Primary cutaneous lymphomas (PCLs) represent a heterogeneous group of extranodal T- and B-cell malignancies. The underlying molecular pathogenesis of this malignancy remains unclear. This study aimed to characterize oncogene abnormalities in PCLs. Using genomic microarray, we detected oncogene copy number gains of RAF1(3p25), CTSB (8p22), PAK1 (11q13), and JUNB (19p13) in 5 of 7 cases of mycosis fungoides (MF)/Sezary syndrome (SS) (71%), gains of FGFR1 (8p11), PTPN (20q13), andBCR (22q11) in 4 cases (57%), and gains ofMYCL1 (1p34), PIK3CA (3q26), HRAS(11p15), MYBL2 (20q13), and ZNF217 (20q13) in 3 cases (43%). Amplification of JUNB was studied in 104 DNA samples from 78 PCL cases using real-time polymerase chain reaction. Twenty-four percent of cases, including 7 of 10 cases of primary cutaneous CD30+ anaplastic large-cell lymphoma (C-ALCL), 4 of 14 MF, 4 of 22 SS, and 2 of 23 primary cutaneous B-cell lymphoma (PCBCL) showed amplification ofJUNB, and high-level amplification of this oncogene was present in 3 C-ALCL and 2 MF cases. JUNB protein expression was analyzed in tissue sections from 69 PCL cases, and 44% of cases, consisting of 21 of 23 SS, 6 of 8 C-ALCL, 5 of 10 MF, and 9 of 21 PCBCL, demonstrated nuclear expression of JUNB by tumor cells. Overexpression of JUNB also was detected in 5 C-ALCL and 2 SS cases. These results have revealed, for the first time, amplification and expression patterns of JUNB in PCL, suggesting thatJUNB may be critical in the pathogenesis of primary cutaneous T-cell lymphomas.
Introduction
Primary cutaneous lymphomas (PCLs) represent a heterogeneous group of extranodal T- and B-cell malignancies with an annual incidence of 0.5-1 per 100 000.1-3 This group of lymphomas has been classified into mycosis fungoides (MF)/Sezary syndrome (SS), accounting for 70% of PCL cases, primary cutaneous B-cell lymphoma (PCBCL), constituting more than 20% of cases, primary cutaneous CD30+ anaplastic large cell lymphoma (C-ALCL), constituting 10% of cases, and blastic natural killer cell lymphoma (NK), accounting for 1% of cases.1-3 These subtypes of PCL have distinctive clinicopathologic and immunophenotypic features.1-3 However, the underlying etiology and pathogenesis remains unclear.
Most malignancies accumulate a series of genetic events including activation of oncogenes and loss of tumor suppressor genes, which lead to a malignant phenotype. Previous studies have shown allelic losses at 9p, 10q, and 17p; microsatellite instability and mutations ofp53 in primary cutaneous T-cell lymphomas (CTCLs);4-6 and hypermethylation of p15 andp16 in both CTCL and PCBCL.7,8 However, little is known about genome-wide genetic alterations in PCL. We have studied a series of PCL cases using comparative genomic hybridization (CGH) and have identified consistent and distinctive patterns of chromosomal imbalances (CI) in both MF and SS and in PCBCL.9,10Genomic microarray is a novel technique of genomic analysis used to rapidly screen for genomic imbalances (GI) in a tumor genome.11 Previous studies have revealed oncogene copy number changes in several epithelial cancers.11-14 We also have detected oncogene gains and losses in PCBCLs using this technique,10 but at present there is no data available in CTCLs.
The aim of this study was to screen for oncogene abnormalities in PCLs. We began with a study of 7 cases of MF/SS using genomic microarray, which showed gains of several oncogenes, including JUNB. We then analyzed 104 DNA samples from 78 PCL patients for JUNBamplification using real-time–polymerase chain reaction (RT-PCR) and correlated these findings with the results of immunohistochemistry (IHC) analysis of formalin-fixed, paraffin-embedded tissue sections from 69 PCL cases.
Materials and methods
Samples
Initially, 7 cases of MF/SS with noticeable CI detected by CGH9 were selected for genomic microarray study (Table1). This included 5 SS (nos. 1 [sample 1524], 3 [1650], 4 [1530], 5 [1278], and 23 [2211]) and 2 MF (nos. 25 [1416] and 32 [1536]) cases (Table 1). Subsequently, 104 DNA samples from 78 patients with PCL, consisting of 41 samples from 22 SS cases, 14 MF samples, 16 samples from 10 C-ALCL cases, 23 PCBCL samples, 7 cutaneous blastic NK cell lymphoma samples, 2 systemic follicular lymphoma (FL) samples, and 1 lymphomatoid papulosis (LyP) sample were selected for RT-PCR study according to specific clinicopathologic features, immunophenotypes, and T-cell receptor/immunoglobulin gene analysis (Table2).2 3 In addition, formalin-fixed, paraffin-embedded tissue sections from 69 of 78 PCL cases analyzed by RT-PCR (23 SS, 10 MF, 8 C-ALCL, 21 PCBCL, 4 cutaneous NK cell lymphoma, 2 systemic FL, and 1 LyP) were selected for IHC study (Table 2). This project was approved by the St Thomas' Hospital Research Ethics Committee for sampling (EC02/089).
A summary of genomic microarray findings in 7 MF/SS cases
| Gene . | Location . | Case (sample) . | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 (1524) . | 3 (1650) . | 4 (1530) . | 5 (1278) . | 23 (2211) . | 25 (1416) . | 32 (1536) . | ||
| FGR | 1p36 | n | n | − | n | n | − | n |
| MYCL1 | 1p34 | + | + | − | + | n | n | n |
| NRAS | 1p13 | n | n | n | n | n | n | n |
| LAMC2 | 1q25 | n | n | n | n | n | n | − |
| NMYC | 2p24 | n | n | n | n | n | + | − |
| REL | 2p13 | n | n | n | n | n | n | n |
| RAF1 | 3p25 | n | n | + | + | + | + | + |
| TERC | 3q26 | n | n | − | n | n | n | n |
| PIK3CA | 3q26 | + | n | n | + | n | + | n |
| PDGFRA | 4q12 | n | n | n | n | n | n | n |
| MYB | 6q22 | n | n | n | n | n | n | n |
| ESR | 6q25 | n | n | n | n | n | n | n |
| EGFR | 7p12 | n | n | n | − | n | n | n |
| PGY1 | 7q21 | n | n | n | − | n | n | n |
| MET | 7q31 | − | n | + | n | n | n | − |
| CTSB | 8p22 | n | n | + | + | + | + | + |
| FGFR1 | 8p11 | n | n | n | + | + | + | + |
| MOS | 8q11 | n | n | − | − | n | n | n |
| MYC | 8q24 | n | n | n | n | n | n | n |
| ABL1 | 9q34 | n | n | + | − | n | n | − |
| FGFR2 | 10q26 | n | n | n | n | n | n | n |
| HRAS | 11p15 | + | n | n | n | n | + | + |
| CCND1 | 11q13 | n | n | n | − | n | n | − |
| FGF4 | 11q13 | n | n | − | n | n | n | n |
| EMS1 | 11q13 | + | n | n | n | n | − | n |
| GARP | 11q13 | n | n | − | − | n | − | n |
| PAK1 | 11q13 | + | n | + | + | + | n | + |
| MLL | 11q23 | n | n | − | − | n | n | n |
| CCND2 | 12p13 | n | n | n | n | n | n | n |
| KRAS2 | 12p12 | + | n | + | n | n | n | n |
| WNT1 | 12q12 | n | n | n | n | n | − | n |
| GLI | 12q13 | n | n | n | + | + | n | n |
| SAS | 12q13 | n | − | n | n | n | n | n |
| MDM2 | 12q14 | n | n | n | n | n | n | n |
| AKT1 | 14q32 | n | n | n | − | n | − | n |
| IGFR1 | 15q25 | n | n | n | + | n | + | n |
| FES | 15q26 | − | n | n | + | + | n | n |
| MRP1 | 16p13 | n | n | n | n | n | n | n |
| TOP2A | 17q2 | n | n | n | − | n | n | n |
| ERBB2 | 17q21 | n | n | n | − | n | n | n |
| RPS6KB1 | 17q23 | n | n | n | n | n | n | − |
| D17S1670 | 17q23 | n | n | + | n | n | n | − |
| YES1 | 18p11 | n | n | n | + | n | + | n |
| BCL23′ | 18q21 | n | n | n | n | n | n | n |
| BCL25′ | 18q21 | n | n | n | n | n | − | n |
| INSR | 19p13 | n | n | + | n | n | n | n |
| JUNB | 19p13 | + | n | n | + | + | + | + |
| CCNE1 | 19q13 | n | n | n | n | n | n | n |
| AIB1 | 20q12 | n | n | + | n | n | n | n |
| STK15 | 20q13 | − | n | n | n | n | n | n |
| CSE1L | 20q13 | n | n | n | n | n | n | n |
| MYBL2 | 20q13 | n | n | + | + | n | + | n |
| PTPN1 | 20q13 | n | n | + | + | n | + | + |
| ZNF217 | 20q13 | n | n | + | + | n | + | n |
| CBFA2 | 21q22 | n | n | n | n | n | + | n |
| BCR | 22q11 | n | n | n | + | + | + | + |
| PDGFB | 22q12 | n | n | n | n | n | n | n |
| AR5′ | Xq11 | + | n | n | n | n | + | n |
| AR3′ | Xq11 | + | n | n | n | n | + | n |
| Gene . | Location . | Case (sample) . | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 (1524) . | 3 (1650) . | 4 (1530) . | 5 (1278) . | 23 (2211) . | 25 (1416) . | 32 (1536) . | ||
| FGR | 1p36 | n | n | − | n | n | − | n |
| MYCL1 | 1p34 | + | + | − | + | n | n | n |
| NRAS | 1p13 | n | n | n | n | n | n | n |
| LAMC2 | 1q25 | n | n | n | n | n | n | − |
| NMYC | 2p24 | n | n | n | n | n | + | − |
| REL | 2p13 | n | n | n | n | n | n | n |
| RAF1 | 3p25 | n | n | + | + | + | + | + |
| TERC | 3q26 | n | n | − | n | n | n | n |
| PIK3CA | 3q26 | + | n | n | + | n | + | n |
| PDGFRA | 4q12 | n | n | n | n | n | n | n |
| MYB | 6q22 | n | n | n | n | n | n | n |
| ESR | 6q25 | n | n | n | n | n | n | n |
| EGFR | 7p12 | n | n | n | − | n | n | n |
| PGY1 | 7q21 | n | n | n | − | n | n | n |
| MET | 7q31 | − | n | + | n | n | n | − |
| CTSB | 8p22 | n | n | + | + | + | + | + |
| FGFR1 | 8p11 | n | n | n | + | + | + | + |
| MOS | 8q11 | n | n | − | − | n | n | n |
| MYC | 8q24 | n | n | n | n | n | n | n |
| ABL1 | 9q34 | n | n | + | − | n | n | − |
| FGFR2 | 10q26 | n | n | n | n | n | n | n |
| HRAS | 11p15 | + | n | n | n | n | + | + |
| CCND1 | 11q13 | n | n | n | − | n | n | − |
| FGF4 | 11q13 | n | n | − | n | n | n | n |
| EMS1 | 11q13 | + | n | n | n | n | − | n |
| GARP | 11q13 | n | n | − | − | n | − | n |
| PAK1 | 11q13 | + | n | + | + | + | n | + |
| MLL | 11q23 | n | n | − | − | n | n | n |
| CCND2 | 12p13 | n | n | n | n | n | n | n |
| KRAS2 | 12p12 | + | n | + | n | n | n | n |
| WNT1 | 12q12 | n | n | n | n | n | − | n |
| GLI | 12q13 | n | n | n | + | + | n | n |
| SAS | 12q13 | n | − | n | n | n | n | n |
| MDM2 | 12q14 | n | n | n | n | n | n | n |
| AKT1 | 14q32 | n | n | n | − | n | − | n |
| IGFR1 | 15q25 | n | n | n | + | n | + | n |
| FES | 15q26 | − | n | n | + | + | n | n |
| MRP1 | 16p13 | n | n | n | n | n | n | n |
| TOP2A | 17q2 | n | n | n | − | n | n | n |
| ERBB2 | 17q21 | n | n | n | − | n | n | n |
| RPS6KB1 | 17q23 | n | n | n | n | n | n | − |
| D17S1670 | 17q23 | n | n | + | n | n | n | − |
| YES1 | 18p11 | n | n | n | + | n | + | n |
| BCL23′ | 18q21 | n | n | n | n | n | n | n |
| BCL25′ | 18q21 | n | n | n | n | n | − | n |
| INSR | 19p13 | n | n | + | n | n | n | n |
| JUNB | 19p13 | + | n | n | + | + | + | + |
| CCNE1 | 19q13 | n | n | n | n | n | n | n |
| AIB1 | 20q12 | n | n | + | n | n | n | n |
| STK15 | 20q13 | − | n | n | n | n | n | n |
| CSE1L | 20q13 | n | n | n | n | n | n | n |
| MYBL2 | 20q13 | n | n | + | + | n | + | n |
| PTPN1 | 20q13 | n | n | + | + | n | + | + |
| ZNF217 | 20q13 | n | n | + | + | n | + | n |
| CBFA2 | 21q22 | n | n | n | n | n | + | n |
| BCR | 22q11 | n | n | n | + | + | + | + |
| PDGFB | 22q12 | n | n | n | n | n | n | n |
| AR5′ | Xq11 | + | n | n | n | n | + | n |
| AR3′ | Xq11 | + | n | n | n | n | + | n |
n indicates normal (balanced); −, copy number loss; and +, copy number gain.
A summary of RT-PCR and IHC analyses of JUNB in PCL
| Case . | Sample* . | Diagnosis† . | RT-PCR* . | IHC . | ||
|---|---|---|---|---|---|---|
| Staining† . | Large cells (%)‡ . | Positive large cells (%)2-153 . | ||||
| 1 | 1524s/1525b | SS | n | + | 20 | 50 |
| 2 | 1528b | SS | n | +++ | 10 | 100 |
| 3 | 4011/1650b | SS | n | +++ | 10 | 100 |
| 4 | 1530s/1603l | SS | n | ++ | 10 | 80 |
| 5 | 1277s/1278b | SS | ++ | ++ | 10 | 90 |
| 6 | 1124s | SS | n/a | ++ | 40 | 90 |
| 7 | 1378b | SS | n | ++ | 20 | 60 |
| 8 | 1598s/702b | SS | ++ | ++ | 70 | 70 |
| 9 | 16041/1586b | SS | n | + | 50 | 50 |
| 10 | 548s/546b | SS | n | − | − | − |
| 11 | 2135s/2131b | SS | n | ++ | 40 | 80 |
| 12 | 1711s/17461/1712b | SS | n | + | 10 | 50 |
| 13 | 1900s/2530b | SS | n | + | 10 | 50 |
| 14 | 2061s/2062b | SS | n | − | 10 | − |
| 15 | 2707s/2708b | SS | + | ++ | 40 | 75 |
| 16 | 2652s/2651b | SS | n | + | 10 | 5 |
| 17 | 2335s/2329b | SS | n | ++ | 20 | 60 |
| 16 | 2724s/2725b | SS | n | ++ | 20 | 60 |
| 19 | 2559s | SS | n | + | 20 | 40 |
| 20 | 2729s/2728b | SS | n | + | 30 | 50 |
| 21 | 2718s/2719b | SS | n | + | 10 | 50 |
| 22 | 2720s/2721b | SS | n | + | 20 | 40 |
| 23 | 2211b | SS | + | ++ | 10 | 80 |
| 24 | 1484I | MF | − | − | 10 | − |
| 25 | 1416s | MF | + | + | 20 | 20 |
| 26 | 1445b | MF | n | − | − | − |
| 27 | 1975s | MF | n | − | − | − |
| 28 | 1394b | MF | n | − | − | − |
| 29 | 650I | MF | n | + | 10 | 5 |
| 30 | 688s | MF | n | + | 10 | 10 |
| 31 | 1487s | MF | +++ | ++ | 25 | 60 |
| 32 | 1536s | MF | n | + | 10 | 50 |
| 33 | 1587s | MF | ++++ | n/a | n/a | n/a |
| 34 | 2609I | MF | n | − | − | − |
| 35 | 1348s | MF | + | n/a | n/a | n/a |
| 36 | 1375s | MF | n | n/a | n/a | n/a |
| 37 | 143s | MF | n | n/a | n/a | n/a |
| 38 | 1455s | C-ALCL | +++ | +++ | 100 | 100 |
| 39 | 2624s/2626b | C-ALCL | ++++ | +++ | 60 | 100 |
| 40 | 2092s/2095b | C-ALCL | + | +++ | 100 | 100 |
| 41 | 2267b/1358b | C-ALCL | ++ | ++ | 20 | 90 |
| 42 | 2065s | C-ALCL | +++ | +++ | 80 | 100 |
| 43 | 1281s/1282I | C-ALCL | ++ | +++ | 90 | 100 |
| 44 | sk273 | C-ALCL | n | − | − | − |
| 45 | 1950s | C-ALCL | n | − | − | − |
| 46 | SK46s/SK47s | C-ALCL | ++ | n/a | n/a | n/a |
| 47 | SK40s/SK44s | C-ALCL | n | n/a | n/a | n/a |
| 48 | 302s | LyP | n | − | − | − |
| 49 | 142s | DLCL | n | − | − | − |
| 50 | 1233b/1234s | DLCL | n | − | 10 | − |
| 51 | 613b | DLCL | n | + | 60 | 5 |
| 52 | 419s | DLCL | n | − | − | − |
| 53 | 1662s | DLCL | n | + | 10 | 50 |
| 54 | 1993s | DLCL | n | − | 20 | − |
| 55 | 1201s | MZL | n | + | 10 | 40 |
| 56 | sk311s | MZL | n | + | 50 | 5 |
| 57 | 1533s | MZL | n | − | − | − |
| 58 | 227s | MZL | n | − | 10 | − |
| 59 | 1848s | MZL | n | − | − | − |
| 60 | 1794b | MZL | n | − | 10 | − |
| 61 | 1797s | MZL | n | − | 10 | − |
| 62 | 928s | MZL | + | + | 10 | 10 |
| 63 | 1475s | MZL | n | n/a | n/a | n/a |
| 64 | 258s | Immunocytoma | n | − | − | − |
| 65 | 1728s | FCCL | n | + | 50 | 5 |
| 66 | 423s | FCCL | − | − | 10 | − |
| 67 | 2047s | FCCL | n | − | 10 | − |
| 68 | 2075s | FCCL | + | ++ | 30 | 60 |
| 69 | 2198s | FCCL | n | ++ | 20 | 90 |
| 70 | 1943b | Unclassified | n | + | 10 | 10 |
| 71 | 1648b | Systemic FL | + | + | 10 | 10 |
| 72 | 1564b | Systemic FL | n | − | 10 | − |
| 73 | 1683s | NK | n | − | − | − |
| 74 | 18021i | NK | n | + | 50 | 5 |
| 75 | 2022s | NK | n | + | 20 | 20 |
| 76 | 2412s | NK | n | − | 60 | − |
| 77 | 1860s | NK | + | n/a | n/a | n/a |
| 78 | 1231s | NK | n | n/a | n/a | n/a |
| 79 | 2634b | NK | − | n/a | n/a | n/a |
| Case . | Sample* . | Diagnosis† . | RT-PCR* . | IHC . | ||
|---|---|---|---|---|---|---|
| Staining† . | Large cells (%)‡ . | Positive large cells (%)2-153 . | ||||
| 1 | 1524s/1525b | SS | n | + | 20 | 50 |
| 2 | 1528b | SS | n | +++ | 10 | 100 |
| 3 | 4011/1650b | SS | n | +++ | 10 | 100 |
| 4 | 1530s/1603l | SS | n | ++ | 10 | 80 |
| 5 | 1277s/1278b | SS | ++ | ++ | 10 | 90 |
| 6 | 1124s | SS | n/a | ++ | 40 | 90 |
| 7 | 1378b | SS | n | ++ | 20 | 60 |
| 8 | 1598s/702b | SS | ++ | ++ | 70 | 70 |
| 9 | 16041/1586b | SS | n | + | 50 | 50 |
| 10 | 548s/546b | SS | n | − | − | − |
| 11 | 2135s/2131b | SS | n | ++ | 40 | 80 |
| 12 | 1711s/17461/1712b | SS | n | + | 10 | 50 |
| 13 | 1900s/2530b | SS | n | + | 10 | 50 |
| 14 | 2061s/2062b | SS | n | − | 10 | − |
| 15 | 2707s/2708b | SS | + | ++ | 40 | 75 |
| 16 | 2652s/2651b | SS | n | + | 10 | 5 |
| 17 | 2335s/2329b | SS | n | ++ | 20 | 60 |
| 16 | 2724s/2725b | SS | n | ++ | 20 | 60 |
| 19 | 2559s | SS | n | + | 20 | 40 |
| 20 | 2729s/2728b | SS | n | + | 30 | 50 |
| 21 | 2718s/2719b | SS | n | + | 10 | 50 |
| 22 | 2720s/2721b | SS | n | + | 20 | 40 |
| 23 | 2211b | SS | + | ++ | 10 | 80 |
| 24 | 1484I | MF | − | − | 10 | − |
| 25 | 1416s | MF | + | + | 20 | 20 |
| 26 | 1445b | MF | n | − | − | − |
| 27 | 1975s | MF | n | − | − | − |
| 28 | 1394b | MF | n | − | − | − |
| 29 | 650I | MF | n | + | 10 | 5 |
| 30 | 688s | MF | n | + | 10 | 10 |
| 31 | 1487s | MF | +++ | ++ | 25 | 60 |
| 32 | 1536s | MF | n | + | 10 | 50 |
| 33 | 1587s | MF | ++++ | n/a | n/a | n/a |
| 34 | 2609I | MF | n | − | − | − |
| 35 | 1348s | MF | + | n/a | n/a | n/a |
| 36 | 1375s | MF | n | n/a | n/a | n/a |
| 37 | 143s | MF | n | n/a | n/a | n/a |
| 38 | 1455s | C-ALCL | +++ | +++ | 100 | 100 |
| 39 | 2624s/2626b | C-ALCL | ++++ | +++ | 60 | 100 |
| 40 | 2092s/2095b | C-ALCL | + | +++ | 100 | 100 |
| 41 | 2267b/1358b | C-ALCL | ++ | ++ | 20 | 90 |
| 42 | 2065s | C-ALCL | +++ | +++ | 80 | 100 |
| 43 | 1281s/1282I | C-ALCL | ++ | +++ | 90 | 100 |
| 44 | sk273 | C-ALCL | n | − | − | − |
| 45 | 1950s | C-ALCL | n | − | − | − |
| 46 | SK46s/SK47s | C-ALCL | ++ | n/a | n/a | n/a |
| 47 | SK40s/SK44s | C-ALCL | n | n/a | n/a | n/a |
| 48 | 302s | LyP | n | − | − | − |
| 49 | 142s | DLCL | n | − | − | − |
| 50 | 1233b/1234s | DLCL | n | − | 10 | − |
| 51 | 613b | DLCL | n | + | 60 | 5 |
| 52 | 419s | DLCL | n | − | − | − |
| 53 | 1662s | DLCL | n | + | 10 | 50 |
| 54 | 1993s | DLCL | n | − | 20 | − |
| 55 | 1201s | MZL | n | + | 10 | 40 |
| 56 | sk311s | MZL | n | + | 50 | 5 |
| 57 | 1533s | MZL | n | − | − | − |
| 58 | 227s | MZL | n | − | 10 | − |
| 59 | 1848s | MZL | n | − | − | − |
| 60 | 1794b | MZL | n | − | 10 | − |
| 61 | 1797s | MZL | n | − | 10 | − |
| 62 | 928s | MZL | + | + | 10 | 10 |
| 63 | 1475s | MZL | n | n/a | n/a | n/a |
| 64 | 258s | Immunocytoma | n | − | − | − |
| 65 | 1728s | FCCL | n | + | 50 | 5 |
| 66 | 423s | FCCL | − | − | 10 | − |
| 67 | 2047s | FCCL | n | − | 10 | − |
| 68 | 2075s | FCCL | + | ++ | 30 | 60 |
| 69 | 2198s | FCCL | n | ++ | 20 | 90 |
| 70 | 1943b | Unclassified | n | + | 10 | 10 |
| 71 | 1648b | Systemic FL | + | + | 10 | 10 |
| 72 | 1564b | Systemic FL | n | − | 10 | − |
| 73 | 1683s | NK | n | − | − | − |
| 74 | 18021i | NK | n | + | 50 | 5 |
| 75 | 2022s | NK | n | + | 20 | 20 |
| 76 | 2412s | NK | n | − | 60 | − |
| 77 | 1860s | NK | + | n/a | n/a | n/a |
| 78 | 1231s | NK | n | n/a | n/a | n/a |
| 79 | 2634b | NK | − | n/a | n/a | n/a |
s indicates skin lesion; b, blood; l, lymph nodes; SS, Sezary syndrome; MF, mycosis fungoides; C-ALCL, primary cutaneous CD30+ anaplastic large cell lymphoma; LyP, lymphomatoid papulosis; DLCL, diffuse large cell lymphoma; MZL, marginal zone lymphoma; FCCL, follicle center cell lymphoma; unclassified, unclassified lymphoma; systemic FL, systemic follicular lymphoma; NK, natural killer cell lymphoma; n, N value <2 (normal); n/a, not available; +, N value >2; ++, N value >4; +++, N value >8; and ++++, N value >16.
indicates N value < 0.5.
indicates negative staining.
Proportion of large atypical cells in total dermal infiltrate; −, no large cell.
Proportion of large atypical cells with positive staining; −, no positive large cell.
Genomic microarray
This experiment was conducted as previously described.10 After probe labeling, hybridization, and posthybridization washes, fluorescent images of the hybridized microarray chips, AmpliOnc I DNA array (Vysis, Downers Grove, IL), which contains 59 clones from 57 oncogenes (BCL2 andAR are represented by both 5′ and 3′ genomic clones) representing genomic regions that have been reported to be amplified in human tumors (Vysis, Downers Grove, IL) were captured and analyzed using the GenoSensor Reader System (Vysis). The fluorescence ratio thresholds for gains and losses of oncogene copy number were set according to our control experiments using test and reference DNA from healthy individuals and lymphoma patients.10 Thus, ratios equivalent to 1 ± 0.2 were set as the level for disomy (normal gene copy number), whereas ratios > 1.25 represented trisomy (gain of gene copy number), and ratios < 0.75 were indicative of monosomy (loss of gene copy number).10
RT-PCR
To further confirm amplification of JUNB in PCL, RT-PCR studies were performed. This experiment, based on the TaqMan assay,15 was carried out using the ABI Prism 7700 Sequence Detector System (ABI/Perkin Elmer, Foster City, CA) as previously reported.10 16 Primer and TaqMan probe sequences were designed using the Primer Express version 1.0 software (ABI/Perkin Elmer) and GenBank sequence numbers were M29039, M12523, and M17987 for JUNB, ALB, and B2M, respectively, with the following primer sequences: JUNB: forward CTACGGGATACGGCCGG, reverse AGGCTCGGTTTCAGGAGTTTG, ALB: forward AGGGTAAAGAGTCGTCGATATGCT, reverse CAATCTCAACCCACTGTCAGCTA,B2M: forward GGAATTGATTTGGGAGAGCATC, reverse CAGGTCCTGGCTCTACAATTTACTAA.
The TaqMan probes were JUNB: 5′-(FAM)-CCCCTGGTGGCCTCTCTCTACACGACTA-(TAMRA)-3′, ALB: 5′-(FAM)-CAAACGCATCCATTCTACCAACTTGAGCAT-(TAMRA)-3′, B2M: 5′-(FAM)-AGTGTGACTGGGCAGATCATCCACCTTC-(TAMRA)-3′.
PCR mixes (25 μL) contained 12.5 μL of 2 × TaqMan Universal PCR master mix, 2.5 μL of each primer, and 2.5 μL of TaqMan probe with 1 to 2.5 μL of DNA. The Universal master mix contains ROX (6-carboxy-X-rhodamine), the passive reference fluorochrome that normalizes for pipetting volume errors. Thermal cycling consisted of 2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Each assay included a “no template” control and a standard curve for JUNB,ALB, and B2M produced by normal placental DNA, and all were carried out in triplicate (96-well maximum). The parameter CT is defined as the fractional cycle number at which the fluorescence generated by cleavage of the probe passes a fixed threshold above baseline. The target gene copy number (JUNB)is quantified by measuring CT and by using a standard curve to determine the starting copy number. The ratio of the target gene copy number to the reference gene copy number normalizes the amount and quality of genomic DNA. The ratio defining the level of increased copy number of the target gene was termed as “N” and was determined as follows: n = copy number of target gene/copy number of reference gene. An N value > 2 was set for gene amplification (+), > 4 (++), > 8 (+++), and > 16 (++++) (Table 2).10,15 16An N value < 0.5 was regarded as decreased copy number (Table 2).
IHC
To further investigate the expression pattern of JUNB in PCL, immunohistochemical studies were performed using the DAKO ChemMate horseradish peroxidase system and DAKO DAB substrate system according to the supplier's instruction (DAKO, Carpinteria, CA).17Briefly, deparaffinized tissue sections were first treated with 3% H2O2 for 10 minutes to inhibit endogenous peroxidase and then microwaved at 700 watts for 18 minutes in 0.01 M sodium citrate buffer solution (pH 6.0) for 30 minutes. After a series of washes, the sections were incubated with primary mouse monoclonal antibody against JUNB (C-11: sc-8051) (Santa Cruz Biotechnology, Santa Cruz, CA) at room temperature for 1 hour at a final antibody concentration of 2 μg/mL diluted in blocking serum solution. After further incubation with universal biotinylated link antibody and peroxidase-labeled streptavidin, the reaction was developed with DAB substrate-chromogen solution for 10 minutes, followed by counterstaining with Harris hematoxylin. To test the specific reactivity of antibody with JUNB, C-11, paraffin sections from colorectal adenocarcinoma, keratosis, and normal lymph nodes (5 samples each) were initially stained with C-11 and an antibody of nonhuman reactive rabbit IgG (Santa Cruz Biotechnology), which was used as the negative control. All colorectal adenocarcinomas and epidermal basal and suprabasal keratinocytes in keratosis showed strong nuclear JUNB expression as previously reported.17,18 However, 5 normal lymph nodes had negative staining for JUNB. In addition, all the samples tested were negative staining for the nonhuman reactive rabbit IgG. To further exclude false-positive and false-negative results in each experiment, a positive control consisting of a colorectal adenocarcinoma and epidermal basal and suprabasal keratinocytes in each sample with known expression of JUNB and a negative control consisting of the rabbit IgG and normal lymph nodes without expression of this oncoprotein were used. The slides were analyzed for the proportion of tumor cells showing nuclear positivity. The level of JUNB expression was qualitatively defined as (+) when 5%-45% of tumor cells were positive, (++) when 50%-90% of tumor cells were positive, and (+++) when 100% of tumor cells were positive.19
Results
Genomic microarray
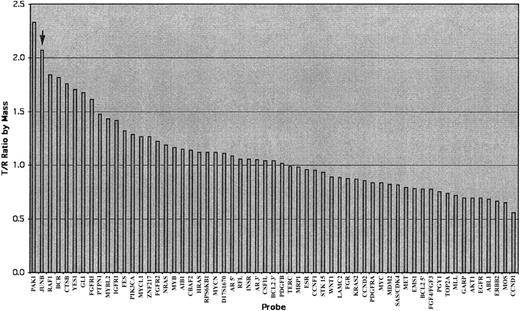
All 7 MF/SS cases studied showed GI (100%). Oncogene copy number gains of RAF1 (3p25), CTSB(8p22), PAK1 (11q13), andJUNB (19p13) were identified in 5 cases (71%), gains ofFGFR1 (8p11), PTPN (20q13),and BCR (22q11) in 4 cases (57%), gains of MYCL1(1p34), PIK3CA (3q26), HRAS(11p15), MYBL2 (20q13), andZNF217 (20q13) in 3 cases (43%), and gains ofKRAS2 (12p12), GLI (12q13), IGFR1 (15q25), FES (15q26),and YES1 (18p11) in 2 cases (29%) (Table 1) (Figure1). Two female patients showed gains ofAR5′ and AR3′ as evidence for hybridization efficiency. There was a similar pattern of GI in SS and MF (Table 1) (Figure 1).
Illustration of an analyzed chart of the AmpliOnc microarray chip hybridized with the DNA sample from a patient with an SS patient (case 5, sample 1278).
The x-axis lists all informative oncogene probes and the y-axis represents the detected fluorescence ratio changes of tumor (T) against reference (R). The T/R ratio of JUNB (arrow) was beyond the threshold of 1.25 indicating gains of these oncogenes.
Illustration of an analyzed chart of the AmpliOnc microarray chip hybridized with the DNA sample from a patient with an SS patient (case 5, sample 1278).
The x-axis lists all informative oncogene probes and the y-axis represents the detected fluorescence ratio changes of tumor (T) against reference (R). The T/R ratio of JUNB (arrow) was beyond the threshold of 1.25 indicating gains of these oncogenes.
RT-PCR
Of 78 PCL cases analyzed with RT-PCR, 19 cases showed amplification of JUNB (24%) (Table 2). This included 7 C-ALCL (70%), 4 MF (29%), 4 SS (18%), 2 PCBCL (9%), 1 NK cell lymphoma (14%), and 1 systemic FL (50%). High-level amplification ofJUNB (+++) was present in 3 C-ALCL (30%) and 2 MF cases (14%) (Table 2). Decreased copy number of JUNB also was seen in 1 MF, 1 FCCL, and 1 NK cell lymphoma each (Table 2). When multiple samples such as blood and skin lesions from the same individual were analyzed, results were consistent. Three cases (nos. 5, 20, 21) showing gain of JUNB detected with genomic microarray also revealed amplification of JUNB using RT-PCR, but case 1 had gain of JUNB by genomic microarray without RT-PCR evidence of amplification of JUNB (Table 2).
IHC
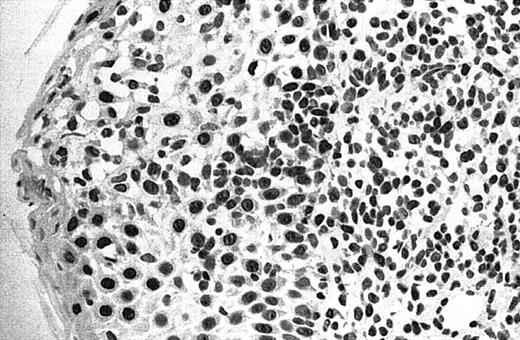
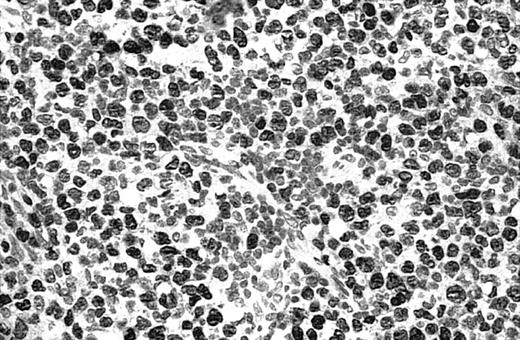
Of 69 PCLs analyzed by IHC, 44 cases showed nuclear JUNB expression in a proportion of tumor cells (64%) (Table 2). This included 21 (91%) of 23 SS, 6 (75%) of 8 C-ALCL, 5 (50%) of 10 MF, 9 (43%) of 21 PCBCL, 2 (50%) of 4 NK cell lymphoma, and 1 (50%) of 2 systemic FL (Table 2). Seven cases (10%) revealed expression of JUNB by all tumor cells (+++) (overexpression), including 5 C-ALCL (63%) and 2 SS cases (9%) (Table 2; Figures2-3). Epidermal basal and suprabasal keratinocytes also expressed JUNB, which represented a useful internal control to indicate the efficiency of immunohistochemistry (Figure 2). All the positively stained PCBCL cases showed only occasional cells expressing JUNB (+) (Table 2), and in this case it is difficult to conclusively establish whether expression is restricted to tumor cells or activated B cells on morphology.
Illustration of IHC staining of JUNB in an SS patient (case 8).
This photograph (original magnification × 40) revealed strong nuclear expression of JUNB by large epidermotropic tumor cells with absent expression in small dermal mononuclear cells. Epidermal basal keratinocytes also expressed JUNB, representing a useful internal control.
Illustration of IHC staining of JUNB in an SS patient (case 8).
This photograph (original magnification × 40) revealed strong nuclear expression of JUNB by large epidermotropic tumor cells with absent expression in small dermal mononuclear cells. Epidermal basal keratinocytes also expressed JUNB, representing a useful internal control.
Illustration of IHC staining of JUNB in a C-ALCL patient (case 38).
This photograph (original magnification × 40) showed strong nuclear expression of JUNB by large anaplastic cells with no expression by small reactive lymphocytes.
Illustration of IHC staining of JUNB in a C-ALCL patient (case 38).
This photograph (original magnification × 40) showed strong nuclear expression of JUNB by large anaplastic cells with no expression by small reactive lymphocytes.
There was a striking concordance between the results of RT-PCR and IHC (Table 2). Fifteen cases (6 C-ALCL, 4 SS, 2 MF, 2 PCBCL, and 1 systemic FL) with amplification of JUNB identified by RT-PCR also showed expression of JUNB by tumor cells (Table 2). In addition, 25 cases (12 PCBCL, 5 MF, 2 SS, 2 C-ALCL, 2 NK cell lymphoma, 1 systemic FL, and 1 LyP) without amplification of JUNB did not express JUNB protein by tumor cells (Table 2).
Discussion
This genomic microarray study of CTCL variants has shown a global picture of oncogene copy number changes in MF and SS. Seventy-one percent of cases revealed gains of JUNB, RAF1,CTSB, and PAK1, and 57% demonstrated gains ofFGFR1, PTPN, and BCR. In addition, a consistent pattern of GI was present in SS and MF, supporting our previous hypothesis that both SS and MF represent part of a spectrum of the same disease as suggested by a similar pattern of CI detected by CGH.9 In contrast, these results are different from our previous observations in PCBCL,10 which is consistent with a different pathogenesis. RT-PCR analysis of PCL showed amplification of JUNB in 70% of C-ALCL, 29% of MF, 18% of SS, 14% of cutaneous NK cell lymphoma, and 9% of PCBCL cases. JUNB expression in more than 50% of CTCL variants was present in a majority of tumor cells. In contrast, expression of JUNB in PCBCL was limited to a minority of the cellular infiltrate. There was general concordance between the results of JUNB detected by these 3 different techniques. For instance, high-level amplification and overexpression of JUNB was consistently detected in C-ALCL, MF, and SS using RT-PCR and IHC. Occasional rare discrepancies were noted and are likely to be due to the different detection sensitivity of these techniques. These findings suggest that amplification and overexpression of JUNB may be a key pathogenetic event in CTCL.
JUNB protein is one of the principal components of the activating protein-1 (AP-1) transcription factor complex, consisting of the JUN (C-JUN, JUNB, and JUND) and FOS (C-FOS, FOSB, FRA1, and FRA2) families, which has been implicated in a variety of biologic processes.20,22-24 For example, previous studies have shown that JUNB is involved in control of the cell cycle by inducing high level expression of the cyclin-dependent kinase inhibitor p16(INK4a) and down-regulating cyclin D expression, producing a decrease in pRb hyperphosphorylation and G1-phase extension leading to premature cell senescence.21,25,26 In addition, in T cells JUNB has been found to promote T–helper-2 cell (TH2) differentiation through up-regulation of interleukin-4 (IL-4).27-29 Transforming growth factor-beta1 (TGFB1) also induces expression of JUNB in human leukemic cells,26 and interestingly, a TH2 immunophenotype and overexpression of TGFB1 are characteristic of CTCL.30-32 The TAX oncoprotein of human T-cell leukemia virus type 1 (HTLV-1) also induces JUNB expression,33,34 although all the cases in this study were HTLV-1 negative, and HTLV-1 is not associated with MF/SS.35 JUNB is expressed in epidermal keratinocytes, consistent with our observation in this study, and is thought to have a role in wound healing, photoaging, and UV-induced skin carcinogenesis.18 It has been suggested that JUNB and JUND are proliferation inhibitor or tumor suppressors.20,22,23,36-39 In contrast, C-JUN functions as a promoter of cell proliferation and as an apoptosis inhibitor due to down-regulation of p53, p21, and p16, and up-regulation of cyclin-dependent kinases.20,22,23,36-39 These studies would appear to suggest that amplification and overexpression ofJUNB would be apoptotic and inhibit tumor cell proliferation in CTCL. However, recent knock-in mouse studies have shown that JUNB can substitute for the absence of C-JUN during mouse development and cell proliferation,39,40 and this is thought to be through the constitutive activation of transcription factor NF-κB, which controls the activation of JUNB.41 This may explain why overexpression of C-JUN and JUNB proteins are detected in human colorectal, ovarian, and cervical cancers but not in normal tissues, whereas JUND protein is present in normal tissues but rarely in tumors.17,42,43 Amplification and/or overexpression ofJUNB also have been described in Hodgkin lymphoma cell lines, cervical cancer cell lines, and endometrial cancers.41,44,45 In this study, we identified copy number gain of JUNB in 5 MF/SS cases using genomic microarray. This was supported by RT-PCR and IHC studies, which revealed amplification and expression of JUNB in 8 and 28 cases, respectively. In addition, high-level amplification and overexpression ofJUNB were identified in CTCL variants. This was in contrast to findings in PCBCL, in which only rare cases showed amplification and expression of JUNB, and in fact decreased copy number ofJUNB was detected in 3 cases.10 Taken together, these results suggest that up-regulation of JUNB is frequently associated with CTCL variants rather than PCBCL. Further studies of C-JUN, C-FOS, and JUND are now required in PCL.
Apart from JUNB, other oncogenes such as RAF1,CTSB, and PAK1 also were frequently identified in MF/SS. RAF1 (v-raf-1 murine leukemia viral oncogene homolog 1) is a mitogen-activated protein kinase that acts downstream of RAS viral oncogene homolog and is regulated by BCL2 and other apoptosis-related proteins.46CTSB encodes cathepsin B, a lysosomal cysteine protease that cleaves amyloid precursor protein and is involved in tumor invasion.46PAK1 (p21/Cdc42/Rac1-activated kinase 1) belongs to the PAK family composed of serine/threonine p21-activating kinases, which are involved in cytoskeleton reorganization and nuclear signaling. PAK1 regulates cell motility and morphology.46 Previous studies have shown amplification of RAF1, CTSB, andPAK1 in breast, esophageal, and urinary bladder carcinomas, respectively.47-49 At present there are no data on alterations of these oncogenes in nodal lymphomas. Therefore, further studies are required to confirm amplification of these oncogenes in PCLs with functional studies to establish the significance of these findings.
In summary, we have found consistent patterns of GI in MF/SS and frequent amplification and overexpression of JUNB in C-ALCL, MF, and SS, suggesting that this oncogene may play an important role in the pathogenesis of CTCL.
Prepublished online as Blood First Edition Paper, October 3, 2002; DOI 10.1182/blood-2002-08-2434.
Supported by grants from the British Skin Foundation, Dermatrust, and St John's Special Purpose Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Xin Mao, Skin Tumour Unit, St John's Institute of Dermatology, St Thomas' Hospital, Lambeth Palace Road, London SE1 7EH, United Kingdom; e-mail: mxmayo@hotmail.com ormxmayo@yahoo.co.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal