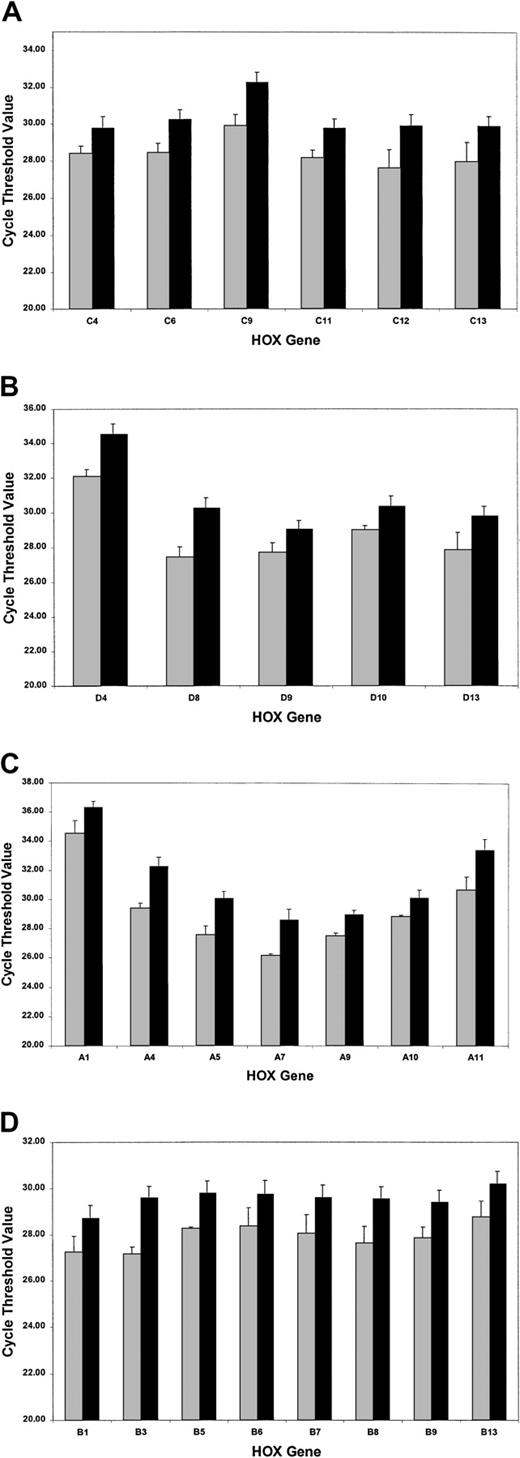
The SMART-HOX platform was applied to cDNAs obtained from 16 PML-RARα+ cases of APL and compared with normal bone marrow cDNA samples (n = 3). SMART-HOXassays were carried out in triplicate on a 96-well plate format with endogenous control genes (18SrRNA and GAPDH) and analyzed with corresponding negative controls (minus template). CT values were normalized to either 18S orGAPDH values, which gave comparable results. 18Snormalized values were plotted (Figure4). APL patients exhibited a global down-regulation (increase in CT value) ofHOX gene expression, within paralogs and across clusters for all 26 genes assayed (HOXC5 was omitted due to insufficient quantity), compared with healthy controls (Figure 4). The degree of down-regulation did not appear to follow any defined pattern. The most significant fold reductions in expression (Figure5) included such divergent genes as HOXA4, HOXA11, HOXB3, HOXC9, and HOXD8. Of the 26 genes assayed, the most significant changes occurred in 71% of theHOXA and HOXC clusters (5 of 7), 37.5% of theHOXB cluster (3 of 8), and 60% of the HOXDcluster (3 of 5). The fold decrease in expression ranged from 1.8(HOXC4) to 7.4 (HOXA4). A pattern of expression throughout the HOXA cluster whereby the low (1 to 4) and high (10 to 11) numbered paralogs were expressed at lower levels than the mid (5 to 9) paralogs was observed in both the healthy and APL patient samples. This pattern of expression has also been noted in other AML and ALL patient samples (data not shown).
Global down-regulation of HOX gene expression in APL patient samples compared with normal bone marrow controls.
SMART-HOX was applied to cDNAs obtained from APL or normal bone marrow samples. Data presented are the mean CTvalues normalized for 18SrRNA and standard errors from triplicate experiments for (A) the HOXA cluster, (B) theHOXB cluster, (C) the HOXC cluster, and (D) theHOXD cluster genes (bone marrow, n = 3; APL, n = 16). ░ indicates bone marrow (BM); and ▪, APL.
Global down-regulation of HOX gene expression in APL patient samples compared with normal bone marrow controls.
SMART-HOX was applied to cDNAs obtained from APL or normal bone marrow samples. Data presented are the mean CTvalues normalized for 18SrRNA and standard errors from triplicate experiments for (A) the HOXA cluster, (B) theHOXB cluster, (C) the HOXC cluster, and (D) theHOXD cluster genes (bone marrow, n = 3; APL, n = 16). ░ indicates bone marrow (BM); and ▪, APL.
Relative fold reduction of a subset of HOXgenes in APL patient samples compared with normal controls.
Relative fold values were obtained from the data in Figure 4using the following formula: fold difference = 2(ΔCT), where ΔCT = CT (APL) − CT (BM). The values are plotted as a measure of the fold decrease in expression for the particular gene observed in the APL subgroup compared with the normal samples. For clarity, only the more significant decreases (at least 3-fold) in expression are shown.
Relative fold reduction of a subset of HOXgenes in APL patient samples compared with normal controls.
Relative fold values were obtained from the data in Figure 4using the following formula: fold difference = 2(ΔCT), where ΔCT = CT (APL) − CT (BM). The values are plotted as a measure of the fold decrease in expression for the particular gene observed in the APL subgroup compared with the normal samples. For clarity, only the more significant decreases (at least 3-fold) in expression are shown.
Discussion
The PML-RARα fusion protein associated with APL functions as an aberrant retinoid receptor in the establishment of a myeloid-specific differentiation block, but the precise molecular consequences of this fusion product remain unclear. Recent studies of forced underexpression or overexpression of particular HOXgenes have shown a marked effect on myeloid cell maturation.43-47 To investigate possible links between the presence of PML-RARα and HOX gene expression, a novel strategy has been applied to APL patient samples and NB4 cells. The SMART-HOX platform permitted rapid and simultaneous measurement of 27 HOX genes. Initially cloning and sequencing individual amplicons validated this system. Subsequently HOX gene expression was measured in 5 distinct leukemic cell lines. Comparative analysis of HOX gene expression normalized to endogenous controls (Figure 1) resulted in refinement of profiles previously reported.35,38,39,48 Significant expression of the HOXA cluster in the myeloid cell lines (HL60 and U937) and the HOXB cluster in the erythroid cell line (HEL) is consistent with data previously obtained by Northern analysis.39 Previous techniques have also suggested that expression of the HOXC cluster is limited to the lymphoid compartment48; so the expression of severalHOXC cluster genes in nonlymphoid cells (U937, NB4, and HEL) reported here is a novel observation (Figure 1). Similarly, constitutively high expression of HOXD13 in several leukemic cell lines (HL60, NB4, and HEL) has not previously been reported.
Leukemic cell lines provide important models of dysregulated hematopoiesis, but immortalization or transformation can directly perturb the expression of genes involved in regulatory processes, such as HOX genes. It is possible that this holds for the NB4 cell line, which showed moderate to high expression of 18 of the 27 genes assayed. NB4 cells were originally isolated from long-term cultures of APL leukemic blasts as a late event and have a complex karyotype.49 The cells do, however, retain thePML-RARα and RARα-PML fusion genes resulting from the t(15;17) translocation and differentiate in response to ATRA, which makes them a valuable model of APL (Figure 2).
ATRA treatment of NB4 cells resulted in moderate up-regulation ofHOXA1 (4- to 5-fold) as previously reported50and down-regulation of several other genes, namely HOXA9, A11, B9, C5, and C9 (Figure 3A-B). A more significant up-regulation of HOXC12 (at least 20-fold) and down-regulation of HOXD13 (at least 5-fold) were also observed (Figure 3C). In both cases the ATRA effect occurred within 4 hours of treatment and was increased at 48 hours when the NB4 cells had begun to differentiate. These results agree with reports that pharmacologic doses of ATRA release the block in myeloid differentiation and suggest that early and sustained modulation of particular HOX genes is involved in this process.
To examine the relationship between HOX genes and APL in vivo, diagnostic PML-RARα+ patient samples were analyzed and compared with normal bone marrow controls. Down-regulation in the expression of 26 HOX genes analyzed was observed (Figure 5). Previous studies of APL patient samples by RNase protection assays showed no detectable signal for HOXA937 or HOXA1014 that corresponds to the high CT value observed here. The RARα gene product is a member of the nuclear hormone receptor superfamily along with its obligate dimerization partner retinoid X receptor (RXR). In the absence of retinoic acid (RA), the RARα/RXR heterodimer can repress transcription by recruiting N-CoR/SMRT/Sin3 and HDACs.51 This phenomenon is mimicked by the PML-RARα fusion protein.52 The presence of RARα/RXR binding-site elements (RAREs) has been established in the promoter regions of several HOX genes (eg, HOXA1, A4, A7, A9, B1, D4).53-57 The present study suggests a model where either a common positive regulator (eg, MLL) or theHOX genes en masse are direct targets of the PML-RARα nuclear corepressor-HDAC complex. Alternatively, the differentiation blockage associated with APL may have resulted in the enrichment of a population of bone marrow cells that express HOX genes at a low level. Regardless of the regulatory pathway involved, SMART-HOX will now provide a practical approach to gain additional functional insights into the differentiation block associated with the PML-RARα repressor complex found in APL.
We thank Dr Steve Picton and Dr Adam Corner (Applied Biosystems) for their expert advice and support, Dr Colin McGuckin and Nico Forraz for providing normal bone marrow samples, and Dr Francisco Lopez-Gordillo for his technical assistance.
Supported by the Northern Ireland Leukaemia Research Fund (A.T., C.M.O.), Leukaemia Research Fund of Great Britain (F.C., D.G.), and Action Cancer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Terence R. J. Lappin, Queen's University Belfast, Department of Haematology, University Floor, Tower Block Belfast City Hospital, Lisburn Rd, Belfast BT97AB, Northern Ireland; e-mail: t.lappin@qub.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal