Platelet adhesion to damaged vessel wall and shear-induced platelet aggregation necessitate binding of the von Willebrand factor (VWF) A1 domain to platelet GPIbα. Blocking this interaction represents a promising approach to the treatment of arterial thrombosis. Comparison of amino acid sequences of the VWF A1 domain in several species, expressing VWF recognized by the blocking monoclonal antibody AJvW-2, suggested 9 residues (His563, Ile566, Asp570, Ala581, Val584, Ala587, Arg616, Ala618, and Met622) to contribute to the epitope for AJvW-2 or to be part of the GPIbα-binding site. Glutathione-S-transferase (GST)–human VWF A1 fusion proteins, in which these amino acids were mutated to their murine counterparts, were tested for their capacity to bind AJvW-2 or heparin, to interfere with botrocetin- or ristocetin-mediated VWF binding to GPIb, or to induce flow-dependent platelet tethering in a perfusion chamber. Thus, mutations His563Arg, Ile566Leu, Asp570Ala, and Ala587Thr, clustered on the outer surface of the A1 domain, dramatically impaired binding of AJvW-2 to A1. The His563Arg, Ile566Leu, and Asp570Ala mutations also impaired the binding of heparin, which competes with AJvW-2 for binding to A1. Perfusion studies revealed that His563, Ile566, Asp570, Arg616, and Ala618 take part in GPIbα binding, their mutation-impairing platelet recruitment. In agreement with the surface distribution of VWF type 2M mutations, this study demonstrates overlapping of the epitope for AJvW-2 and the GPIbα-binding site, located around the front pocket of the A1 domain and defined by strands β3, β4, and helix α3, and it provides a mechanistic basis for VWF neutralization by this antibody.
Introduction
The interaction between the A1 domain of von Willebrand factor (VWF) and the glycoprotein Ibα (GPIbα) chain of its platelet receptor, the GPIb-IX-V membrane complex (GPIb), is critical in primary hemostasis and thrombosis.1,2 It is thought that this interaction requires a conformational transition in VWF,3 achieved by VWF binding to subendothelial ligands such as collagen,4,5 or by its exposure to high shear stress generated by the rapid blood flow encountered in stenosed arteries and in the microcirculation,6 initiating shear-induced platelet aggregation (SIPA) in fluid phase.7In contrast, on vascular damage, circulating blood platelets tether on immobilized VWF and initiate rolling.8 Translocating platelets undergo inside-out signaling, leading to subsequent activation of the GPIIb/IIIa receptor and stable aggregate formation.9
The A1 domain of VWF extends from amino acid residues 497 to 716 and is structurally characterized by a disulfide bridge between cysteine residues 509 and 695.10,11 X-ray diffraction studies of crystals of the A1 domain, in complex with the neutralizing anti-VWF antibody NMC-4,12 and of the recombinant chymotrypsin-treated A1 domain13 have revealed an A1 domain conformation with a globular shape comprising a central core constituted of 6 hydrophobic β-strands, surrounded by 6 amphipathic α-helices. The conformations of the VWF A1 and A3 domains are similar13,14 and comply with the structure of other type A domains such as in complement receptor 3 (CR3) and lymphocyte function-associated antigen-1 (LFA-1).15
Peptide-docking studies have anticipated a central front groove on the A1 domain, next to strand β3, to be part of the binding site for GPIbα.16 On the other hand, spontaneously occurring missense mutations in VWF have been found, associated with decreased affinity of VWF for GPIbα. These so-called type 2M loss-of-function mutations, with preserved degree of VWF-multimerization, have identified mutations located on the front of the VWF A1 domain (International Society on Thrombosis and Haemostasis, Scientific and Standardization Committee (ISTH SSC VWF) database:www.shef.ac.uk/vwf/). Likewise, mutagenesis studies using recombinant full-length VWF17,18 or recombinant VWF A1 domain mutants16,19 20 have further identified a number of front residues involved in VWF binding to GPIbα (see “Discussion”).
We have previously shown that the murine anti-VWF antibody AJvW-2 is a potent inhibitor of shear-induced platelet activation and of VWF-dependent thrombosis. This antibody binds to the A1 domain of VWF derived from several species, recognizing an epitope conserved in all these species.21-25 We have used this property to identify the amino acid residues recognized by AJvW-2 and to study the interaction between VWF and GPIbα in more detail. By using a series of recombinant A1 domain mutants, which were designed following sequence comparison between murine, human, canine, porcine, and rabbit VWF, we found that AJvW-2 does indeed bind to the front side of strand β3 of the A1 domain, involving sequences that are part of both the binding pocket for GPIbα and the heparin-binding site. Analysis of the dynamic interactions between the A1 domain mutants and platelets and comparison of the location in the A1 domain of our loss-of-function mutations to the known type 2M missense mutations led to the conclusion that AJvW-2 shields the GPIb binding pocket of VWF A1, comprised of strands β3 and β4, along with helix α3.
Materials and methods
Materials
Glycocalicin (GPIbα extracellular domain) was isolated from fresh platelets after calpain digestion, as described.26Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with anti-GPIbα monoclonal antibodies (mAbs) raised in the laboratory confirmed a homogeneous glycocalicin band (not shown). Human VWF was purified from plasma cryoprecipitate by gel filtration on a Sepharose 4B-CL column.27 The neutralizing murine anti-VWF mAb AJvW-2 was from Ajinomoto (Kawasaki, Japan).21 The neutralizing murine anti-GPIbα mAb G19H1028 (immunoglobulin G1 [IgG1]), the anti-GST mAb 21C11, and the conformation-dependent anti-VWF A1 domain mAbs A21H3 and A7C6 were raised in our laboratory. Antibodies were purified by Protein A–Sepharose chromatography.
Peroxidase-conjugated rabbit anti-VWF polyclonal antibody (poly–anti-VWF), conformation-dependent for the A1 domain of VWF, was from DAKO (Glostrup, Denmark). Botrocetin was isolated from the venom of Bothrops jararaca (Sigma, St Louis) as reported.29 The GPIIb/IIIa antagonist tirofiban was from Merck (Whitehouse Station, NJ). Calcein-am was purchased from Molecular Probes (Leiden, The Netherlands), and heparin was from Rhône-Poulenc Rorer (Brussels, Belgium).
Expression and purification of glutathione-S-transferase–VWF A1 domain fusion proteins
Glutathione-S-transferase (GST)–VWF A1 wild-type (wt) and mutant domain fusion proteins were made as follows. cDNA fragments were amplified from the pSP8800VWF vector, encoding mature VWF,30 introducing a 5′ BamHI site and a 3′SalI site flanking the nucleotide sequence encoding the VWF A1 domain (residues 499-729). Amplification by polymerase chain reaction (PCR) was performed in a PTC-100 Programmable Thermal Controller (MJ Research, Watertown, MA). First, a 5′ sense primer corresponding to the BamHI site (5′-GTG GAG GGA TCC TCG GAA CCG CCG TTG CAC) and a 3′ antisense oligonucleotide primer with the desired nucleotide substitution were combined. Separately, the 5′ sense primer corresponding to the SalI site (5′-AGA ACC GTC GAC TTC CTC TTG GGC CCC AG) and a 3′ primer containing the desired mutation were combined. PCR was performed as follows: heating at 94°C for 5 minutes; 25 cycles of amplification at 94°C for 20 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; then an additional cycle of 10 minutes at 72°C. The resultant DNA fragments of both reactions were mixed to produce the VWF-A1 mutant cDNA, and a third PCR was performed using the 5′ terminal and 3′ terminal primers (heating at 94°C for 5 minutes; then 25 cycles of amplification at 94°C for 30 seconds, 58°C for 45 seconds, and 72°C for 45 seconds; and an additional cycle of 10 minutes at 72°C). PCR products were digested by BamHI and SalI, inserted into the pGEX4T-2 vector containing the DNA sequence for GST (Amersham Pharmacia Biotech, Roosendaal, The Netherlands), and cloned. Mutant plasmids were sequenced to confirm the desired mutations and were transformed inEscherichia coli BL21 cells.
Fusion protein expression was induced with 500 μM isopropyl-B-D-thiogalactopyranoside (IPTG) for 3 hours at 30°C. Bacteria were centrifuged at 3500g for 30 minutes, and the pellet was lysed at room temperature (RT), in 50 mM Tris (tris[hydroxymethyl]aminomethane)–HCl buffer, pH 8.0, containing, 100 mM KCl, 400 mM NaCl, 5 mM dithioerythritol (DTE), 1 mg/mL lysosome, and a cocktail of protease inhibitors (Complete; Boehringer Mannheim, Germany). The lysate was sonicated 2 × 2 minutes, incubated for 30 minutes at RT with 1% Triton X-100, and centrifuged at 20 000g for 15 minutes at 4°C, and the supernatant was incubated overnight at 4°C with 4% (vol/vol) glutathione–Sepharose 4B beads (Pharmacia, Uppsala, Sweden). After washing the beads in phosphate-buffered saline (PBS; 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, pH 7,4) the GST-fusion proteins were eluted with 100 mM Tris-HCl buffer, pH 8.1, containing 20 mM reducing glutathione and 120 mM NaCl. Samples were filtered at 0.22 μm and dialyzed overnight at 4°C in 20 mM Tris-HCl buffer, pH 8.0, containing 50 mM NaCl. Samples were loaded onto a Q-Sepharose column (Pharmacia), equilibrated in the same buffer, and eluted with a 50 to 500 mM NaCl gradient. GST-A1 wild-type A1 (wt-A1) and mutants eluted between 250 and 300 mM NaCl (flow rate, 1 mL/min).
Binding of AJvW-2 to wild-type A1 and mutants
The binding of AJvW-2 and of the conformation-dependent poly–anti-VWF, A21H3, and A7C6 to A1 fusion proteins were tested by enzyme-linked immunosorbent assay (ELISA). The wt-A1 and mutants were coated overnight at 4°C in microtiter plates (Costar, Corning, NY; high binding) at 2 μg/mL in 100 μL PBS. After blocking the plates with 1% (wt/vol) bovine serum albumin (BSA) in PBS, antibodies (2 to 10 μg/mL) were deposited in the wells in PBS supplemented with 0.002% (vol/vol) Tween 80 (PBS-T), containing 0.1% BSA, for 2 hours at RT. Bound mAbs were revealed with secondary horseradish peroxidase-conjugated goat antimouse IgG (DAKO A/S; dilution 1/3000), and o-phenylenediamine, whereas the poly–anti-VWF was directly revealed by the chromogenic substrate. Equal amounts of wt-A1 and mutants were found to adsorb onto the wells, as measured by using the anti-GST antibody 21C11.
Heparin-binding assays
Fifty-microliter–packed heparin-Sepharose beads (CL-6B; Pharmacia) were incubated in microcentrifuge tubes with 50 μL of 20 mM Tris-HCl buffer, pH 7.3, 150 mM NaCl containing 5 μg of wt-A1 or mutants, for 2 hours at RT. Samples were then centrifuged at 10 000g for 2 minutes, and unbound ligands in the supernatants were measured using the Bio-Rad protein assay (Munich, Germany). A GST-VWF-A3 domain fusion was used as a negative control.
The competition between heparin and AJvW-2 for binding to wt-A1 was investigated using Biospecific Interaction Analysis in a BIACore 1000 instrument (BIACore, Uppsala, Sweden). AJvW-2 was immobilized on CM5 chips and superfused with 25 μg/mL wt-A1 in the presence of increasing concentrations of unfractionated heparin.
Inhibition of botrocetin-induced binding of VWF to glycocalicin
Microtiter plates coated with 2 μg/mL glycocalicin in 200 μL PBS overnight at 4°C were saturated with 0.5% (wt/vol) casein in 10 mM Tris-HCl buffer, pH 7.3, containing 0.9% NaCl, for 1 hour at RT. Wells were washed with PBS-T and incubated for 2 hours at RT with 0.5 μg/mL VWF and 2 μg/mL botrocetin in the presence of increasing concentrations (0-20 μg/mL) of wt-A1 or mutants in PBS-T containing 0.5% casein, following which bound VWF was detected with the poly–anti-VWF antibody and o-phenylenediamine. The fraction of absorbance resulting from binding of the poly–anti-VWF to the A1 mutants represented less than 10% of the signal produced by the full-length VWF and could be neglected.
Inhibition of ristocetin-induced platelet agglutination by wild-type A1 and mutants
The capacity of each A1 mutant to inhibit ristocetin-induced platelet agglutination (RIPA) was measured in the platelet-rich plasma (PRP) from 3 different donors, adjusted to 250 000 platelets per microliter with platelet-poor plasma using a 4-channel Chrono-Log aggregometer (Kordia, Leiden, The Netherlands). First the concentration of wt-A1 leading to 50% inhibition of the initial slope of platelet agglutination induced by 0.8 or 0.9 mg/mL ristocetin was determined at 37°C, with constant stirring at 1200 rpm. The A1 mutants were then tested at the same concentration—20 or 30 μg/mL, depending on the PRP donor—and the initial slopes of platelet agglutination were analyzed.
Platelet interaction with wild-type A1 and mutants under flow conditions
Preparation of reconstituted blood.
PRP prepared from blood collected on 1:6 acid-citrate-dextrose (ACD; 93 mM trisodium citrate, 7 mM citric acid, 140 mM dextrose, pH 6.5) and 1 μM tirofiban was incubated for 20 minutes at 37°C with 5 μM calcein-am, an acetoxymethyl ester, which is fluorescent once cleaved by nonspecific esterases inside the cell, with no detectable effect on platelet function in our perfusion studies. PRP was then centrifuged at 700g for 25 minutes, and fluorescent platelets were resuspended in HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)–Tyrode buffer (5 mM HEPES, 137 mM NaCl, 2.7 mM KCl, 12 mM NaHCO3, 0.36 mM NaH2PO4, 1% (wt/vol) glucose, pH 7.3) containing 1% (vol/vol) human serum albumin (HSA). Just before perfusion, platelets were added at 10 000/μL to washed red blood cells (RBCs) adjusted to a hematocrit level of 45%, in the presence of 1 μM tirofiban. RBCs were prepared from packed RBCs (group O-negative; Red Cross Blood Bank, Leuven, Belgium) diluted 1:3 with HEPES-Tyrode buffer in the presence of 1:6 volume of ACD. On centrifugation at 450g for 25 minutes at 23°C, the supernatant and residual white blood cells on top of the sedimented RBCs were removed. This procedure was repeated twice. Finally, RBCs were centrifuged at 1300g for 25 minutes and were resuspended in HEPES-Tyrode buffer containing 1 mM CaCl2, 1 mM MgCl2, and 1% HSA.
Glass coverslip coating with A1 fusion proteins.
24 × 50-mm glass coverslips (CSs) were incubated with 250 μL fusion proteins at a concentration of 300 μg/mL in Tris-buffered saline (TBS; 50 mM Tris, 137 mM NaCl, 2.7 mM KCl, pH 7.3), for 2 hours at 37°C and subsequently were saturated with HEPES-Tyrode buffer containing 1% HSA for 30 minutes at RT. When specified, wt-A1–coated CSs were further incubated for 30 minutes at 37°C in TBS containing 1 mM CuSO4 or 10 mM DTE, before the saturation step. The degree of adhesion for wt-A1 and mutants onto CSs was measured by incubating 20 × 20-mm glass coverslips coated with the A1 fusion proteins in 6-well plates (Falcon; Becton Dickinson Labware, Franklin Lakes, NJ), presaturated with 1% HSA overnight at 4°C, with the anti-GST antibody 21C11, in an ELISA-type assay. The wt-A1 and mutants were found to adsorb onto the CSs with comparable efficiencies (SD ± 5.0%). Reconstituted blood was also perfused over CSs coated with 250 μL native VWF at 50 μg/mL in PBS for 2 hours at 37°C.
Flow chamber and perfusion studies
Dynamic interactions between platelets and A1 mutants or native VWF were analyzed in a parallel-plate perfusion chamber.28 The coated coverslip constituted the bottom of the chamber, and the actual chamber was formed by a 254-μm–high silicon rubber gasket designed with a conically shaped flow path, thus resulting in a 3-fold increase of wall shear rate from the inlet of the chamber to the outlet. Maintaining a flow rate of 0.5 or 2.5 mL/min with an inverted syringe pump (Harvard Instruments, South Natick, MA), wall shear rates ranged from 200 seconds−1 to 1500 seconds−1 throughout the flow path. HEPES-Tyrode buffer containing 1% HSA was aspirated at 37°C for 5 minutes to warm the chamber and perfuse the coverslip, after which reconstituted blood at 37°C was perfused. When used, blocking antibodies AJvW-2 or G19H10 were added to reconstituted blood 2 minutes before the onset of perfusion. By mounting the flow chamber on the table of an inverted epifluorescence microscope (Diaphot; Nikon, Melville, NY), coupled to a charge-coupled device video camera (COHU, San Diego, CA), images were read into the memory of an attached computer.28 Captured images were digitized with a Scion LG3 frame grabber (Scion, Frederick, MD). Stored images were then analyzed as real-time movies using the NIH Image program version 6.1.
Quantitation of platelet interaction with GST-A1 proteins or native VWF.
This analysis was carried out according to Miyata and Ruggeri.31 Three minutes after the beginning of the perfusion, 10 images of 205 × 330-μm fields were captured randomly at positions in the flow path corresponding to chosen wall shear rates (200, 1000, or 1500 seconds−1). The number of platelets interacting with the surface was counted, applying an arbitrary gray-level threshold to distinguish platelets from background fluorescence. Movies of 12 seconds (10 images per second) were recorded, and the velocity of platelet translocation at a wall shear rate of 1500 seconds−1 was determined by measuring the distance traveled by platelets rolling over the protein-coated surface during 1 second of flow.
Statistics.
Data are expressed as means ± SDs of at least 3 independent assays and were analyzed using the Student t test (significance for P < .05 or P < .01).
Results
Design, production, and characterization of GST-A1 domain fusion proteins
AJvW-2 reacts with human, canine, porcine, and rabbit VWF. The amino acid sequence alignment of the VWF A1 domain for these species (Table 1) reveals a high degree of homology. AJvW-2 is raised in the mouse and does not react with murine VWF. Hence, identification of residues conserved between human, canine, porcine, and rabbit VWF A1 domains, but different from the corresponding murine A1 domain residues, disclosed some 9 positions, potentially involved in antibody binding: His563, Ile566, Asp570, Ala581, Val584, Ala587, Arg616, Ala618, and Met622. In view of the inhibitory properties of AJvW-2, these residues are potentially operational in VWF binding to GPIbα. Two residues, at positions 581 and 622, are not entirely conserved between human, canine, porcine, and rabbit VWF.
Alignment of VWF A1 domain amino acids
| Mouse (1) DTP EPPLHNFYCS KLLDLVFLLD GSSMLSEAEF-530 |
| Human (2) DIS EPPLHDFYCS RLLDLVFLLD GSSRLSEAEF |
| Dog (3) DTP EPPLHDFHCS RLLDLVFLLD GSSKLSEDEF |
| Pig (4) DTP EPPLHDFFCS KLLDLVFLLD GSDKLSEADF |
| Rabbit (5) DTP EPPLHDFYWS NLMDLVFLLD GSAQLSEAEF |
| (1) EVLKAFVVGM MERLHISQKR TRVAVVEYHD GSRAYLELKA-570 |
| (2) EVLKAFVVDM MERLRISQKW VRVAVVEYHD GSHAYIGLKD |
| (3) EVLKVFVVGM MEHLHISQKR IRVAVVEYHD GSHAVIELKD |
| (4) EALKVFVVGM MEHLHISQKH IRVAVVEYHD GPHAVISLQD |
| (5) GVLKAFVVSV MERLHISQKR IRVAVVEYHD GSHSYISLKD |
| (1) RKRPSELRRI TSQIKYTGSQ VASTSEVLKY TLFQIFGKID-610 |
| (2) RKRPSELRRI ASQVKYAGSQ VASTSEVLKY TLFQIFSKID |
| (3) RKRPSELRRI TSQVKYAGSE VASTSEVLKY TLFQIFGKID |
| (4) RKRPSELRRI ASQVKYAGSE VASISEVLKY TLFQIFGRVD |
| (5) RKRPSELRRI ASQVKYAGGP VASTSEVLKY TLFHIFSNVD |
| (1) RPEASHITLL LTASQEPPRM ARNLVRYVQG LKKKKVIVIP-650 |
| (2) RPEASRIALL LMASQEPQRM SRNFVRYVQG LKKKKVIVIP |
| (3) RPEASRIALL LMASQEPSRL ARNLVRYVQG LKKKKVIVIP |
| (4) RPEASRIALL LMASQEPRRL AQNLARYLQG LKKKKVIVIP |
| (5) RPEASRIALL LSASQETPRM VRNLVRYAQG LKKEKVIVIP |
| (1) VGIGPHASLK QIRLIEKQAP ENKAFLLSGV DELEQRRDEI-690 |
| (2) VGIGPHANLK QIRLIEKQAP ENKAFVLSSV DELEQQRDEI |
| (3) VGIGPHASLK QIHLIEKQAP ENKAFVFSGV DELEQRRDEI |
| (4) VGIGPHVSLK QIRLIEKQAP ENKAFVVSGV DELEQRKNEI |
| (5) VGIGPHVSLR QIHLIEKQAP ENKAFVLSGV DELEQRRDEI |
| (1) VSYLCDLAPE APAP T QPPOV AHV T VSPGIA GI S-723 |
| (2) VSYLCDLAPE APPP T LPPHN AQV T VGPGLL GV S |
| (3) INYLCDLAPE APAP T QHPPM AQV T VGSELL GV S |
| (4) ISYLCDLAPE VPAP T RRPLV AQV T VAPELP GV S |
| (5) ISYLCDLGPE APVP T QRPPT ARV T VSPGQL GV S |
| Mouse (1) DTP EPPLHNFYCS KLLDLVFLLD GSSMLSEAEF-530 |
| Human (2) DIS EPPLHDFYCS RLLDLVFLLD GSSRLSEAEF |
| Dog (3) DTP EPPLHDFHCS RLLDLVFLLD GSSKLSEDEF |
| Pig (4) DTP EPPLHDFFCS KLLDLVFLLD GSDKLSEADF |
| Rabbit (5) DTP EPPLHDFYWS NLMDLVFLLD GSAQLSEAEF |
| (1) EVLKAFVVGM MERLHISQKR TRVAVVEYHD GSRAYLELKA-570 |
| (2) EVLKAFVVDM MERLRISQKW VRVAVVEYHD GSHAYIGLKD |
| (3) EVLKVFVVGM MEHLHISQKR IRVAVVEYHD GSHAVIELKD |
| (4) EALKVFVVGM MEHLHISQKH IRVAVVEYHD GPHAVISLQD |
| (5) GVLKAFVVSV MERLHISQKR IRVAVVEYHD GSHSYISLKD |
| (1) RKRPSELRRI TSQIKYTGSQ VASTSEVLKY TLFQIFGKID-610 |
| (2) RKRPSELRRI ASQVKYAGSQ VASTSEVLKY TLFQIFSKID |
| (3) RKRPSELRRI TSQVKYAGSE VASTSEVLKY TLFQIFGKID |
| (4) RKRPSELRRI ASQVKYAGSE VASISEVLKY TLFQIFGRVD |
| (5) RKRPSELRRI ASQVKYAGGP VASTSEVLKY TLFHIFSNVD |
| (1) RPEASHITLL LTASQEPPRM ARNLVRYVQG LKKKKVIVIP-650 |
| (2) RPEASRIALL LMASQEPQRM SRNFVRYVQG LKKKKVIVIP |
| (3) RPEASRIALL LMASQEPSRL ARNLVRYVQG LKKKKVIVIP |
| (4) RPEASRIALL LMASQEPRRL AQNLARYLQG LKKKKVIVIP |
| (5) RPEASRIALL LSASQETPRM VRNLVRYAQG LKKEKVIVIP |
| (1) VGIGPHASLK QIRLIEKQAP ENKAFLLSGV DELEQRRDEI-690 |
| (2) VGIGPHANLK QIRLIEKQAP ENKAFVLSSV DELEQQRDEI |
| (3) VGIGPHASLK QIHLIEKQAP ENKAFVFSGV DELEQRRDEI |
| (4) VGIGPHVSLK QIRLIEKQAP ENKAFVVSGV DELEQRKNEI |
| (5) VGIGPHVSLR QIHLIEKQAP ENKAFVLSGV DELEQRRDEI |
| (1) VSYLCDLAPE APAP T QPPOV AHV T VSPGIA GI S-723 |
| (2) VSYLCDLAPE APPP T LPPHN AQV T VGPGLL GV S |
| (3) INYLCDLAPE APAP T QHPPM AQV T VGSELL GV S |
| (4) ISYLCDLAPE VPAP T RRPLV AQV T VAPELP GV S |
| (5) ISYLCDLGPE APVP T QRPPT ARV T VSPGQL GV S |
Residues, numbered according to the sequence of the mature human VWF, are aligned for murine, human, canine, porcine, and rabbit VWF A1 domain (residues 498-723). The critical disulfide bridge residues delineating the AI domain (Cys509 and Cys695) are underlined. The 9 residues at positions 563, 566, 570, 581, 584, 587, 616, 618, and 622, indicated with boldfacing, show partial to complete homology between human, canine, porcine, and rabbit VWF AI domains and no homology with murine residues.
We produced a series of fusion proteins between A1 and GST in which all these residues were individually substituted by their murine homologues. Following their isolation through glutathione-Sepharose beads, fusion proteins were further purified by Q-Sepharose chromatography (Figure 1A) as a single peak, eluted between 250 and 300 mM NaCl. SDS-PAGE (Figure 1B) showed that the fusion proteins were largely recovered in an oxidized form, suggestive of preservation of the critical intramolecular disulfide bond Cys509-Cys695 of the A1 domain. On reduction, a characteristic shift to an apparently higher molecular mass occurred (Figure 1B), as observed before.32 Platelet perfusion studies at 1500 seconds−1 over wt-A1, just after pretreating the coverslips with either CuSO4 or DTE, revealed that the number of platelets interacting with the CuSO4-treated wt-A1 was similar to that found for untreated wt-A1, but coverslip pretreatment with DTE almost abolished platelet tethering (Figure2A). Perfusions of blood platelets over wt-A1 and native VWF were compared at different wall shear rates (Figure 2B). The wt-A1 exhibited functional characteristics closely comparable to those of native VWF. Indeed, most (more than 95%) of the platelets interacting with the immobilized fusion protein translocated in a continuous rolling motion over the surface in the direction of the flow (video sequences showing platelet translocation over wt-A1 are available on request). Furthermore, the number of platelets interacting with wt-A1 went up with increasing shear rates, as was found for perfusions over VWF. Finally, platelet rolling and adhesion on wt-A1 were specifically inhibited by the anti–VWF-A1 mAb AJvW-2 and by the anti-GPIbα G19H10 mAbs (Figure 2B). We previously reported that the epitope for AJvW-2 on the A1 domain is highly conformational because the antibody recognizes neither a heat-denatured nor a reduced VWF A1 domain.22 Likewise, AJvW-2 does not interact with heat-denatured (boiling for 5 minutes) or reduced wt-A1 (results not shown), suggesting that the fusion protein was correctly folded, enabling further functional studies. Detection of coated wt-A1 by a nonconformational anti-GST mAb during these experiments confirmed that heat denaturation did not affect the coating efficacy of heated wt-A1.
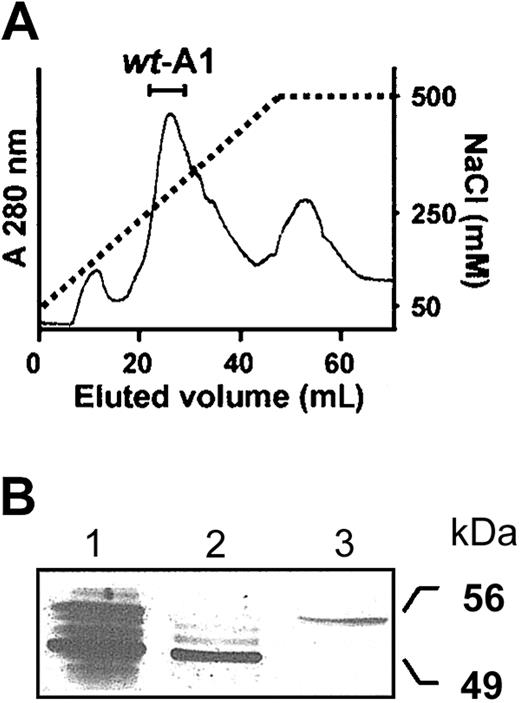
Purification of the wt-A1 fusion protein.
Wild-type A1, prepurified with glutathione-Sepharose beads, was dialyzed (24 hours, 4°C) against 20 mM Tris-HCl, pH 8.0 buffer containing 50 mM NaCl and loaded onto a Q-Sepharose column. (A) wt-A1 was eluted with a 50- to 500-mM NaCl gradient and collected as indicated. (B) SDS-PAGE and silver staining analysis of the collected wt-A1. Lane 1: prepurified wt-A1 after elution from the glutathione-Sepharose beads; lane 2: wt-A1 eluted from the Q-Sepharose. The fusion protein exhibits an apparent Mr of 49 kDa, which increases to 56 kDa after reduction with 100 mM DTE (lane 3).
Purification of the wt-A1 fusion protein.
Wild-type A1, prepurified with glutathione-Sepharose beads, was dialyzed (24 hours, 4°C) against 20 mM Tris-HCl, pH 8.0 buffer containing 50 mM NaCl and loaded onto a Q-Sepharose column. (A) wt-A1 was eluted with a 50- to 500-mM NaCl gradient and collected as indicated. (B) SDS-PAGE and silver staining analysis of the collected wt-A1. Lane 1: prepurified wt-A1 after elution from the glutathione-Sepharose beads; lane 2: wt-A1 eluted from the Q-Sepharose. The fusion protein exhibits an apparent Mr of 49 kDa, which increases to 56 kDa after reduction with 100 mM DTE (lane 3).
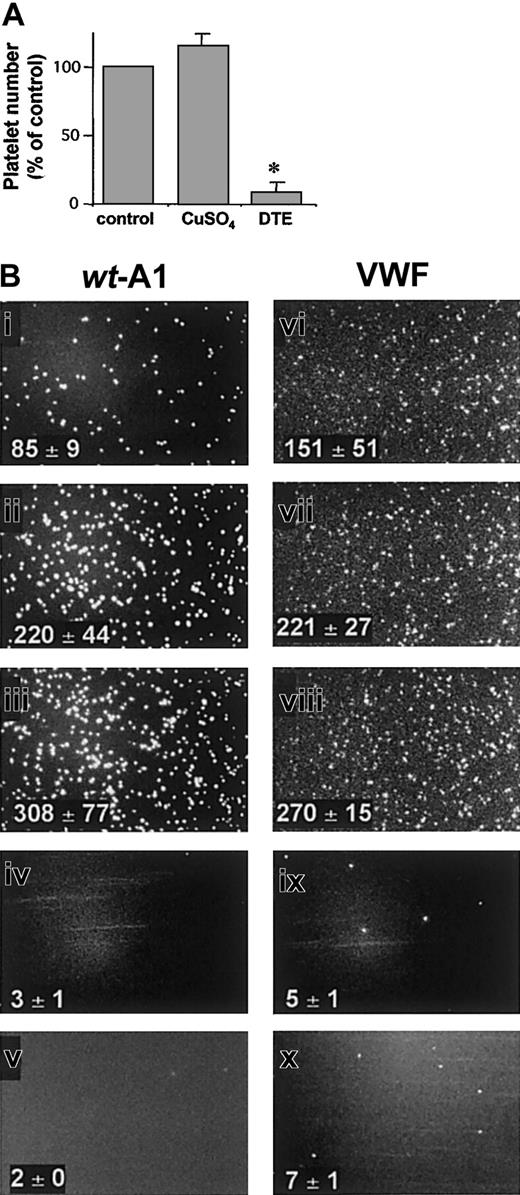
Functional evaluation of the wt-A1 in flow-dependent platelet tethering.
(A) Glass coverslips coated with purified wt-A1 (300 μg/mL) were untreated (control) or treated for 30 minutes at 37°C with 1mM CuSO4 or 10 mM DTE as indicated and were perfused with platelets in reconstituted blood at a wall shear rate of 1500 seconds−1. After 3 minutes, interacting platelets were counted. Results are expressed as percentages of interacting platelets relative to the nontreated wt-A1 (*P < .01). (B) Images (205 × 330 μm) show the flow-dependent tethering of fluorescently labeled platelets on glass coverslips coated with wt-A1 (300 μg/mL) or native VWF (50 μg/mL) at 200 seconds−1(i,vi), 1000 seconds−1 (ii,vii), and 1500 seconds−1 (iii,viii) and the inhibitory effect of 30 μg/mL G19H10 (iv,ix) and 20 μg/mL AJvW-2 (v,x) at 1500 seconds−1. Corresponding platelet counts ± SDs are provided in subpanels.
Functional evaluation of the wt-A1 in flow-dependent platelet tethering.
(A) Glass coverslips coated with purified wt-A1 (300 μg/mL) were untreated (control) or treated for 30 minutes at 37°C with 1mM CuSO4 or 10 mM DTE as indicated and were perfused with platelets in reconstituted blood at a wall shear rate of 1500 seconds−1. After 3 minutes, interacting platelets were counted. Results are expressed as percentages of interacting platelets relative to the nontreated wt-A1 (*P < .01). (B) Images (205 × 330 μm) show the flow-dependent tethering of fluorescently labeled platelets on glass coverslips coated with wt-A1 (300 μg/mL) or native VWF (50 μg/mL) at 200 seconds−1(i,vi), 1000 seconds−1 (ii,vii), and 1500 seconds−1 (iii,viii) and the inhibitory effect of 30 μg/mL G19H10 (iv,ix) and 20 μg/mL AJvW-2 (v,x) at 1500 seconds−1. Corresponding platelet counts ± SDs are provided in subpanels.
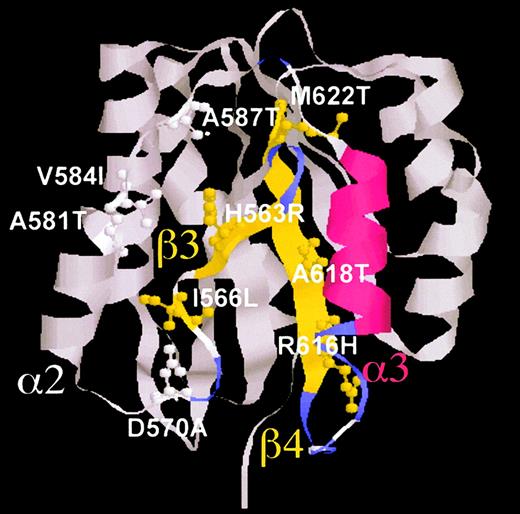
We therefore produced 9 A1 mutants, in which the 9 residues identified in Table 1 were mutated to their murine counterparts—His563Arg-, Ile566Leu-, Asp570Ala-, Ala581Thr-, Val584Ile-, Ala587Thr-, Arg616His-, Ala618Thr-, and Met622Thr-A1—plus an additional double mutant, His563Arg, Ile566Leu-A1. Figure 3 shows the topographic position of the 9 mutated residues in the 3-dimensional structure representation of the A1 domain. These residues cover a vast area within the VWF A1 domain. According to the numbering by Celikel et al,12 His563, Ile566, and Asp570 are located in strand β3 and succeeding loop β3-α2; Ala581, Val584, and Ala587 are in helix α2 and succeeding loop α2-α3; and Arg616, Ala618, and Met622 are in strand β4.
Ribbon representation of the human VWF A1 domain and location of the mutated residues.
The tridimensional structure front view of the A1 domain, based on the 3-dimensional coordinates by Celikel et al,12 is shown with the spatial distribution of all 9 mutated residues, displayed in ball-and-stick representation. Critical secondary structural elements were colored for clarity. For space considerations, single-letter codes were used for amino acids. The helix α3 is shown in red; strands β3 and β4 are shown in yellow; β-turn sequences are displayed in violet. Pictures were created with RasMol v2.6 (Glaxo Research and Development, London, United Kingdom).
Ribbon representation of the human VWF A1 domain and location of the mutated residues.
The tridimensional structure front view of the A1 domain, based on the 3-dimensional coordinates by Celikel et al,12 is shown with the spatial distribution of all 9 mutated residues, displayed in ball-and-stick representation. Critical secondary structural elements were colored for clarity. For space considerations, single-letter codes were used for amino acids. The helix α3 is shown in red; strands β3 and β4 are shown in yellow; β-turn sequences are displayed in violet. Pictures were created with RasMol v2.6 (Glaxo Research and Development, London, United Kingdom).
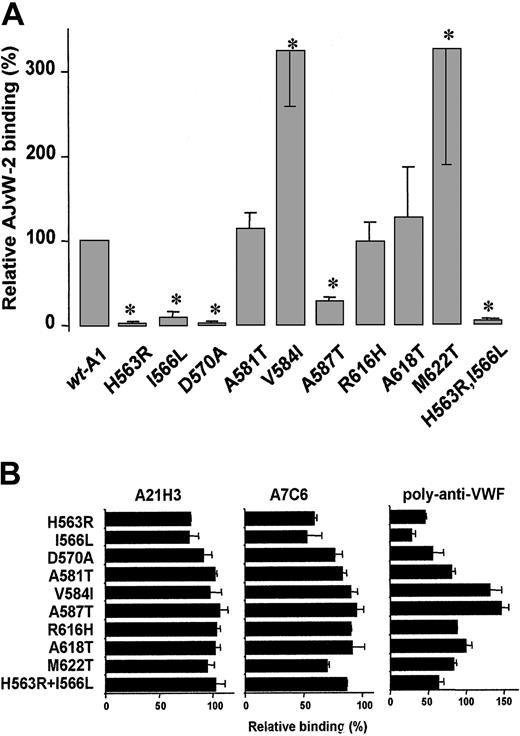
Epitope mapping of AJvW-2
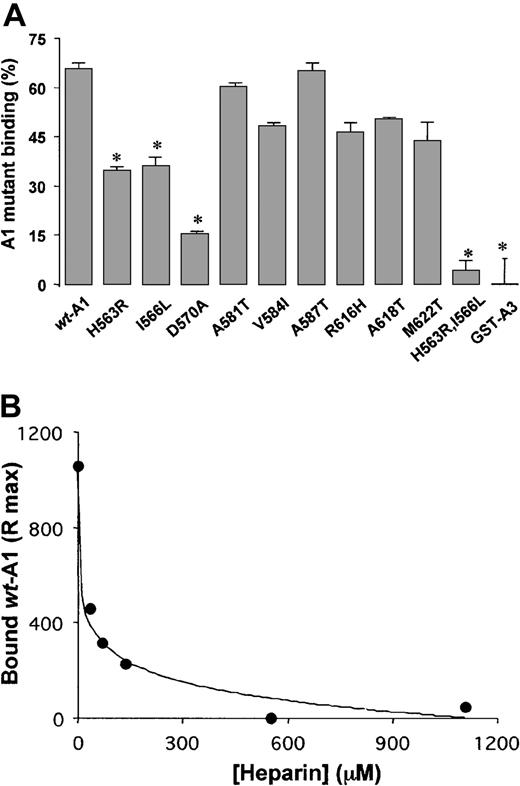
The ability of AJvW-2 to bind to wt-A1 and the mutants was tested in ELISA (Figure 4A). Compared with wt-A1, AJvW-2 bound normally to Ala581Thr-A1, Arg616His-A1, and Ala618Thr-A1, but it presented almost no binding to the mutants His563Arg-A1, Ile566Leu-A1, Asp570Ala-A1, and His563Arg, Ile566Leu-A1 (less than 10% of the binding to wt-A1) and an impaired binding to Ala587Thr-A1 (29% ± 3%). Binding to Val584Ile-A1 and Met622Thr-A1 was elevated (more than 300%). All loss-of-binding mutations are located on the surface of the domain, colocalized on the front of the molecule (Figure 3). Their close spatial arrangement indicates that these combined residues may constitute the epitope for AJvW-2 (see “Discussion”). To ascertain that the mutations His563Arg, Ile566Leu, Asp570Ala, and Ala587Thr specifically affected binding of AJvW-2 without altering the tridimensional folding of the A1 domain, we measured the binding of the conformation-dependent antibodies poly–anti-VWF, A21H3, and A7C6. Binding to the A1 domain is abolished by heat denaturation of wt-A1 (results not shown). As shown in Figure4B, each mutant reacted similarly to the wt-A1 (wt-A1 value ± 30%) with at least one antibody, indicative of no gross misfolding in the mutated A1 domains. This finding suggests that the increased binding of AJvW-2 to Val584Ile-A1 and Met622Thr-A1 was attributed to local alterations in the A1 conformation, increasing the accessibility for AJvW-2. The different results found for A21H3 and A7C6 on the one hand and the poly–anti-VWF on the other reflect the presence in the poly–anti-VWF antibodies of immunoglobulins that recognize epitopes involving His563, Ile566, and Asp570.
Binding of AJvW-2 to wt-A1 and mutants.
Plateau values for the binding of saturating concentrations of AJvW-2 (A) and the conformation-dependent anti-VWF A1 antibodies, poly–anti-VWF, A21H3, and A7C6 (B), to microtiter plate-coated A1 mutants (2 μg/mL), were determined by ELISA. Results are expressed as the mean percentages ± SDs of triplicate determinations (*P < .01), relative to binding to wt-A1. For space considerations, single-letter codes were used for amino acids.
Binding of AJvW-2 to wt-A1 and mutants.
Plateau values for the binding of saturating concentrations of AJvW-2 (A) and the conformation-dependent anti-VWF A1 antibodies, poly–anti-VWF, A21H3, and A7C6 (B), to microtiter plate-coated A1 mutants (2 μg/mL), were determined by ELISA. Results are expressed as the mean percentages ± SDs of triplicate determinations (*P < .01), relative to binding to wt-A1. For space considerations, single-letter codes were used for amino acids.
Wild-type A1 and mutants binding to heparin
The binding of wt-A1 and mutants to heparin was assessed by incubating heparin-Sepharose beads with 50 μg/mL of each mutant and by measuring the unbound protein fraction in the supernatant. After 2 hours at RT, 65.5% ± 1.2% of the wt-A1 was bound to heparin (Figure 5A). Nonspecific binding was negligible, as evidenced by the absence of binding of a fusion protein between GST and the A3 domain of VWF. Among the 9 single mutants tested, His563Arg-A1, Ile566Leu-A1, and Asp570Ala-A1 exhibited reduced binding to heparin, corresponding to 52.7% ± 2.0%, 54.9% ± 4.6%, and 23.5% ± 0.6%, respectively, of the wt-A1 binding. Interestingly, the double mutation His563Arg, Ile566Leu almost abolished binding to heparin (6.5% ± 6.3% of the wt-A1 value).
Heparin binding.
(A) Binding of wt-A1 and mutants to heparin. Fusion proteins (50 μg/mL) were incubated with heparin-Sepharose beads for 2 hours at 37°C. The bound fraction of the proteins was calculated from the concentration of unbound material measured after centrifugation of the beads. Results are expressed as mean percentages ± SDs of protein bound to the beads for triplicate determinations (*P < .01). A GST/human VWF A3 domain fusion protein was tested as a control for nonspecific binding to heparin-Sepharose beads. For space considerations, single-letter codes were used for amino acids. (B) Inhibition of wt-A1 binding to immobilized AJvW-2 by soluble unfractionated heparin during perfusions in a BIACore 1000 instrument.
Heparin binding.
(A) Binding of wt-A1 and mutants to heparin. Fusion proteins (50 μg/mL) were incubated with heparin-Sepharose beads for 2 hours at 37°C. The bound fraction of the proteins was calculated from the concentration of unbound material measured after centrifugation of the beads. Results are expressed as mean percentages ± SDs of protein bound to the beads for triplicate determinations (*P < .01). A GST/human VWF A3 domain fusion protein was tested as a control for nonspecific binding to heparin-Sepharose beads. For space considerations, single-letter codes were used for amino acids. (B) Inhibition of wt-A1 binding to immobilized AJvW-2 by soluble unfractionated heparin during perfusions in a BIACore 1000 instrument.
Mutations His563Arg, Ile566Leu, and Asp570Ala impair AJvW-2 and heparin binding to the A1 domain, suggesting that AJvW-2 and heparin share a common binding region on VWF- 1. This assumption was confirmed by competition assays between AJvW-2 and heparin for binding to wt-A1 (Figure 5B). Soluble unfractionated heparin completely inhibited the binding of wt-A1 to immobilized AJvW-2, with an IC50 of 16.7 ± 4.6 μM.
Functional evaluation of wild-type A1 and mutants under static conditions
Botrocetin assay.
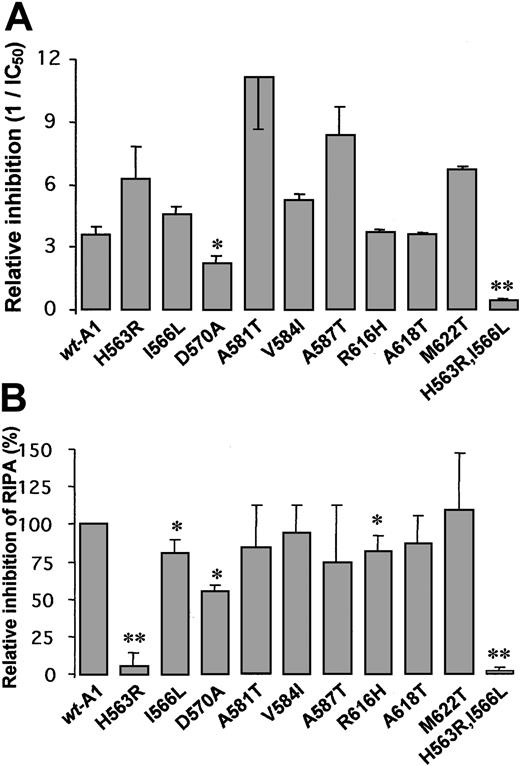
A1 mutants were tested in a static assay for their ability to compete with the botrocetin-mediated binding of multimeric VWF to glycocalicin. In this assay, 5 μg/mL wt-A1 completely inhibited the binding of 0.5 μg/mL VWF to glycocalicin, with an IC50 of 0.28 ± 0.03 μg/mL (Figure 6A). Of the 9 single mutants tested, only Asp570Ala-A1 exhibited a significantly reduced inhibitory capacity (IC50 = 0.46 ± 0.09 μg/mL). Because full-length recombinant VWF comprising the mutation Asp570Ala binds normally to sodium iodide 125I botrocetin,18 our finding underscores that the Asp570Ala mutation results in the direct impairment of VWF binding to glycocalicin and is not the result of a disturbed recognition of botrocetin by the mutant. All other mutations had no impact or improved binding to glycocalicin. It is likely that the binding of the exogenous modulator botrocetin to the A1 domain overrules the structural impact of some single mutations, as also suggested by others.18 31 This is illustrated in this study for the mutations His563Arg and Ile566Leu. Separately, these mutants normally inhibit the botrocetin-induced VWF binding, but the double mutant His563Arg, Ile566Leu-A1 is almost inactive, with a 10-fold elevated IC50 (2.4 ± 0.46 μg/mL), suggesting that the double mutation affects the conformation of the GPIbα binding site in A1 to the extent that this local structural deficiency can no longer be overcome by botrocetin binding to His563Arg, Ile566Leu-A1.
Inhibition of mediator-induced GPIb-VWF interaction by wt-A1 and mutants.
(A) Botrocetin assay. Reciprocal IC50 values for the competitive binding of the indicated A1 mutants, in comparison with wt-A1, during the binding of 0.5 μg/mL VWF to microtiter plate-coated glycocalicin, mediated by 2 μg/mL botrocetin. Results are expressed as relative inhibition (1/IC50). (B) Inhibition of RIPA by wt-A1 and mutants conducted in PRP. Means ± SDs of triplicate determinations (*P < .05; **P < .01). For space considerations, single-letter codes were used for amino acids.
Inhibition of mediator-induced GPIb-VWF interaction by wt-A1 and mutants.
(A) Botrocetin assay. Reciprocal IC50 values for the competitive binding of the indicated A1 mutants, in comparison with wt-A1, during the binding of 0.5 μg/mL VWF to microtiter plate-coated glycocalicin, mediated by 2 μg/mL botrocetin. Results are expressed as relative inhibition (1/IC50). (B) Inhibition of RIPA by wt-A1 and mutants conducted in PRP. Means ± SDs of triplicate determinations (*P < .05; **P < .01). For space considerations, single-letter codes were used for amino acids.
Ristocetin assay.
We further investigated the impact of the selected mutations on the GPIb/VWF-A1 interaction through their capacity to inhibit the RIPA (Figure 6B). The mutants His563Arg-A1, Ile566Leu-A1, Asp570Ala-A1, Arg616His-A1, and His563Arg, Ile566Leu-A1 had weakly to strongly impaired capacity to inhibit RIPA, exhibiting residual inhibition levels relative to wt-A1 of 5.7% ± 8.1%, 80.7% ± 8.9%, 55.0% ± 4.6%, 81.6% ± 10.3%, and 1.6% ± 2.8%, respectively. The mutations Ala581Thr, Val584Ile, Ala587Thr, Ala618Thr, and Met622Thr had no effect.
Functional evaluation of wild-type A1 and mutants under dynamic conditions
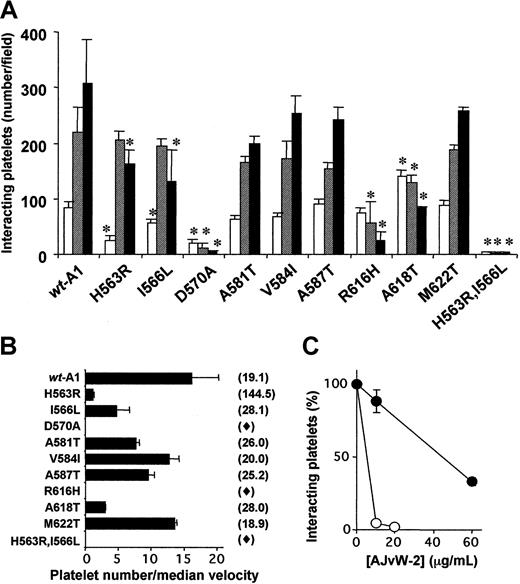
In a first set of experiments, we measured the number of interacting platelets while reconstituted blood was flowing over the A1 mutants (Figure 7A). The degree of adhesion and the shear rate dependence of this adhesion obtained with Ala581Thr-, Val584Ile-, Ala587Thr-, and Met622Thr-A1–coated coverslips were comparable to those for wt-A1. Mutants Asp570Ala-A1 and Arg616His-A1 displayed a strongly reduced platelet tethering at 1500 seconds−1 with, respectively, 1.2% ± 1.2% and 7.8% ± 5.6% of the wild-type value, whereas mutations His563Arg and Ile566Leu had a moderate effect on the number of interacting platelets with, respectively, 53.3% ± 8.4% and 42.8% ± 18.0% of the wild-type value at 1500 seconds−1. However, the combined mutation His563Arg, Ile566Leu abolished platelet tethering at all shear rates tested. At a shear rate of 200 seconds−1, only 2 single mutations, His563Arg and Asp570Ala, strongly decreased the number of platelets interacting with the A1 mutants with, respectively, 29.0% ± 11.0% and 24.9% ± 7.7% of the wild-type value. Interestingly, the Ala618Thr-A1 mutant interacted with a significantly higher number of platelets, at 200 seconds−1(166% ± 13.4% of the wild-type value) but presented an impaired capacity to induce platelet tethering at 1000 and 1500 seconds−1 (58.4% ± 6.2% and 27.7% ± 1.0% of the wild-type value, respectively). To further characterize the effect of the selected mutations on the functionality of the A1 domain toward platelet GPIb in flow, we recorded real-time movies of platelets rolling over the A1 mutants at 1500 seconds−1 and measured the corresponding median velocities of the translocating platelets (Figure 7B). Ala581Thr-A1, Val584Ile-A1, Ala587Thr-A1, and Met622Thr-A1 mutants triggered median velocity values comparable with those of the wild type (19.1 μm/sec), in agreement with the similar numbers of platelets interacting with these mutants (Figure7A). Asp570Ala-A1, Arg616His-A1, and His563Arg, Ile566Leu-A1 failed to generate sufficiently long platelet contact times to allow platelet velocity measurement (the average platelet contact time was less than 0.1 second), whereas mutations Ile566Leu and Ala618Thr induced a moderate increase in platelet median velocity. Interestingly, the mutation His563Arg, which only moderately reduced the number of interacting platelets (Figure 7A), induced a 7.6-fold increase in platelet rolling speed (144.5 μm/sec). The ratios of the platelet count and the median velocity of platelets interacting with the wt-A1 and mutants (at a shear rate of 1500 seconds−1) is reported in Figure 7B. These affinity-weighted ratios revealed that among the 9 mutations tested, His563Arg, Ile566Leu in strand β3, Asp570Ala in loop β3-α2, and Arg616His and Ala618Thr in strand β4 induced a loss of platelet GPIb binding.
Shear-dependent interaction of platelets with immobilized wt-A1 and mutants.
Reconstituted blood containing fluorescently labeled platelets (10 000/μL) was perfused over wt-A1 or mutant fusion proteins, immobilized onto glass coverslips coated at 300 μg/mL in a flow chamber (see “Materials and methods”). (A) Number of platelets translocating over the A1 domain mutants per field (205 × 330 μm), after 3 minutes of perfusion, at 200 seconds−1 (■), 1000 seconds−1 (▨), and 1500 seconds−1 (▪). Data are expressed as the means ± SDs of at least 3 separate assays (*P < .05). (B) Median velocities of platelet translocation on immobilized wt-A1 and mutants were determined from real-time movies by measuring the distance traveled by single platelets during 1 second of perfusion at 1500 seconds−1. For each mutant, the velocity of 50 individual platelets was measured in one typical experiment. Ratios of the platelet count to the median velocity of platelets interacting with the mutants are represented as black bars. Median platelet velocity (μm/sec) is indicated in parentheses. ♦ indicates a contact time that was too short to allow velocity measurement. For space considerations, single-letter codes were used for amino acids. (C) Platelets in reconstituted blood were perfused at 1500 seconds−1 over wt-A1 (○) and over Ala587Thr-A1 (●) in the presence of increasing concentrations of AJvW-2. After 3 minutes of perfusion, interacting platelets were counted. Results are expressed as the percentages of platelet numbers in the absence of AJvW-2.
Shear-dependent interaction of platelets with immobilized wt-A1 and mutants.
Reconstituted blood containing fluorescently labeled platelets (10 000/μL) was perfused over wt-A1 or mutant fusion proteins, immobilized onto glass coverslips coated at 300 μg/mL in a flow chamber (see “Materials and methods”). (A) Number of platelets translocating over the A1 domain mutants per field (205 × 330 μm), after 3 minutes of perfusion, at 200 seconds−1 (■), 1000 seconds−1 (▨), and 1500 seconds−1 (▪). Data are expressed as the means ± SDs of at least 3 separate assays (*P < .05). (B) Median velocities of platelet translocation on immobilized wt-A1 and mutants were determined from real-time movies by measuring the distance traveled by single platelets during 1 second of perfusion at 1500 seconds−1. For each mutant, the velocity of 50 individual platelets was measured in one typical experiment. Ratios of the platelet count to the median velocity of platelets interacting with the mutants are represented as black bars. Median platelet velocity (μm/sec) is indicated in parentheses. ♦ indicates a contact time that was too short to allow velocity measurement. For space considerations, single-letter codes were used for amino acids. (C) Platelets in reconstituted blood were perfused at 1500 seconds−1 over wt-A1 (○) and over Ala587Thr-A1 (●) in the presence of increasing concentrations of AJvW-2. After 3 minutes of perfusion, interacting platelets were counted. Results are expressed as the percentages of platelet numbers in the absence of AJvW-2.
The Ala587Thr-A1 mutant supports a normal GPIb recognition (Figure 7A-B), despite reduced AJvW-2 binding (Figure 4). Figure 7C shows that AJvW-2 still is capable of inhibiting platelet binding to Ala587Thr-A1, albeit at higher concentrations than those required to inhibit platelet binding to wt-A1. These experiments confirm that the residual binding of AJvW-2 to the front loop of Ala587Thr-A1 prevents GPIbα-VWF A1 domain interactions. Neither A21H3 nor A7C6 mAb inhibited platelet rolling over wt-A1, in agreement with recognition of different epitopes on wt-A1 for these antibodies and AJvW-2 (results not shown).
Discussion
Numerous animal studies have documented the importance of VWF neutralization in arterial and venous thrombosis.21-25 The humanized form of the blocking mAb AJvW-2 thus constitutes a potential therapeutic agent for the prevention of acute thrombosis in clinical practice. In this study, we tested the hypothesis that the antibody reacts with a highly conserved epitope on human VWF A1, critical for binding to GPIbα.
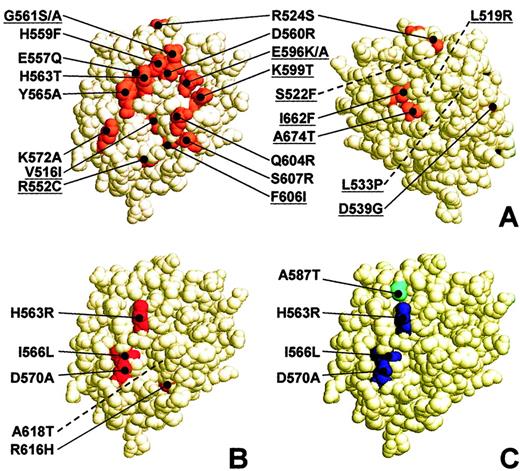
Based on the GPIbα-peptide docking model,16 residues Gly561, Tyr565, Glu596, and Lys599 were claimed to play a direct role in GPIb binding. These residues are represented in Figure8A. In this tridimensional space-filling model of the VWF A1 domain, all reported mutations that result in impaired flow-dependent GPIbα binding are grouped. These mutations include those generating type 2M von Willebrand disease (VWF database) and those shown in different studies to abolish platelet interactions with the VWF A1 domain in flow.16,19,20 The present localization of these mutations on the VWF A1 domain supports the model proposed by Vasudevan et al.16 Among the 21 mutated residues identified, 14 are located at the front of the A1 domain. Eleven of these frontal mutations were located either in strand β3 and flanking loops (His559Phe, Asp560Arg, Gly561Ser/Ala, His563Thr, Tyr565Ala, and Lys572Ala) or in helix α3 and flanking loops (Glu596Ala, Lys599Thr, Gln604Arg, Phe606Ile, and Ser607Arg). The back of the A1 domain model identifies a few substitutions capable of impairing binding to GPIbα, although they are located in distant areas.
Location of amino acid mutations impairing GPIbα, heparin, and AJvW-2 binding to VWF A1.
Space-filling, 3-dimensional representation of the human VWF A1 domain. (A) Front (left) and back (right) view of the A1 domain. Surface residues of mutations previously shown to abolish shear-dependent platelet interactions with the recombinant VWF A1 domain16,19 20 or to cause VWD type 2M (ISTH SSC VWF database: www.shef.ac.uk/vwf/) are indicated in orange. Location of buried residues is indicated by dotted lines. Amino acid substitutions detected in type 2M VWD patients are underlined. For space considerations, single-letter codes were used for amino acids. (B) Surface residues of mutations in the present study that impaired the shear-dependent interaction of the VWF A1 domain with platelet GPIbα are indicated in red. The location of the buried residue Ala618 is indicated by a dotted line. (C) Residues of mutations in the present study that impaired heparin and AJvW-2 recognition are indicated in blue. Amino acid Ala587, whose mutation only impairs AJvW-2 binding, is indicated in cyan. Pictures were created with RasMol v2.6.
Location of amino acid mutations impairing GPIbα, heparin, and AJvW-2 binding to VWF A1.
Space-filling, 3-dimensional representation of the human VWF A1 domain. (A) Front (left) and back (right) view of the A1 domain. Surface residues of mutations previously shown to abolish shear-dependent platelet interactions with the recombinant VWF A1 domain16,19 20 or to cause VWD type 2M (ISTH SSC VWF database: www.shef.ac.uk/vwf/) are indicated in orange. Location of buried residues is indicated by dotted lines. Amino acid substitutions detected in type 2M VWD patients are underlined. For space considerations, single-letter codes were used for amino acids. (B) Surface residues of mutations in the present study that impaired the shear-dependent interaction of the VWF A1 domain with platelet GPIbα are indicated in red. The location of the buried residue Ala618 is indicated by a dotted line. (C) Residues of mutations in the present study that impaired heparin and AJvW-2 recognition are indicated in blue. Amino acid Ala587, whose mutation only impairs AJvW-2 binding, is indicated in cyan. Pictures were created with RasMol v2.6.
In this study, we constructed fusion proteins between GST and mutated A1 domains. We demonstrated that the GST/wild-type A1 construct specifically inhibited RIPA and supported platelet tethering and rolling in a shear rate–dependent manner as effectively as native VWF multimers. We identified 5 residues, the mutations of which impaired the dynamic platelet-binding to A1 fusion proteins: His563, Ile566, Asp570, Arg616, and Ala618 (Figure 7B). These findings closely fit with the above model because His563, Ile566, and Asp570 are located within strand β3 and the succeeding loop β3-α2, at the surface of the VWF A1 domain (Figure 8B). The effects of mutation Ile566Leu had not been reported before. Our results with His563Arg-A1 are in accordance with those by Cruz et al.19 For residue Asp570, Celikel et al20 have proposed that its negatively charged side chain could constitute a repulsive site for the negatively charged GPIbα residues. Because we showed that the substitution of Asp570 by a noncharged residue (alanine) almost abolishes platelet tethering in flow without altering the overall structure of the A1 domain, we believe that Asp570 is involved in the local intramolecular stabilization of the GPIbα-binding domain.
Interestingly, this study highlights the importance of the central-strand β4 residues Arg616 and Ala618 (Figure 3). An Arg616Cys mutation has recently been reported in a patient with unclassified von Willebrand disease (VWD) (ISTH SSC VWF database); moreover, normal botrocetin but impaired ristocetin-induced VWF binding to GPIb has been found for an Arg616Ala mutation.18 We found here that the mutation Arg616His has a mild effect in RIPA and in platelet tethering in the perfusion chamber at low shear rate (200 seconds−1) but strongly reduces platelet tethering at moderate (1000 seconds−1) and high (1500 seconds−1) shear rates. The mutation of Arg616 might, therefore, generate a type 2M phenotype. The region of the A1 domain surrounding Arg616 appears to play an important role in GPIb binding because the closely located mutations Phe606Ile and Ser607Arg cause VWD type 2M and impaired GPIb binding, respectively.19,33 The Arg616 residue is positively charged and partially exposed on the surface of the A1 domain, where it could take part in GPIbα binding. The mutation Ala618Thr also abolished platelet GPIb binding in flow. At variance with Arg616, Ala618 is buried in the hydrophobic core of the A1 domain, suggesting that the Ala618Thr mutation would rather destabilize the correct folding of the GPIbα binding site on the A1 domain, at least at high shear forces. Indeed, at low shear rate, the Ala618Thr substitution even enhanced VWF A1-GPIb interactions. These findings suggest that high shear rates are capable of inducing conformational changes within the A1 domain, a process interfered with by mutations such as Ala618Thr. Ala618Thr has been described as a natural polymorphism34 and has been linked to the occurrence of VWD. The present findings suggest that at high shear rates the Ala618Thr polymorphism is associated with a mild loss of VWF function.
The RIPA inhibition assay yielded results comparable to those in the shear-dependent platelet tethering assay, when analyzed at low (200 seconds−1) rather than at high (1500 seconds−1) shear rate. Indeed the former assay showed that His563Arg and Asp570Ala mutations impair GPIb binding, but it failed to detect the loss-of-function mutation Ala618Thr and hardly detected the loss-of-function mutations Ile566Leu and His616Arg. Only at a shear rate of 1500 seconds−1 were all 5 loss-of-function mutations recognized by the combined analysis of interacting platelet numbers and their translocation velocity during perfusion studies.
The structural integrity of strand β3, the preceding loop β2-β3, and the succeeding loop β3-α2 (residues 559-573) appear to be essential because the mutation of residues within this structure is invariably associated with reduction or loss of GPIbα binding, as shown by others16,19,20 and by this study. The independent confirmation, however, that shielding this structural region of the A1 domain would inhibit the binding to GPIbα was still missing. In the present study, we demonstrated that the blocking mAb AJvW-2 binds to an area of the VWF A1 domain comprising the strand β3. We identified the residues His563, Ile566, Asp570, and, to a lesser degree, Ala587 as essential for AJvW-2 binding (Figure 8C). The 4 residues are distant from each other in the amino acid sequence, but spatially they cluster on the outer-front surface of the tri-dimensionally folded A1 domain, consistent with our finding that the epitope of the antibody is conformational.22 The neutral effect of mutation Ala581Thr on AJvW-2 binding to the A1 domain was expected because mouse and canine VWF contain a threonine at position 581, and the antibody was recently reported to have an inhibitory effect in a canine model of thrombosis.25 Further restricting AJvW-2 binding to strand β3 in the Ala587Thr-A1 mutant still leads to the blockade of VWF A1-GPIb interactions, compatible with frontal shielding of the A1 domain in strand β3. The same mutations that affect AJvW-2 binding also affect GPIbα binding, indicative of overlapping binding sites on or near this strand.
In contrast to the blocking mAb NMC-4, whose epitope overlaps with the botrocetin binding site comprising residues Arg632 and Arg636 in helix α4,15 21 AJvW-2 interacts with VWF A1 residues directly involved in GPIbα binding.
It has been reported that the main heparin-binding site on the A1 domain is located between residues Tyr565 and Ala587 on strand β3 and helix α2.35 36 In this study, we showed that the frontal residues His563, Ile566, and Asp570 are part of the area involved in heparin binding and that heparin competes with AJvW-2 for binding to wt-A1, indicating that heparin and AJvW-2 binding sites on VWF A1 overlap (Figure 8C).
In conclusion, our finding that the binding site in VWF for AJvW-2 comprises residues in the strand β3 of the A1 domain, critical for binding to GPIbα, further adds to our understanding of the dynamic interaction between platelets and von Willebrand factor and provides a molecular basis for its inhibition.
Prepublished online as Blood First Edition Paper, October 10, 2002; DOI 10.1182/blood-2002-06-1818.
Supported by the FWO Vlaanderen (project no. G.0376.01). A.B. is the recipient of a Marie Curie Fellowship of the European Commission.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Note
When this study was completed, the crystal structure of a complex between the gain-of-function mutants GPIbα-Met239Val and A1-Arg543Gln, associated with platelet-type and type 2B von Willebrand disease, respectively,37 became available. This crystal, resulting from enhanced affinity for complex formation, highlights two crucial areas in the A1 domain involved in the interaction with GPIbα—that is, the strand β3, the helix α3, and residues in the loop β3-α2. The conclusions of that structural study and of our own functional analysis involving selective mutants of A1 are in complete agreement and independently emphasized the importance of the strand β3, the helix α3, and the loop β3-α2 in the von Willebrand factor A1 domain during its interaction with GPIbα.
Author notes
Marc Hoylaerts, Center for Molecular and Vascular Biology, University of Leuven, Campus Gasthuisberg, Herestraat 49, B-3000 Leuven, Belgium; e-mail:marc.hoylaerts@med.kuleuven.ac.be.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal