Children with Down syndrome (DS) with acute myeloid leukemia (AML) have significantly higher event-free survival rates compared to those with non-DS AML, linked to greater cytosine arabinoside (ara-C) sensitivity and higher transcript levels of the chromosome 21–localized gene, cystathionine-β-synthase(CBS), in DS myeloblasts. In this study, we examined the transcriptional regulation of the CBS gene in the DS megakaryocytic leukemia (AMkL) cell line, CMK, characterized by significantly higher CBS transcripts compared with the non-DS AMkL cell line, CMS. Rapid amplification of 5′-cDNA ends (5′-RACE) analysis demonstrated exclusive use of the CBS−1b promoter in the cell lines, and transient transfections with the full-length CBS −1b luciferase reporter gene construct showed 40-fold greater promoter activity in the CMK than CMS cells. Electrophoretic mobility shift assays showed enhanced binding of the transcription factors Sp1/Sp3 to 2 GC/GT-box elements (GC-f and GT-d) in the upstream regions of the CBS −1b promoter in CMK nuclear extracts and undetectable binding in CMS cells. Mutation of the GC-f– or GT-d–binding site resulted in an approximately 90% decrease of theCBS −1b promoter activity in transient transfections of CMK cells. Chromatin immunoprecipitation assays confirmed in vivo binding of Sp3, USF-1, and nuclear factor YA (NF-YA) to theCBS −1b promoter region in chromatin extracts of CMK and CMS cells. Decreased binding of Sp1/Sp3 in CMK nuclear extracts following treatment with calf alkaline phosphatase suggested a role for phosphorylation of Sp1/Sp3 in regulating CBS promoter activity and in the differential CBS expression between CMK and CMS cells. The results of this study with clinically relevant cell line models suggest potential mechanisms for disparate patterns ofCBS gene expression in DS and non-DS myeloblasts and may, in part, explain the greater sensitivity to chemotherapy shown by patients with DS AML.
Introduction
Acute myeloid leukemia (AML) is the second most common form of leukemia in the pediatric population and the most common form of acute leukemia in adults. The 7 established subtypes of AML differ based on morphologic appearance, surface antigen expression, and characteristic leukemia karyotypes. Acute megakaryocytic leukemia (AMkL; M7) was the most recently classified AML subtype by the French-American-British (FAB) group (in 1985), more than 50 years after its initial description by Von Boros et al in 1931.1,2With current techniques, AMkL can be readily diagnosed based on the identification of surface expression of platelet-associated membrane antigens (glycoprotein IIb/IIIa) and associated bone marrow fibrosis.3 AMkL is estimated to represent 3% to 14.6% of pediatric AML cases and 1% of adult AML cases.4-6 The majority of AMkL cases are not characterized by particular chromosomal alterations, although infant AMkL cases frequently contain the translocation, t(1;22)(p13;q13).7 AMkL cases may occur as secondary leukemias and have also been associated with mediastinal germ cell tumors.5 8
Children with Down syndrome (DS) have a significantly increased risk of developing both acute lymphoblastic leukemia (ALL) and AML compared to children without DS.9 DS AML cases have unique biologic features including a predominance of the AMkL phenotype and significantly higher event-free survival (EFS) rates and lower relapse rates compared to non-DS AML patients treated with cytosine arabinoside (ara-C)–based protocols.9 Zipursky et al estimated that DS children have a 500-fold greater risk of developing AMkL compared with non-DS children.10
We previously described a relationship between the expression of the gene, cystathionine-β-synthase (CBS; localized to 21q22.3) and increased in vitro and in vivo ara-C sensitivity of DS myeloblasts over non-DS myeloblasts.11,12CBScatalyzes the condensation of serine and homocysteine to form cystathionine, and increased CBS activity may have downstream effects on reduced folate and S-adenosylmethionine pathways, resulting in increased sensitivity to ara-C. CBS transcript levels in DS myeloblasts were a median 12-fold higher (far exceeding levels predicted by the gene dosage of 3 copies of the CBSgene in DS cells) than for non-DS myeloblasts, and CBStranscripts correlated with the generation of ara-C triphosphate (ara-CTP), the active intracellular ara-C metabolite, following in vitro incubations with 3H-ara-C.12 These results were recently replicated in a CBS-transfected leukemia cell line (CCRF-CEM) model.13
The human CBS gene spans over 30 kb consisting of 23 exons and the CBS polypeptide is encoded by exons 1-14 and 16.14Our recent studies began to explore the transcriptional regulation of the CBS gene in non-AML cells and demonstrated important transactivating roles for critical transcription factors, including Sp1/Sp3, NF-Y, and USF1.15 16 In this study, we extended these results to clinically relevant DS and non-DS AMkL cell lines to better understand the molecular bases for variations in CBSgene expression in AML and, potentially, the high EFS rates of DS AML patients. Identifying mechanisms of differential expression of chromosome 21–localized genes in DS cells could also provide insights into leukemogenesis in DS and the DS phenotype, in general.
Materials and methods
Chemicals and reagents
γ-[32P] adenosine triphosphate (ATP; 3000 Ci/mmol [10 000 × 1010 Bq]) was purchased from Dupont-New England Nuclear (Boston, MA). Synthetic oligonucleotides were purchased from Genosys Biotechnologies (The Woodlands, TX). Restriction and modifying enzymes, reporter gene vectors (pGL3-Basic, pRLSV40), and other molecular biologicals were purchased from Promega (Madison, WI). [5-3H]-Cytosine-β-arabino-furanoside was obtained from Moravek Biochemicals (Brea, CA). Unlabeled ara-C was obtained from Sigma Chemical (St Louis, MO). Tissue culture reagents and supplies were purchased from assorted vendors.
Cell culture
The HepG2 human hepatocellular carcinoma cell line was obtained from American Type Culture Collection (ATCC; Manassas, VA) and cultured as previously described.16 The DS AMkL cell line, CMK (established from a 1-year-old DS boy with AMkL),17was obtained from the German Collection of Microorganisms and Cell Cultures; the non-DS AMkL cell line, CMS (established from a 2-year-old non-DS girl with AMkL),17 was a gift from Dr A. Fuse (National Institute of Infectious Diseases, Tokyo, Japan). The CMK and CMS cell lines were maintained in RPMI 1640 medium containing 10% heat-inactivated iron-supplemented calf serum (Hyclone Labs, Logan, UT), 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified atmosphere at 37°C in the presence of 5% CO2/95% air. The megakaryocytic phenotypes of the CMK and CMS cell lines were confirmed by flow cytometric analysis.
RT-PCR analysis of CBS transcripts
Total RNAs were isolated from 5 × 106 HepG2, CMK, and CMS cells using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA). Reverse transcription–polymerase chain reaction (RT-PCR) was performed as previously described.16 Levels of CBS transcripts were normalized to the levels of 18S ribosomal RNA (localized to chromosome 13; Ambion, Austin, TX).
Rapid amplification of 5′-cDNA ends (5′-RACE) assay
5′-RACE was performed using a 5′-RACE system (version 2.0; Life Technologies) and total RNAs from the leukemia cell lines, CMK and CMS, as previously described.16
In vitro drug cytotoxicity assay
For the determination of cytotoxicity, the DS and non-DS leukemia cell lines were cultured in complete medium with dialyzed calf serum in 24-well plates at a density of 50 000 cells/mL media. Cells were cultured continuously with a range of concentrations of ara-C at 37°C and the cell numbers were counted after 4 days with trypan blue staining. The 50% inhibitory concentration (IC50) values were calculated as the concentration of drug necessary to inhibit 50% growth compared to control cells grown in the absence of drug.
Ara-C incubations and measurement of ara-CTP
Incubation of leukemia cells with 3H-ara-C and the measurement of intracellular ara-CTP levels were performed as previously described.11
Construction of luciferase plasmids and site-directed mutagenesis
The full-length CBS −1b promoter reporter gene construct pCBSb-4046/-3565, and the CAAT-box, E-box, GC- and GT-box–mutated CBS −1b promoter constructs were generated as previously described.15 16 Mutation primers are summarized in Table 1.
Summary of oligonucleotides for gel shifts and mutagenesis
| Oligonucleotide . | Sequence . |
|---|---|
| FPA (−3994 to −3967) | GCTGGAACCCGGGGCGGGCGGGCTCAGG |
| FPA-GC-g mt | GCTGGAACCCGGGGCtGtCGGGCTCAGG |
| FPB (−3957 to −3933) | AGGCGGGGGAGGGGGCGGCGACCCG |
| FPB-GC-f mt | AGGCGGGGGAtGGttCGGCGACCCG |
| FPC (−3876 to −3845) | CATGCCGGGGTCCCGGGGAGGGCGGGGGTCGG |
| FPD (−3816 to −3787) | GGGTGGGGAAGGGGTGGGGGGGAGGGGCCC |
| FPD-GT-d mt | GGGTGGGGAAGGGtTGGGGGGGAGGGGCCC |
| −3766/−3725 | GGGGCTGGGCGGGCGGGTGCTCTGCCACGAGACATTGGCCG |
| USF-1 mt | GGGGCTGGGCGGGCGGGTGCTCTGCCAtcAGACATTGGCCG |
| −3724/−3682 | CGCCGGCCTCCGCCCCCGCCTCTCCGCCAATCACCGCCCGCCT |
| NF-Y mt | CGCCGGCCTCCGCCCCCGCCTCTCCGCtAtTCACCGCCCGCCT |
| Oligonucleotide . | Sequence . |
|---|---|
| FPA (−3994 to −3967) | GCTGGAACCCGGGGCGGGCGGGCTCAGG |
| FPA-GC-g mt | GCTGGAACCCGGGGCtGtCGGGCTCAGG |
| FPB (−3957 to −3933) | AGGCGGGGGAGGGGGCGGCGACCCG |
| FPB-GC-f mt | AGGCGGGGGAtGGttCGGCGACCCG |
| FPC (−3876 to −3845) | CATGCCGGGGTCCCGGGGAGGGCGGGGGTCGG |
| FPD (−3816 to −3787) | GGGTGGGGAAGGGGTGGGGGGGAGGGGCCC |
| FPD-GT-d mt | GGGTGGGGAAGGGtTGGGGGGGAGGGGCCC |
| −3766/−3725 | GGGGCTGGGCGGGCGGGTGCTCTGCCACGAGACATTGGCCG |
| USF-1 mt | GGGGCTGGGCGGGCGGGTGCTCTGCCAtcAGACATTGGCCG |
| −3724/−3682 | CGCCGGCCTCCGCCCCCGCCTCTCCGCCAATCACCGCCCGCCT |
| NF-Y mt | CGCCGGCCTCCGCCCCCGCCTCTCCGCtAtTCACCGCCCGCCT |
DNA sequences for wild-type double-stranded oligonucleotides were used as probes or competitive oligonucleotides in gel shift assays. Single-stranded mutant oligonucleotides were used in the preparation of mutant reporter gene constructs for transient transfection experiments. The consensus sequences of the GC/GT-box, E-box, and CAAT-box elements are underlined. Lowercase letters indicate the bases mutated.
Transient transfections and luciferase assays
CBS-luciferase reporter gene constructs in pGL3-Basic or the promoterless vector (5 μg) were cotransfected with 250 ng pRLSV40 (Promega) into 5 × 106 CMK and CMS cells (in 1 mL Opti-MEM) using Lipofectin reagent (Invitrogen Life Technologies) in accordance with the manufacturer's protocols. Lipofectin treatments were for 24 hours and, after an additional 48 hours of incubation in complete medium, cells were harvested and lysates prepared. Firefly luciferase activities were assayed with a Dual-Luciferase Reporter Assay System (Promega) in a Turner TD20/20 luminometer and normalized to Renilla luciferase activity.
Preparation of nuclear extracts and electrophoretic mobility shift assays
Nuclear extracts from CMK and CMS cells were prepared as previously described.15 16 Complementary single-stranded oligonucleotides (Table 1) were annealed, end-labeled with32P, and purified using Sephadex G-25 quick spin columns (Boehringer-Mannheim, Indianapolis, IN). Nuclear proteins were preincubated in a reaction buffer containing 20 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, pH 7.9, 2 mM MgCl2, 1 mM EDTA (ethylenediaminetetraacetic acid), 50 mM KCl, 0.5 mM dithiothreitol, 10% glycerol, 0.1% Nonidet P-40, and 2 μg poly(dI-dC). After 10 minutes, the32P-end–labeled duplex oligonucleotide (2 × 105 cpm) was added, and the reaction was incubated for another 20 minutes on ice. DNA/protein complexes were separated on 5% nondenaturing polyacrylamide gels in 0.5 × Tris-borate-EDTA (TBE, pH 8.4) at 4°C and 35 mA. The gels were dried and the complexes were visualized by autoradiography.
Chromatin immunoprecipitation assay
We used a modification of the technique for the chromatin immunoprecipitation assay (ChIP) described by Boyd and Farnham.18 Formaldehyde (Fisher Scientific, Pittsburg, PA) was added directly to cell culture media at a final concentration of 1% followed by a room temperature incubation for 10 minutes. Glycine was added to a final concentration of 125 mM, and the cells were incubated for an additional 5 minutes at room temperature. Cells were collected by centrifugation, washed twice with cold phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), and swelled in 5 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; pH 8.0), 85 mM KCl, 0.5% NP40, 0.5 mM PMSF, and 100 ng/mL leupeptin and aprotinin, incubated on ice for 10 minutes, and lysed with a Dounce homogenizer. Nuclei were collected by microcentrifugation at 5000 rpm, resuspended in sonication buffer (1% sodium dodecyl sulfate [SDS], 10 mM EDTA, 50 mM Tris-HCl [pH 8.0], 0.5 mM PMSF, and 100 ng/mL leupeptin and aprotinin) and incubated on ice for 10 minutes. The nuclei were sonicated on ice to an average length of 500 to 1000 bp and then clarified at 14 000 rpm for 15 minutes. The chromatin solution was adjusted to a concentration of 100 A260 U/mL. Equal aliquots were used for individual ChIP reactions, and the remaining aliquots were stored frozen at −80°C. The chromatin solution was precleared by addition of protein A or G beads (80 μL each, Santa Cruz Biotechnologies, Santa Cruz, CA) for 1 hour at 4°C. Precleared chromatin (100 A260 U) was incubated with 30 μg Sp1 antibody (mouse monoclonal antibody, Santa Cruz Biotechnologies), or 10 μg each of Sp3, USF-1, or nuclear factor YA (NF-YA) antibodies (rabbit polyclonal antibody, Santa Cruz Biotechnologies) at 4°C for 12 hours. Immunoprecipitation, washing, and elution of immune complexes were carried out as described.18 Cross-links were reversed by addition of NaCl to a final concentration of 200 mM followed by incubation at 65°C for 5 hours. Samples were extracted with phenol/chloroform and then precipitated at −20°C by the addition of 2 volumes of ethanol and then pelleted by microcentrifugation. Samples were resuspended in 50 μL sterile H2O, and 2 to 4 μL was used in each PCR. Total input samples were resuspended in 100 μL sterile H2O and then diluted 1:100 before PCR.
Standard PCR for the CBS −1b promoter region spanning the basal and the upstream elements was performed with a sense primer (5′-GAATCAACAGGGCGTGGGAATGGGG-3′; positions −3897 to −3921) and semi-nested antisense primers (5′-AGGCGGAGACGGCGACCCC-3′ and 5′-GCGAGGGGAGCAGGCGGGCGGTGATTG-3; positions −3625 to −3643 and −3671 to −3697, respectively). Final PCR products were analyzed on agarose gels (2%) with ethidium bromide staining.
Western blot analysis
Nuclear proteins were isolated from CMK and CMS cells, as described. Then, 50-μg aliquots of each nuclear protein or 100-μg aliquots of each cellular extract were fractionated on a polyacrylamide gel (7.5% for Sp1 and Sp3; 12% for USF1 and NF-YA; 10% for CBS) with SDS and electroblotted onto a polyvinylidene difluoride (PVDF) membranes. The blots were blocked overnight at room temperature in TTBS (Tween Tris-based saline with 0.1% Tween 20, pH 7.5) containing 1% fat-free dried milk powder and were then incubated with Sp1 (rabbit polyclonal antibody, Geneka, Montreal, QC, Canada), Sp3, USF-1 or NF-YA (rabbit polyclonal antibody, Santa Cruz Biotechnologies) antibodies or CBSantiserum in TTBS containing 0.5% fat-free dried milk powder for 2 hours at room temperature. The blots were washed with TTBS and incubated with a second antibody (goat antirabbit IgG linked to horseradish peroxidase conjugate, diluted 1:5000 in TTBS-0.5% milk powder) for 1 hour at room temperature, and detected by Lumi-Light Western Blotting Substrate (Roche Diagnostics, Indianapolis, IN).
Dephosphorylation of nuclear extracts
Nuclear extracts of CMK cells (100 μg) were incubated with or without 20 U calf alkaline phosphatase (Promega) for 1 hour at 37°C in a total volume of 30 μL. The reactions were stopped by the addition of a mixture of phosphatase inhibitors.15 Ten micrograms of the treated nuclear extract were used for electrophoretic mobility shift assay (EMSA), as described.
Results
CBS gene expression in CMK and CMS cells
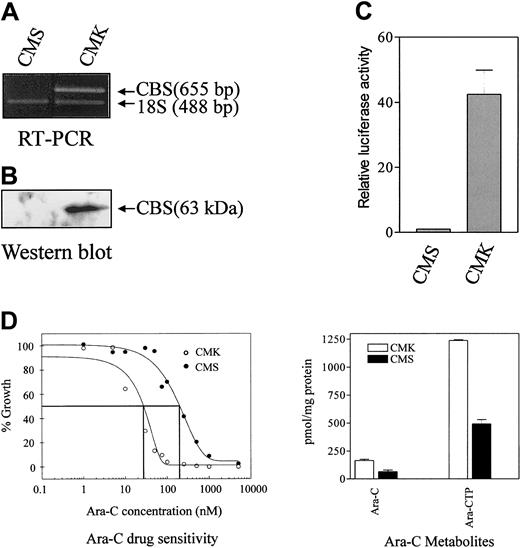
Our earlier studies documented the differential expression ofCBS transcribed exclusively from the CBS −1b promoter in clinical DS and non-DS myeloblasts.19 The DS AMkL cell line, CMK, and non-DS AMkL cell line, CMS, appeared to closely resemble the primary AML specimens from patients because CBS transcripts were readily detected by RT-PCR in the CMK cells, though they were undetectable in the CMS cells (Figure1A). Similar results were obtained by Western blot analysis of CBS protein levels in the CMK and CMS cells (Figure 1B).
Analysis of CBS transcripts by RT-PCR, protein levels by Western blot, transient transfections of CBS −1b promoter and ara-C sensitivities/metabolism in clinically relevant AML cell lines.
(A) Results are shown for an RT-PCR analysis of CBStranscripts in the CMK (DS) and CMS (non-DS) AMkL cell lines normalized to 18S RNA transcript levels, demonstrating CBS transcripts in the DS CMK cell line and a lack of transcripts in the non-DS CMS cell line. (B) A Western blot analysis of CBS protein levels in CMS and CMK cells, as described in “Materials and methods.” (C) Data are shown for transient transfections of the CMS and CMK cell lines with the full-length CBS luciferase reporter gene construct (positions −4046 to −3565) demonstrating approximately 40-fold greater promoter activity in the CMK compared with CMS cell line. The data are presented as means ± SE from 3 independent experiments performed in duplicate. (D) In vitro ara-C sensitivities of the CMK and CMS cell lines measured by the percentage growth inhibition of the cell lines incubated for 4 days with ara-C. (Left) Following in vitro incubation of the cell lines with 5 μM 3H-ara-C for 3 hours, the 3H-ara-CTP generation was assessed by high-performance liquid chromatography (HPLC). The metabolites are the mean of 3 independent experiments ± SE (right).
Analysis of CBS transcripts by RT-PCR, protein levels by Western blot, transient transfections of CBS −1b promoter and ara-C sensitivities/metabolism in clinically relevant AML cell lines.
(A) Results are shown for an RT-PCR analysis of CBStranscripts in the CMK (DS) and CMS (non-DS) AMkL cell lines normalized to 18S RNA transcript levels, demonstrating CBS transcripts in the DS CMK cell line and a lack of transcripts in the non-DS CMS cell line. (B) A Western blot analysis of CBS protein levels in CMS and CMK cells, as described in “Materials and methods.” (C) Data are shown for transient transfections of the CMS and CMK cell lines with the full-length CBS luciferase reporter gene construct (positions −4046 to −3565) demonstrating approximately 40-fold greater promoter activity in the CMK compared with CMS cell line. The data are presented as means ± SE from 3 independent experiments performed in duplicate. (D) In vitro ara-C sensitivities of the CMK and CMS cell lines measured by the percentage growth inhibition of the cell lines incubated for 4 days with ara-C. (Left) Following in vitro incubation of the cell lines with 5 μM 3H-ara-C for 3 hours, the 3H-ara-CTP generation was assessed by high-performance liquid chromatography (HPLC). The metabolites are the mean of 3 independent experiments ± SE (right).
Rapid amplification of 5′-cDNA ends (5′-RACE) assay was performed to confirm whether the CBS transcripts were transcribed from the CBS −1b promoter, as with the primary patient samples. Although there was some heterogeneity in the cDNA 5′ termini, essentially identical results for both lines were obtained, with all the transcripts containing 5′-untranslated regions (UTRs) with exon −1b sequence (data not shown). There were no sequence differences between the 5′-RACE products and the published GenBank sequence for human CBS (accession number AF042836). The different levels of CBS transcripts between CMK and CMS cells were likely due to differences in promoter activity because transient transfections with the full-length CBS −1b luciferase reporter gene construct resulted in approximately 40-fold greater promoter activity in the CMK cells than in CMS cells (Figure1C).
Ara-C metabolism and sensitivity in CMK and CMS cells
For the CMK cells, overexpression of CBS was accompanied by about 10-fold greater in vitro ara-C sensitivity measured by the growth inhibition assay compared with the CMS cell line (Figure 1D). The increased ara-C sensitivity of the CMK cell line was accompanied by a 2.4-fold increased generation in vitro of3H-ara-CTP after 3 hours of incubation with3H-ara-C compared with the CMS cell line (Figure1D).
These results validate the CMK and CMS sublines as clinically relevant AML models to study the transcriptional regulation of the humanCBS gene and the mechanisms that result in the differentialCBS expression in DS and non-DS AML samples.
In vitro differential binding of Sp1/Sp3 to CBS −1b promoter in CMS and CMK cells
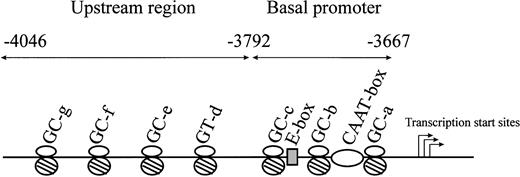
We previously characterized the CBS −1b minimal promoter (positions −3792 to −3667) in HepG2 cells and found that Sp1/Sp3, NF-Y, and USF-1 were all involved in the regulation of basal promoter activity.16 Additional studies examined the critical cis elements and transcription factors in the CBS −1b upstream region (positions −4046 to −3792) in HT1080 and HepG2 cells and identified transcriptionally important roles for Sp1/Sp3 binding to 3 GC-boxes (GC-e, positions: −3851 to −3857; GC-f, −3940 to −3945; and GC-g, −3973 to −3983) and 1 GT-box (GT-d, −3799 to −3805).15 A schematic representation of the major transcriptional elements of theCBS −1b promoter is shown in Figure2.
Schematic representation of the CBS −1b promoter and upstream regions of the promoter from positions −3667 to −4046.
Major transcription factor–binding sites in the CBS −1b basal promoter region from positions −3667 to −3792 (3 GC-boxes, 1 E-box, 1 CAAT-box), and in the upstream regions from positions −3792 to −4046 (3 GC-boxes, 1 GT-box) are represented.
Schematic representation of the CBS −1b promoter and upstream regions of the promoter from positions −3667 to −4046.
Major transcription factor–binding sites in the CBS −1b basal promoter region from positions −3667 to −3792 (3 GC-boxes, 1 E-box, 1 CAAT-box), and in the upstream regions from positions −3792 to −4046 (3 GC-boxes, 1 GT-box) are represented.
EMSAs were used to establish whether there were differences in transcription factor binding to the basal and upstream regions of theCBS −1b promoter that account for differentialCBS expression in CMK and CMS cells. Synthetic FPA, FPB, FPC, FPD double-stranded oligonucleotides (Table 1) were designed based on our previous findings of protected regions on DNase I footprints of the CBS −1b promoter in HepG2 cells.15Additional oligonucleotides (−3766/−3725 and −3724/−3682; Table 1) were designed for the basal CBS −1b promoter region.16 Oligonucleotides were labeled by γ-32P-ATP and incubated with nuclear extracts prepared from CMK and CMS cells. Nuclear extracts from the HepG2 cells, in which DNA/protein complexes were previously identified by oligonucleotide competitions and supershift assays,15 16 were used as controls. Competitions with unlabeled Sp1, USF1, or NF-Y consensus oligonucleotides were also performed in CMK and CMS cells to confirm the identities of the specific complexes (data not shown).
On EMSAs, transcription factor binding was increased with nuclear extracts prepared from HepG2 cells from that with CMK and CMS cells (Figure 3), reflecting the higher CBS transcripts in HepG2 cells (data not shown). For the FPA, FPC and the −3766/−3725 and −3724/−3682 basal oligonucleotides, there were no significant differences in transcription factor binding (eg, Sp1/Sp3, USF-1, NF-Y) between CMK and CMS cells to account for differential CBS expression between the lines (Figure 3A,C,E-F). However, significant differences in transcription factor (ie, Sp1/Sp3) binding were detected with FPB and FPD oligonucleotides. For the FPB probe, corresponding to the GC-f box, undetectable binding of Sp1/Sp3 was found for CMS cells compared with CMK cells (Figure 3B). For the FPD probe, including the GT-d box, Sp1/Sp3 binding was, likewise, undetectable in the CMS nuclear extracts (Figure 3D).
EMSAs with CMK and CMS nuclear extracts spanning the upstream and basal regions of the CBS −1b promoter.
Gel shift assays were performed with CMK and CMS nuclear extracts and the 32P-labeled FPA, FPB, FPC, FPD oligonucleotide probes (designated A-D, respectively), spanning upstream regions of theCBS promoter (positions −4046 to −3792), andCBS −1b basal promoter probes (−3766/−3725 and −3724/−3682; designated E and F, respectively). Major DNA/protein complexes were previously identified in the HepG2 cell line (control) and are indicated.16 17 NS indicates a nonspecific complex.
EMSAs with CMK and CMS nuclear extracts spanning the upstream and basal regions of the CBS −1b promoter.
Gel shift assays were performed with CMK and CMS nuclear extracts and the 32P-labeled FPA, FPB, FPC, FPD oligonucleotide probes (designated A-D, respectively), spanning upstream regions of theCBS promoter (positions −4046 to −3792), andCBS −1b basal promoter probes (−3766/−3725 and −3724/−3682; designated E and F, respectively). Major DNA/protein complexes were previously identified in the HepG2 cell line (control) and are indicated.16 17 NS indicates a nonspecific complex.
In vivo binding of the transcription factors to the CBS −1b promoter in chromatin extracts of the CMK and CMS cells was confirmed by the ChIP assay (Figure 4). Both CMK and CMS cells exhibited in vivo binding of Sp3, USF-1 and NF-YA, consistent with the in vitro binding patterns on EMSAs.
In vivo binding of Sp1/Sp3, USF-1, and NF-YA to the CBS −1b promoter.
Chromatin extracts of the CMK, CMS, and HepG2 (control) cell lines bound to transcription factor complexes were immunoprecipitated with Sp1, Sp3, USF-1, and NF-YA antisera. PCR amplifications were performed with seminested primers for the CBS −1b promoter region (positions −3921 to −3625), as described in “Materials and methods,” and the amplicons visualized on 2% agarose gels.
In vivo binding of Sp1/Sp3, USF-1, and NF-YA to the CBS −1b promoter.
Chromatin extracts of the CMK, CMS, and HepG2 (control) cell lines bound to transcription factor complexes were immunoprecipitated with Sp1, Sp3, USF-1, and NF-YA antisera. PCR amplifications were performed with seminested primers for the CBS −1b promoter region (positions −3921 to −3625), as described in “Materials and methods,” and the amplicons visualized on 2% agarose gels.
Functional analysis of transcription factor–binding sites by site-directed mutagenesis
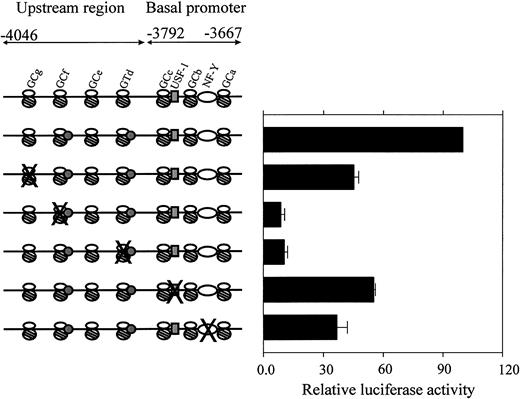
Based on the EMSA results, the relevant Sp1/Sp3 consensus elements in the CBS −1b upstream region and the critical USF-1 and NF-Y consensus elements in the CBS −1b basal promoter region were mutated individually using the single-stranded mutant oligonucleotides as primers (Table 1). The mutant CBS −1b promoter reporter gene constructs were transiently transfected into CMK cells. Luciferase activities for the mutant promoter constructs were compared to that of the wild-type full-length promoter construct pCBSb −4046/−3565.
Mutation of GC-g (FPA-GC-g mt; Table 1) resulted in approximately 50% decreased CBS −1b promoter activity in the CMK cells (Figure 5). Significant losses (approximately 90%) of CBS −1b promoter activity accompanied mutations of the GC-f (FPB-GC-f mt) and GT-d (FPD-GT-d mt) elements. Lesser decreases accompanied mutations of the essential basal elements, including USF-1 (promoter activity decreased by 50%) and NF-Y (decrease by 65%). These results confirm important transactivating roles for the GC-f and GT-d elements in the upstream regions of the CBS −1b promoter in CMK cells.
Functional analysis of mutated regulatory sequences in theCBS −1b basal promoter and upstream regions.
CMK cells were transfected with the wild-type and mutant CBS−1b promoter reporter gene constructs spanning the basal (positions −3792 to −3667) and upstream regions (positions −4046 to −3792) of the CBS −1b promoter, as described in “Materials and methods.” The oligonucleotides used for preparing the mutant CBS −1b promoter GC-g, GC-f, GT-d, USF-1, and NF-Y constructs were previously described.16 17 Luciferase activities of the mutant constructs were normalized toRenilla luciferase activities and compared with that of the −4046/−3565 wild-type construct. Data are presented as the means ± SE from 3 independent experiments performed in duplicate.
Functional analysis of mutated regulatory sequences in theCBS −1b basal promoter and upstream regions.
CMK cells were transfected with the wild-type and mutant CBS−1b promoter reporter gene constructs spanning the basal (positions −3792 to −3667) and upstream regions (positions −4046 to −3792) of the CBS −1b promoter, as described in “Materials and methods.” The oligonucleotides used for preparing the mutant CBS −1b promoter GC-g, GC-f, GT-d, USF-1, and NF-Y constructs were previously described.16 17 Luciferase activities of the mutant constructs were normalized toRenilla luciferase activities and compared with that of the −4046/−3565 wild-type construct. Data are presented as the means ± SE from 3 independent experiments performed in duplicate.
Intracellular levels of Sp1/Sp3 and phosphorylation of Sp1/Sp3 as potential factors in differential binding to the CBS −1b promoter in CMK and CMS cells
The differential binding of Sp1/Sp3 to the CBS−1b promoter in CMK versus CMS cells may, in part, explain differences in the levels of CBS transcripts and CBS −1b promoter activity between the lines. Although differences in binding could conceivably reflect variable intracellular levels of critical transcription factors, there were comparable levels of Sp1, Sp3, USF-1, and NF-YA proteins on Western blots of CMK and CMS nuclear extracts (Figure 6A).
Sp1, Sp3, USF-1, and NF-YA levels in CMK and CMS nuclear extracts and the role of Sp1/Sp3 phosphorylation of in vitro binding to the CBS −1b promoter.
(A) Fifty micrograms of nuclear extract proteins from CMS, CMK, and HepG2 (control) cells were fractionated on 7.5% (Sp1, Sp3) or 12% (USF1, NF-YA) polyacrylamide gels with SDS and electroblotted onto PVDF membranes. Immunoreactive Sp1, Sp3, USF-1, and NF-YA proteins were detected with Sp1, Sp3, USF-1, and NF-YA antibodies and Lumi-Light Western blotting substrate. Comparable protein levels of the transcription factors were found for both the CMK and CMS cells. (B) Nuclear extracts from CMK cells were pretreated with calf intestinal alkaline phosphatase (CIP), 32P-labeled FPD probe was added, and following incubation on ice for 30 minutes, the DNA protein complexes were electrophoresed, as described in “Materials and methods.” Ten micrograms of the treated nuclear extracts were used for the gel shift assays. Decreased binding of Sp1/Sp3 to the32P-labeled FPD probe was observed following CIP treatment. NA indicates no addition.
Sp1, Sp3, USF-1, and NF-YA levels in CMK and CMS nuclear extracts and the role of Sp1/Sp3 phosphorylation of in vitro binding to the CBS −1b promoter.
(A) Fifty micrograms of nuclear extract proteins from CMS, CMK, and HepG2 (control) cells were fractionated on 7.5% (Sp1, Sp3) or 12% (USF1, NF-YA) polyacrylamide gels with SDS and electroblotted onto PVDF membranes. Immunoreactive Sp1, Sp3, USF-1, and NF-YA proteins were detected with Sp1, Sp3, USF-1, and NF-YA antibodies and Lumi-Light Western blotting substrate. Comparable protein levels of the transcription factors were found for both the CMK and CMS cells. (B) Nuclear extracts from CMK cells were pretreated with calf intestinal alkaline phosphatase (CIP), 32P-labeled FPD probe was added, and following incubation on ice for 30 minutes, the DNA protein complexes were electrophoresed, as described in “Materials and methods.” Ten micrograms of the treated nuclear extracts were used for the gel shift assays. Decreased binding of Sp1/Sp3 to the32P-labeled FPD probe was observed following CIP treatment. NA indicates no addition.
A role for Sp1 phosphorylation in CBS −1b promoter binding and transactivation in HepG2 cells was implied by treatments with calf alkaline phosphatase (CIP) that decreased Sp1 binding by EMSAs.15 Likewise, decreased Sp1/Sp3 binding to the FPD probe in CMK nuclear extracts treated with CIP suggested an important role of Sp factor phosphorylation in the differential CBSexpression between CMK and CMS cells (Figure 6B).15However, treatment of CMS nuclear extracts with protein kinase A catalytic subunit did not result in increased Sp1/Sp3 binding (data not shown).
Discussion
Identifying the biologic basis for the extremely high EFS rates of DS AML patients treated with ara-C–based protocols may lead to improvements in the treatment of AML, which has the worse prognosis of all childhood leukemias. Our prior studies provided compelling evidence that the chromosome 21–localized gene, CBS, plays a crucial role contributing to the enhanced ara-C sensitivity of DS myeloblasts and CBS-transfected leukemia cell line models.11-13 Pronounced variations in CBS expression between DS and non-DS myeloblasts,12 exceeding levels predicted by gene copy number in trisomy 21 (DS) cells,20 highlight its unique biologic and pharmacologic roles and its complex transcriptional regulation in leukemia cells.
In view of the limitations in performing gene transcription studies with primary clinical leukemia samples, identification of relevant cell line models representative of clinical AML is essential. In this study, we selected 2 pediatric AMkL cell lines as representative leukemia models to examine the transcriptional regulation of CBS based on the significant differences in CBS transcripts between the DS and non-DS AML and the high frequency of the AMkL phenotype in DS AML cases.10 Contrary to the clinical outcome of DS AMkL cases, non-DS AMkL cases have an extremely poor prognosis (cure rates < 25%) as reported by both the Children's Cancer Group (CCG) and St Jude Children's Research Hospital,5,21 thus highlighting distinct biologic differences between DS and non-DS AMkL cases. The DS cell line, CMK, generated 2.4-fold higher levels of ara-CTP and exhibited a 10-fold greater ara-C sensitivity compared with the non-DS CMS cell line, thus closely approximating our findings with DS and non-DS clinical AML samples and those with aCBS-transfected CCRF-CEM cell line.12 13
We previously characterized the CBS −1b minimal (basal) promoter (−3792 to −3667) in HepG2 hepatocarcinoma cells and demonstrated that the transcription factors Sp1/Sp3, NF-Y, and USF-1 were important transactivators of the basal promoter and that the synergistic activation of the promoter involved Sp1/Sp3 and NF-Y. The critical cis elements and transcription factors in the upstream region were also examined in HT1080 and HepG2 cell line models, characterized by significant differences in CBStranscript levels, to better identify determinants of cell-specific expression of CBS.15 16 The results of these studies were extrapolated herein to the study of clinically relevant AMkL cell lines. Analysis of the upstream region of the CBS−1b promoter by EMSAs demonstrated significant differences in binding of Sp1/Sp3 to the GC-f and GT-d elements in CMK cells compared with CMS cells. The significance of these 2 binding sites was confirmed by mutation analysis, which resulted in approximately 90% decreasedCBS −1b promoter activity. There were no significant differences in transcription factor binding to the basal CBS−1b promoter between the cell lines to account for the differences inCBS expression.
Based on prior studies of Sp factor phosphorylation in CBS−1b promoter transactivation, we assessed the effects of treating CMK nuclear extracts with CIP on Sp1/Sp3 binding in in vitro assays. Indeed, CIP treatment markedly decreased Sp factor binding, strongly suggesting that the differential binding to the CBS −1b promoter is due to differences in phosphorylation of this critical transcription factor. Although we did not find increased binding of Sp1/Sp3 to the CBS promoter in CMS nuclear extracts following treatment with protein kinase A, other phosphorylation pathways or alternative mechanisms may be involved.
Interestingly, abnormal phosphorylation of a variety of proteins has been demonstrated in a number of DS studies and at least 2 genes have been identified on chromosome 21 that may lead to altered patterns of protein phosphorylation.22,23 The human homologue of theDrosophila minibrain gene, DYRK (dual specificity tyrosine phosphorylation regulated kinase), is localized to 21q22.2 and is believed to be involved in abnormal neurologic development of DS and can phosphorylate serine/threonine residues in protein substrates.22 In DS cerebellum, elevated protein levels of the voltage-dependent anion-selective channel proteins (VDADs), which regulate fluxes of anionic metabolites including ATP, were identified.23
Although it has been postulated that the DS phenotype is due to the presence of genes on the long arm of chromosome 21 (known collectively as the Down syndrome critical region), which are expressed in trisomy 21 cells at 1.5-fold higher levels compared with diploid cells, a variety of genes in DS are expressed at levels not based on a gene dosage effect.24-26 Our finding of increasedCBS expression in CMK cells demonstrates that the interaction of multiple transcription factors and, possibly, the role of protein phosphorylation are significant additional factors to consider in understanding patterns of chromosome 21 gene expression. It is conceivable that variants of the CBS gene (eg, 844ins68 CBS gene polymorphism)27 may also be involved in the modulation of ara-C sensitivity of DS cells. By characterizing the transcriptional regulation of the CBSgene as a representative chromosome 21–localized gene, this will further advance our knowledge of the biology of DS including the mechanisms of chemotherapy sensitivity of DS AML patients and leukemogenesis in DS.
Prepublished online as Blood First Edition Paper, October 17, 2002; DOI 10.1182/blood- 2002-07-2337.
Supported by grant RO1 CA92308 from the National Cancer Institute, National Institutes of Health, the Leukemia and Lymphoma Society, Children's Research Center of Michigan, Art Gagnon Memorial Fund, BenePro/Concepts Technologies (BPCT) and Justin's Gift Golf Charities, and Leukemia, Research, Life, Inc. J.W.T. is a Scholar in Clinical Research of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jeffrey W. Taub, Children's Hospital of Michigan, 3901 Beaubien Blvd, Detroit, MI 48201; e-mail:jtaub@med.wayne.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal