Abstract
Integrin CD11b is a differentiation marker of the myelomonocytic lineage and an important mediator of inflammation. Expression of theCD11b gene is transcriptionally induced as myeloid precursors differentiate into mature cells, then drops as monocytes further differentiate into macrophages. Previous studies have identified elements and factors involved in the transcriptional activation of the CD11b gene during myeloid differentiation, but no data exist regarding potential down-regulatory factors, especially in the later stages of differentiation. Using 2 copies of a GC-rich element (−141 to −110) in the CD11bpromoter, we probed a cDNA expression library for interacting proteins. Three clones were identified among 9.1 million screened, all encoding the DNA-binding domain of the zinc finger factor ZBP-89. Overexpression of ZBP-89 in the monocyte precursor cell line U937 reducedCD11b promoter-driven luciferase activity when U937 cells were induced to differentiate into monocytelike cells using phorbol esters. To identify the differentiation stage at which ZBP-89 repression of the CD11b gene is exerted, the protein level of ZBP-89 was correlated with that of CD11b mRNA in differentiating U937 as well as in normal human monocytes undergoing in vitro differentiation into macrophages. A clear inverse relationship was observed in the latter but not the former state, suggesting that ZBP-89 represses CD11b gene expression during the further differentiation of monocytes into macrophages.
Introduction
CD11b is the α subunit of the integrin αβ heterodimer CD11b/CD18, 1 of 4 members of the β2integrins.1 It is expressed on myelomonocytic cells and mediates essential adhesion-dependent functions. These include adhesion, migration, phagocytosis, degranulation, antibody-dependent cytotoxicity, and release of proteolytic enzymes during the inflammatory response.1 Deficiency of CD11b and the other β2 integrins in the inherited disease leukocyte adhesion molecule deficiency2 predisposes to life-threatening bacterial infections. Increased expression of CD11b, on the other hand, leads to poorly coordinated adhesive functions, thus contributing to the genesis of several pathologic states such as ischemia-reperfusion injury and immune complex diseases.3-6
CD11b is a myeloid-specific marker. During hematopoiesis, it is first detected at the myelocytic and monoblastic stages,7,8 and its expression is significantly increased as cells differentiate into monocytes and granulocytes.9 Subsequently, CD11bgene expression is down-regulated in cells proceeding toward later stages of monocytic differentiation.9 The drop in expression of CD11b can be reproduced in vitro in human peripheral blood monocytes as they differentiate into macrophages.10-13 Although some of the elements and factors underlying the induction of the CD11b gene are known, those contributing to its down-regulation thereafter remain to be defined.
Transcriptional regulation of CD11b is an important determinant in expression of the receptor.14 The proximal promoter of CD11b spanning nucleotides −242 to +71 appears responsible for the tissue-specific and lineage-specific expression of the gene in vitro. The CD11b promoter contains near its transcription start site elements for the transcription factors Sp1, Sp3, and PU.1, which have been shown to prime the CD11b gene for expression.15-17 Inducible expression ofCD11b during monocytic lineage differentiation appears to be mediated by an as yet uncloned nuclear factor, named MS-2.18 No information is available on the nature of factors that mediate transcriptional repression of the CD11bgene. We have used yeast one-hybrid screens to identify a Krüppellike zinc finger transcription factor, ZBP-89, which binds to the CD11b promoter and acts as a transcriptional repressor of this gene as normal monocytes differentiate into macrophages in vitro. The significance of these findings is discussed.
Materials and methods
Cells
Cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA). The promonocytic U937 cell line (ATCC CRL-1539.2) was cultured in RPMI 1640 (Sigma, St Louis, MO) supplemented with 10% heat-inactivated fetal calf serum (FCS; Atlanta Biologicals, Norcross, GA) in an atmosphere of 5% CO2. Treatment of U937 with phorbol 12-myristate 13-acetate (PMA; Sigma) was used as an in vitro model system to assess early stage monopoiesis19; these cells did not express the macrophage marker CD1620 for up to 96 hours following PMA treatment (not shown). A pool of U937 cells stably expressing theCD11b promoter (sU937-11b-luc) spanning nucleotides −242 to +71 fused to the luciferase gene was generated by transfection with a CD11b lucI-pCMV/Zeo (Invitrogen, Carlsbad, CA) construct using electroporation. Cells were selected and maintained in the presence of 200 μg/mL zeocin (Invitrogen). The Drosophila embryo cell line SL2 (ATCC CRL-1963) was grown at 25°C in SchneiderDrosophila medium (Gibco BRL, Grand Island, NY) supplemented with 10% heat-inactivated FCS specified for insect cells (Sigma). Late-stage monocytic differentiation into macrophages was carried out using a technique that relies on mitogenically activated T-cell contact with monocytes, thus mimicking the in vivo conversion of monocytes to macrophages by antigen-activated T cells. Briefly, normal human monocytes were isolated from leukopacks of healthy donors by Ficoll-Hypaque gradient centrifugation,10 21 followed by adherence to Petri dishes in Iscove medium plus10% human AB serum (Sigma) for 90 minutes at 37°C and in 5% CO2. Adherent cells were stimulated with concanavalin A (ConA; 10 μg/mL; Sigma) for 20 hours in 60/30 media (60% AIM V, 30% Iscove medium; Gibco BRL) plus 10% human AB serum. The following day, cells were extensively washed, and the adherent monocytes (day 1) were cultured without any cytokine or mitogen for 12 days. Monocytes were identified by flow cytometric analysis using a fluorescence-activated cell sorting (FACS) Vantage instrument and Cell Quest software (Becton Dickinson, San Jose, CA); more than 90% of these cells were CD14+ positive and less than 3% CD3+.
Cloning of ZBP-89 by yeast one-hybrid screening
The yeast strain YM4271 (MATa, ura3-52, his3-200, ade2-101, lys2-801, leu2-3, 112, trp1-901, tyr1-501, gal4-Δ512, gal80-Δ538, ade5::hisG) was used to make a reporter strain. The bait, consisting of 2 copies of the GC-rich region of the CD11b gene spanning nucleotides −141 to −105 (GC: 5′ GGGTCAGGAAGCTGGGGAGGAAGGGTGGGCAGGCTGT 3′; Figure 1A-B), was subcloned into the plasmid pHISi-1 (Clontech, Palo Alto, CA). The resulting pHISi-1-GC plasmid was digested with XhoI, then transformed into yeast to create the wild-type reporter strain. The empty vector pHISi-1 and the mutant pHisi-1-GCm (5′ GGGTCCTTAATCTGGGGCTTAATGGTGGGCCCTCTTT 3′) plasmids (Figure 1B) were also transformed into yeast after digestion with XhoI to create a background and mutant reporter strains, respectively. Plasmid integration into the yeast genome was analyzed by plating on synthetic dropout medium lacking histidine (SD-H) but supplemented with various concentrations of 3-aminotriazole (3-AT). The resulting reporter strains showed growth inhibition at 45 mM 3-AT. We screened a total of 9.1 million independent colonies from a human HeLa matchmaker cDNA library (Clontech). Plasmid DNAs were recovered from those colonies that were able to grow in the presence of 45 mM 3-AT and synthetic dropout medium without histidine and leucine (SD-HL). The plasmid DNAs were then transformed into the wild-type, mutant, and background yeast strains to confirm the specificity of the interaction between the plasmid-encoded protein and the bait DNA.
Cloning of ZBP-89.
(A) Nucleotide sequence of the proximal human CD11bpromoter. The major transcription initiation site (arrow) is numbered +1. The GC-rich region is in bold and underlined. (B) Nucleotide sequence of GC (wild-type) and GCm (mutant) oligonucleotides (sense strand). Mutations are indicated in bold underlined letters. (C) Schematic of the primary structure of human ZBP-89 and p25 (see text for details). The vertical numbers indicate the amino acid numbers. The acidic domain is shown as a shaded bar; 2 basic domains as solid bars; 4 zinc finger domains as hatched bars, and a PEST sequence as a dotted bar. A single nucleotide insertion in p25 results in an in-frame premature termination codon after amino acid 284 of ZBP-89.
Cloning of ZBP-89.
(A) Nucleotide sequence of the proximal human CD11bpromoter. The major transcription initiation site (arrow) is numbered +1. The GC-rich region is in bold and underlined. (B) Nucleotide sequence of GC (wild-type) and GCm (mutant) oligonucleotides (sense strand). Mutations are indicated in bold underlined letters. (C) Schematic of the primary structure of human ZBP-89 and p25 (see text for details). The vertical numbers indicate the amino acid numbers. The acidic domain is shown as a shaded bar; 2 basic domains as solid bars; 4 zinc finger domains as hatched bars, and a PEST sequence as a dotted bar. A single nucleotide insertion in p25 results in an in-frame premature termination codon after amino acid 284 of ZBP-89.
Plasmid construction
A reporter plasmid, pALbWt, was used, which contains theCD11b promoter spanning nucleotides −242 to +71 fused to the luciferase gene.18 To introduce the transversion mutation (G>T) at nucleotide −117 within theCD11b promoter, a 2-step polymerase chain reaction (PCR) ligation was carried out. The first PCR reaction was performed with primer 1 (5′ CTAGAGTCGACCTCGAGCCAAGC 3′) and primer 2 (5′ CCACAGCCTGCCCAACCTTCCTCCCC 3′), and the second PCR was carried out with primer 3 (5′ GGGGAGGAAGGTTGGGCAGGCTGTGG 3′) and primer 4 (5′ TAGAATGGCGCCGGCCT 3′). The third PCR fragment was generated with primers 1 and 4, using the first and the second PCR fragments as a template.22 The third PCR fragment was digested withPstI and NarI and subcloned into pALbWt. After confirming the mutation by DNA sequencing, the resulting plasmid was designated pALbM4a. Plasmids pCMVSport3-ZBP-89 (which constitutively expresses human ZBP-89 in eukaryotic cell lines) and pGEX5T-ZBP-89 (which expresses ZBP-89 fused to glutathione S-transferase [GST]) were kindly provided by Dr J. L. Merchant.23 The plasmid pCMVSport3-Z (which expresses p25 in eukaryotic cell lines) was generated by subcloning a 0.62-kb PCR product digested withSmaI and XbaI into the SmaI andXbaI sites of pCMVSport3 (Gibco BRL). The fusion polypeptide GST-Z was generated from pGEX4T-2-Z created by insertion of a PCR product encoding p25 digested with SmaI and XhoI into the SmaI and XhoI sites of pGEX4T-2 (Pharmacia Biotech, Piscataway, NJ). The plasmid CD11b lucI-pCMV/Zeo was generated by subcloning a 1247-bp fragment of pCMV/Zeo (Invitrogen) digested with PstI and XhoI into theSalI and PstI sites of pALbWt. The expression plasmid pPacSp1 and the parental plasmid pPacO were kindly provided by Dr Robert Tjian.24 Both pPacZBP-89 and pPacZ, which express human ZBP-89 and p25, respectively, were constructed by insertion of BamHI-XhoI fragments of PCR products into the BamHI and XhoI sites of pPacO. The PCR-generated fragments in each plasmid construct were confirmed by DNA sequencing.
Nuclear extract and total protein lysate preparation
Nuclear extracts were prepared using a method modified from that described by Dignam et al.25 Briefly, cultured cells (1.2 × 107) were harvested and washed twice with cold phosphate-buffered saline (PBS). Cell pellets were resuspended with 2 mL buffer A (20 mM Tris [tris(hydroxymethyl)aminomethane]–-HCl, pH 7.8, 50 mM KCl, 80 mM NaCl, 0.2 mM EDTA [ethylenediaminetetraacetic acid], 0.1 mM EGTA [ethyleneglycoltetraacetic acid], 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 4% glycerol, and the protease inhibitor mixture “Complete”; Roche Molecular Biochemicals, Indianapolis, IN) and incubated on ice for 10 minutes. After addition of 10 μL 10% Nonidet P-40, the cell suspension was vortexed. Disruption of the cell membranes was monitored under a light microscope by Trypan blue staining. When more than 90% of cells were Trypan blue positive, they were spun at 6000 rpm at 4°C for 5 minutes to pellet the nuclei. The pellet was resuspended in 500 μL buffer B (20 mM Tris-HCl, pH 7.8, containing 300 mM KCl, 0.5 mM dithiothreitol [DTT], 0.2 mM EDTA, 0.1 mM EGTA, 0.2 mM PMSF, 25% glycerol, and the protease inhibitor mixture “Complete”) and mixed gently at 4°C for 30 minutes. After spinning the suspension in a microcentrifuge at 16 000g for 15 minutes at 4°C, the supernatant was aliquoted into 20 μL fractions, snap-frozen in liquid nitrogen, and stored at −80°C. For the preparation of the total protein lysate from human monocytes/macrophages, cells were harvested and washed with cold PBS. Cell pellets were incubated with the lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% triton X-100, 0.5% NP-40, 1 mM DTT, and the protease inhibitor mixture “Complete”; Roche Molecular Biochemicals) for 10 minutes at room temperature. After spinning the suspension in a microcentrifuge at 13 000 rpm for 15 minutes at 4°C, the supernatant was aliquoted into 20-μL fractions, snap-frozen in liquid nitrogen, and stored at −80°C.
EMSA and immunoblot analysis
Electrophoretic mobility shift assays (EMSAs) were conducted as previously described,18 using the double-stranded oligonucleotides GC (sense strand: 5′-GGGTCAGGAAGCTGGGGAGGAAGGGTGGGCAGGCTGT-3′) or GC1 (sense strand: 5′-AGCTGGGGAGGAAGGGTGGGCAGGCTGT-3′). For electrophoretic mobility supershift assay (EMSSA), 1 μL of either anti–ZBP-89 antibody (provided by Dr J. L. Merchant26) or anti-Sp1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was preincubated with nuclear extract for 20 minutes at 4°C, followed by the addition of the DNA probe and further incubation for 20 minutes at 4°C. The oligonucleotides used in the analyses are shown in Figure 3A. The other double-stranded oligonucleotides used were: Sp1, 5′-ATTCGATCGGGGCGGGGCGAG-3′; OCT-1, 5′-TGTCGAATGCAAATCACTAGAA-3′; and htβ, 5′-GATCTGGGGGTGGGGTGGGGGTGGGGTGGGGGTGGGG-3′ (containing 4 copies of the CACCC recognition sequence of ZBP-89).27 For immunoblot analysis, nuclear protein (30 μg) was electrophoretically separated by 6% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto a nitrocellulose membrane (Bio-Rad, Hercules, CA). A chemiluminescence Western blot detection kit and protocol (Roche Molecular Biochemicals) were used for antigen detection. The anti–ZBP-89 antibody was diluted 1000-fold and used as the primary antibody.
Transfection
U937 cells were transfected by electroporation in incomplete medium at 280 V and 960 μF using Gene Pulser Cuvettes (Bio-Rad). Cells in the early log phase of growth were harvested by centrifugation, washed once with medium, and resuspended at room temperature. About 4 × 107 cells were transfected with 25 μg luciferase test plasmid together with 5 μg of the plasmid pRSV-β (Promega, Madison, WI), which contains the lacZgene. Each transfection of pALbWt and pALbM4a was performed in parallel with a transfection of the promoterless luciferase plasmid pATLuc. Induction of promoter activity was assessed by treating the electroporated cells for 16 hours with PMA at 100 ng/mL prior to harvesting. Cells were subsequently pelleted, washed twice with PBS, and lysed in 150 μL reporter lysis buffer (Promega). Luciferase and β-galactosidase activities were assessed using assay reagents purchased from Promega and 100 μL and 20 μL lysate, respectively. Luciferase activity was measured using a Monolight 2010 Luminometer, which integrated peak luminescence 10 seconds after injection of assay buffer, and the values were normalized against β-galactosidase activity. Fold above background for each transfection was calculated by dividing the luciferase activity of each test plasmid by that of pATLuc. The fold above background derived from transfection of pALbWt was assigned an arbitrary value of 100%.
Transactivation by ZBP-89 was assessed in cotransfections in which 8 μg pATLuc or pALbWt or pALbM4a was mixed with 1 μg pRSV-β and 16 μg of either pCMVSport3 or pCMVSport3-ZBP-89. After normalization of transfection efficiency against β-galactosidase activity, the level of luciferase activity directed by pALbWt in the presence of pCMVSport3-ZBP-89 was divided by that of luciferase activity directed by pATLuc (also in the presence of pCMVSport3-ZBP-89). This calculation yielded the fold above background activity of pALbWt in the presence of ZBP-89. Nonspecific effects caused by the backbone of the ZBP-89 were corrected by equivalent calculations of luciferase activity but in the presence of pCMVSport3. The resulting figures represented the fold above background activity of pALbWt in the absence of ZBP-89. This value is given as 100%; then the fold above background values obtained in the presence of ZBP-89 was calculated to determine the degree of transactivation. All transfections were repeated independently more than 3 times with at least 2 different DNA preparations for each plasmid.
sU937-11b-luc cells were transfected with 25 μg of either pCMVSport3 or pCMVSport3-ZBP-89 with 1 μg of the plasmid pRSV-β using electroporation, then treated with PMA at 100 ng/mL for 16 hours before harvesting. After normalization of luciferase activity by β-galactosidase activity, the level of luciferase in the presence of pCMVSport3 was assigned a value of 100, and the effect of ZBP-89 on luciferase activity was expressed as a percentage of this value.
For transfection of the Drosophila cell line SL2, cells were plated at a density of 3 × 106/60-mm dish for 18 hours and washed with PBS twice before transfection. A total of 2.6 μg DNA and 10 μL Superfect (Qiagen, Valencia, CA) were mixed in 150 μL incomplete Schneider Drosophila medium. After 10 minutes at room temperature, 0.6 mL medium supplemented with 10% heat-inactivated FCS (tested for use with insect cells; Sigma) was added to the DNA-Superfect complexes. The suspension was then added drop-wise to the culture dishes. After a 3-hour incubation at room temperature, cells were washed with PBS twice and 2 mL complete medium was added. Cells were harvested 24 hours later, washed 2 times with PBS, and lysed in 150 μL reporter lysis buffer (Promega). Generally, 10 μL cell lysate was used in the β-galactosidase assay (Promega) and 50 μL lysate was assayed for luciferase activity. pIE3LacZ, which expresses β-galactosidase, was used in all transfections to correct for transfection efficiency.
Real-time RT-PCR
Total RNA from U937 cells (1.0 × 107), which were treated with PMA (100 ng/mL) for 0 hour, 24 hours, 48 hours and 72 hours, was isolated using the Trizol reagent (Gibco BRL) followed by DNase I treatment (Ambion, Austin, TX) as described by the manufacturer. First-strand cDNA was prepared by extension of an oligo-d(T)16 primer with Moloney-murine leukemia virus reverse transcriptase (RT, Clontech). After incubation at 42°C for 1 hour, the reaction was stopped by heating to 95°C for 5 minutes. Equal amounts of cDNA were subjected to PCR to detectCD11b, ZBP-89, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts.28 PCR primers and TaqMan probes for CD11b and ZBP-89 were designed using Primer Express 1.0 Software program and purchased from Applied Biosystems (Foster City, CA). TaqMan probes forCD11b and ZBP-89 were labeled with a reporter fluorescent dye, FAM (6-carboxyfluorescein), at the 5′ end and a fluorescent dye quencher TAMRA (6-carboxy-tetramethyl-rhodamine) at the 3′ end. For GAPDH, the predeveloped TaqMan assay reagent (PDAR), which has forward and reverse primers, and a probe labeled with a fluorescent dye VIC at the 5′ end and a fluorescent dye quencher TAMRA at the 3′ end, was used (Applied Biosystems). PCR conditions were 2 minutes at 50°C (for AmpErase UNG incubation to remove any uracil incorporated into the cDNA), 10 minutes at 95°C (for AmpliTaq gold activation), followed by 40 cycles at 95°C for 15 seconds and at 60°C for 1 minute. Primers and probes for detecting ZBP-89 and CD11b are as follows: ZBP89-forward, 5′-CCGAGCCTTAACTTTGTGACTGAT-3′; ZBP89-reverse, 5′-ATGGTGGCATAGACCTGCTTGT-3′; ZBP89-probe, 5′-6FAM-CCCAAATCAGCCAGCATTCTCTTCC-TAMRA-3′; CD11b-forward, 5′-AGTTGCCGAATTGCATCGA-3′; CD11b-reverse, 5′-GGCGTTCCCACCAGAGAGA-3′; CD11b probe, 5′-6FAM-AGCCCCATTGTGCTGCGCCT-TAMRA-3′.
Plasmid DNAs pCDNACD11b29 and pCMVSport3-ZBP-89 were used to generate a standard curve for the determination of the copy number of CD11b and ZBP-89, respectively. To construct the GAPDH-plasmid, a 604-bp fragment was amplified using the forward primer 5′-TCTGCTCCTCCTGTTCGACA-3′ and reverse primer 5′-GACGTACTCAGCGCCAGCAT-3′, and a cDNA from U937 cell total RNA as a template. The PCR reaction was performed with the following conditions: 45 seconds at 94°C, 45 seconds at 60°C, and 1 minute at 72°C for 30 cycles. This fragment was cloned using TA cloning kits (Invitrogen) and the resulting plasmid sequenced to confirm its identity. All reactions were performed in the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Test samples, standards, and no template controls were run in triplicate and the data were analyzed using the Sequence Detector V 1.6 program. A standard curve was plotted in cycle threshold (Ct) versus the known copy numbers of the templates from each plasmid. The final copy number of CD11b or ZBP-89 was presented, respectively, as a CD11b/GAPDH or ZBP-89/GAPDH ratio.
Results
Identification of ZBP-89 using yeast one-hybrid screens
The proximal GC-rich element spanning nucleotides −141 to −105 (with respect to the transcriptional start site, Figure 1A, underlined), is required for expression of the CD11b gene in differentiating U937 cells.18 Two copies of this GC-rich element were used as bait in a yeast one-hybrid screen to identify interacting factors. A mutant form of the GC-rich element (Figure 1B) was used as a negative control to assess specificity. After 7.1 × 106 cells in a first and 2 × 106cells in a second screen of a HeLa cell cDNA library were analyzed, 174 positive colonies grew on restreaking onto 45-mM 3-AT SD-H plates. Among these, 43 did not show PCR products, and 19 were redundant (based on the digestion pattern of DNA with HaeIII and AluI). The rest of the candidates were retransformed into the yeast reporter strain and 22 grew on the selection medium. Of these, only 3 grew in the selection medium after transformation into the wild-type reporter strain but not into a background strain (which does not carry the bait), or the mutant reporter strain. Sequence analysis revealed that each of these 3 clones encodes a 206-amino-acid protein of about 25 kDa. This we named p25. Blast searches demonstrated that p25 has 100% amino acid sequence identity, except for the last 2 amino acids, with the N-terminal half of the zinc finger transcription factor, ZBP-89. As shown in Figure 1C, a single nucleotide insertion in the cDNA encoding p25 caused an in-frame stop codon immediately after the 4 zinc finger motifs, resulting in premature termination. To test the possibility that this truncated form of ZBP-89 is a physiologic product generated by alternative splicing in vivo, RT-PCR was performed using total RNA extracted from HeLa and U937 cells and U937 cells treated with PMA for 24 hours. Sequencing of the resulting PCR products demonstrated that none contained the nucleotide insertion, suggesting that the premature termination in p25 represents an in vitro artifact in the cDNA library (data not shown).
ZBP-89 binds to the GC-rich element of the CD11bpromoter
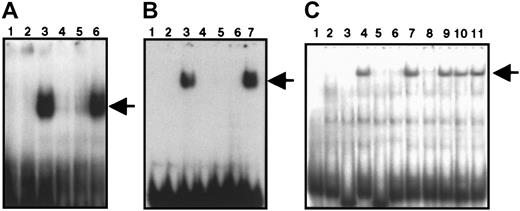
To confirm that p25 actually binds to the bait sequence, EMSA analyses were carried out with a radiolabeled probe (GC, the double-stranded GC-rich element used for the yeast one-hybrid screen) and GST-Z, which represents the protein p25 fused to GST. The fusion proteins were expressed in soluble form in Escherichia coliBL21 (DE3) and used for EMSA. As shown in Figure2A, the GST protein alone does not bind to GC (lane 2), whereas GST-Z formed a complex with GC (lane 3). This complex was competed with unlabeled GC (lane 4) or with the oligonucleotide htβ (lane 5), which represents the ZBP-89–binding site in the T-cell receptor promoter.27 A Sp1 consensus oligonucleotide did not compete (lane 6). We also performed EMSA with the fusion protein GST-ZBP-89, which represents the full-length ZBP-89 protein fused to GST and obtained similar results (Figure 2B). This suggests that ZBP-89 binds to the double-stranded GC-rich element of the CD11b promoter.
EMSAs showing the DNA-binding activity of recombinant GST-Z, GST-ZBP-89, and endogenous ZBP-89.
(A) Lane 1, radiolabeled GC oligonucleotide alone; lanes 2 and 3, labeled GC in the presence of GST or GST-Z, respectively. In panels A-C, the arrow indicates the position of the complex. This complex was competed with the unlabeled GC oligonucleotide (lane 4) or oligonucleotide htβ (lane 5). Lane 6 shows the lack of competition by a consensus Sp1 oligonucleotide. (B) Lane 1, radiolabeled GC alone; lanes 2 and 3, labeled GC in the presence of GST or GST-ZBP-89 fusion protein, respectively. The resulting complex was competed with unlabeled GC (lane 4) or with htβ (lane 5); lanes 6 and 7 show the effect of anti–ZBP-89 antibody (lane 6) and preimmune serum (lane 7), respectively, on the ZBP-89 complex. (C) Lane 1, radiolabeled GC alone. In lanes 2 and 3, binding reactions containing uninduced U937 nuclear extract. Lanes 4 to 11 show binding reactions containing PMA-induced U937 nuclear extract. ZBP-89 binding to DNA (lane 4) was competed with a 250-fold molar excess of unlabeled GC (lanes 3 and 5) or htβ (lanes 4 and 6). A consensus Oct-1 oligonucleotide (lane 7) and a consensus Sp1 oligonucleotide were ineffective (lane 10) in competition. Lanes 8, 9, and 11 represent binding reactions preincubated with anti–ZBP-89 antibody (lane 8), preimmune sera (lane 9), and anti-Sp1 antibody (lane 11), respectively.
EMSAs showing the DNA-binding activity of recombinant GST-Z, GST-ZBP-89, and endogenous ZBP-89.
(A) Lane 1, radiolabeled GC oligonucleotide alone; lanes 2 and 3, labeled GC in the presence of GST or GST-Z, respectively. In panels A-C, the arrow indicates the position of the complex. This complex was competed with the unlabeled GC oligonucleotide (lane 4) or oligonucleotide htβ (lane 5). Lane 6 shows the lack of competition by a consensus Sp1 oligonucleotide. (B) Lane 1, radiolabeled GC alone; lanes 2 and 3, labeled GC in the presence of GST or GST-ZBP-89 fusion protein, respectively. The resulting complex was competed with unlabeled GC (lane 4) or with htβ (lane 5); lanes 6 and 7 show the effect of anti–ZBP-89 antibody (lane 6) and preimmune serum (lane 7), respectively, on the ZBP-89 complex. (C) Lane 1, radiolabeled GC alone. In lanes 2 and 3, binding reactions containing uninduced U937 nuclear extract. Lanes 4 to 11 show binding reactions containing PMA-induced U937 nuclear extract. ZBP-89 binding to DNA (lane 4) was competed with a 250-fold molar excess of unlabeled GC (lanes 3 and 5) or htβ (lanes 4 and 6). A consensus Oct-1 oligonucleotide (lane 7) and a consensus Sp1 oligonucleotide were ineffective (lane 10) in competition. Lanes 8, 9, and 11 represent binding reactions preincubated with anti–ZBP-89 antibody (lane 8), preimmune sera (lane 9), and anti-Sp1 antibody (lane 11), respectively.
To investigate the binding characteristics of ZBP-89 on theCD11b promoter during myeloid differentiation, EMSA was carried out by incubating labeled GC with nuclear extracts prepared from untreated U937 cells or U937 cells treated with PMA for 24 hours to induce their differentiation into monocytelike cells. Figure 2C shows that a DNA-protein complex was formed using extracts prepared from induced cells (lane 4) but not from uninduced U937 cells (lane 2). Unlabeled GC or htβ oligonucleotides competed for the complex (lanes 5 and 6, respectively); however, oligonucleotides Oct-1 (lane 7) and Sp1 (lane 10) did not compete. Incubation with an antibody, which specifically recognizes ZBP-89, resulted in the disappearance of the complex (lane 8), but incubation with preimmune serum (lane 9) or a Sp1-specific antibody (lane 11) did not. These results indicate that the endogenous ZBP-89 protein is inducible on differentiation of U937 cells and that it binds to the GC-rich element of the CD11bgene.
Identification of the residues in the CD11b promoter that are critical for ZBP-89 binding
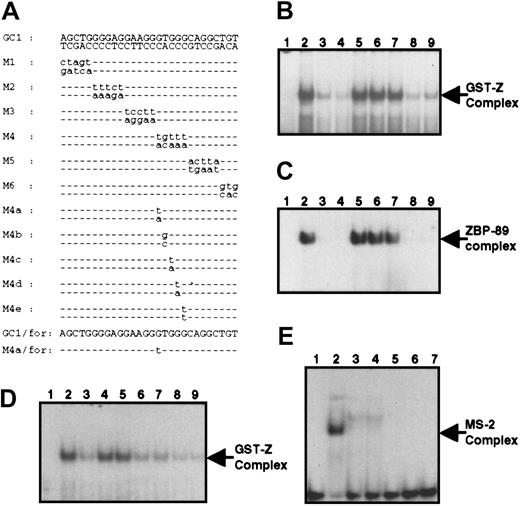
To identify the minimal region required for p25 binding, competitive gel shift assays were performed using the oligonucleotide GC1, which is 9 nucleotides shorter than GC (Figure3A). As shown in Figure 3B, a GST-Z–containing complex was formed with radiolabeled GC1 and this was competed efficiently with its unlabeled equivalent (lane 3). Next, a series of oligonucleotides (M1-M6) representing sequential mutation of 3 or 5 nucleotides in GC1 were used as competing oligonucleotides. The oligonucleotides M2-M4 lost their ability to compete for binding to GST-Z (lanes 5-7), whereas oligonucleotides M1, M5, and M6 effectively competed (lanes 4, 8, and 9, respectively). We also performed the same competition experiment with the fusion protein GST-ZBP-89 and obtained similar results (Figure 3C). These results demonstrate that p25 and ZBP-89 bind to the double-stranded oligonucleotide spanning nucleotides −127 to −113 of the CD11b promoter, thus delineating the DNA-binding segment for ZBP-89.
Characterization of the ZBP-89–binding site within theCD11b gene.
(A) Sequence of the double- or single-stranded oligonucleotides used in EMSA are shown. Mutations incorporated into the GC1 oligonucleotide are shown in lowercase letters, whereas wild-type nucleotides are represented by dashed lines. (B) Lane 1, radiolabeled GC1; lanes 2 to 9, binding reaction with GST-Z before (lane 2) or after competing with the following oligonucleotides: unlabeled GC1 (lane 3), M1 (lane 4), M2 (lane 5), M3 (lane 6), M4 (lane 7), M5 (lane 8), M6 (lane 9). (C) Same as in panel B except that GST-ZBP-89 was used instead of GST-Z. (D) Competition analysis of GC1 binding to GST-Z using mutant oligonucleotides. Lane 1, radiolabeled GC1 alone. Lane 2, binding of GC1 to GST-Z. Competition by the M4b-e oligonucleotides (lanes 6, 7, 8, and 9) was approximately equivalent to that of GC1 (lane 3), whereas competition by M4 (lane 4) and M4a (lane 5) was minimal. (E) Binding of the purine-rich single-stranded GC1/for oligonucleotide to MS-2. Lane 1, radiolabeled GC1/for single-stranded oligonucleotide alone; lane 2, the MS-2 complex produced by mixing the labeled GC1/for oligonucleotide with nuclear extract from U937 cells treated with PMA for 24 hours; lanes 3 and 4, competition in the presence of unlabeled GC1/for (lane 3) or M4a/for (lane 4). Lanes 5 to 7, lack of binding of recombinant GST-ZBP-89 to radiolabeled GC1/for in the absence (lane 5) or presence of unlabeled GC1/for (lane 6) or unlabeled M4a/for (lane 7).
Characterization of the ZBP-89–binding site within theCD11b gene.
(A) Sequence of the double- or single-stranded oligonucleotides used in EMSA are shown. Mutations incorporated into the GC1 oligonucleotide are shown in lowercase letters, whereas wild-type nucleotides are represented by dashed lines. (B) Lane 1, radiolabeled GC1; lanes 2 to 9, binding reaction with GST-Z before (lane 2) or after competing with the following oligonucleotides: unlabeled GC1 (lane 3), M1 (lane 4), M2 (lane 5), M3 (lane 6), M4 (lane 7), M5 (lane 8), M6 (lane 9). (C) Same as in panel B except that GST-ZBP-89 was used instead of GST-Z. (D) Competition analysis of GC1 binding to GST-Z using mutant oligonucleotides. Lane 1, radiolabeled GC1 alone. Lane 2, binding of GC1 to GST-Z. Competition by the M4b-e oligonucleotides (lanes 6, 7, 8, and 9) was approximately equivalent to that of GC1 (lane 3), whereas competition by M4 (lane 4) and M4a (lane 5) was minimal. (E) Binding of the purine-rich single-stranded GC1/for oligonucleotide to MS-2. Lane 1, radiolabeled GC1/for single-stranded oligonucleotide alone; lane 2, the MS-2 complex produced by mixing the labeled GC1/for oligonucleotide with nuclear extract from U937 cells treated with PMA for 24 hours; lanes 3 and 4, competition in the presence of unlabeled GC1/for (lane 3) or M4a/for (lane 4). Lanes 5 to 7, lack of binding of recombinant GST-ZBP-89 to radiolabeled GC1/for in the absence (lane 5) or presence of unlabeled GC1/for (lane 6) or unlabeled M4a/for (lane 7).
MS-2, a putative transcription factor, binds to the GC-rich element of the CD11b promoter and might be involved in induction of theCD11b gene during myeloid cell differentiation.18 In Figure 3E (lane 2), we show that MS-2 is a purine-rich ssDNA-binding factor. Because the binding sites for p25/ZBP-89 and MS-2 overlap, it was necessary to identify a mutation that disrupts ZBP-89 but not MS-2 binding to assess the specific function of ZBP-89. A competitive gel shift assay was performed using GC1 as a probe, and a series of mutant double-stranded oligonucleotides (M4a-e) were used as competitors, each of which represented a different point mutation within GC1 (Figure 3A). Figure3D shows that the M4a mutation, which has a transversion of G>T at nucleotide −117 does not compete with the radiolabeled wild-type double-stranded probe GC1 (lane 4), indicating that G at −117 is crucial for ZBP-89 binding. The purine-rich sense strand of GC1 (GC1/for) formed an MS-2 complex (Figure 3E, lane 2). This complex was competed equally well with unlabeled single-stranded wild-type (GC1/for) and mutant (M4a/for) oligonucleotides (lanes 3 and 4, respectively), indicating that G at −117 is not essential for MS-2 binding. Therefore, the M4a mutation abolishes ZBP-89 binding while retaining the ability to bind MS-2. When GST-ZBP-89 was incubated with radiolabeled purine-rich single-stranded GC1/for (lanes 5-7), no complex was observed in the absence (lane 5) or in the presence of wild-type GC1/for (lane 6) or mutant M4a/for (lane 7). Thus, although ZBP-89 and MS-2 share a common DNA-binding site, they are distinct factors with different binding and DNA-recognition properties.
Overexpression of ZBP-89 represses CD11b gene expression
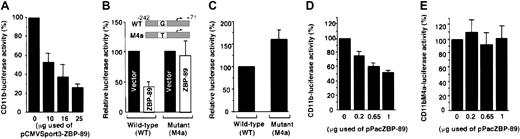
To study the role of ZBP-89 in regulating CD11bpromoter activity, increasing amounts of pCMVSport3-ZBP-89 were transfected into U937 in which the wild-type CD11b promoter fused to the luciferase gene was stably incorporated. Transfection with pCMVSport3 served as a negative control. As can be seen in Figure 4A, ZBP-89 induced repression of PMA-induced CD11b in a dose-dependent manner, reaching 75% at the maximal amount of transfected DNA tested (Figure4A). The effect was also observed in U937 in which pCMVSport3-ZBP-89 was cotransfected with a reporter plasmid containing theCD11b promoter fused to the luciferase gene (Figure 4B, left panel); ZBP-89 decreased PMA-induced wild-typeCD11b (CD11b-wt) gene expression by 55%; the reduced repression seen may be a reflection of the lower amount of plasmid DNA allowable in the cotransfection studies. The effect of repression by ZBP-89 was specific because no such repression occurred in U937 cells cotransfected with pCMVSport3-ZBP-89 and CD11b-M4a promoter plasmids (Figure 4B, right panel).
Transcriptional repression of CD11b by overexpression of ZBP-89 in differentiating U937 cells and in
Drosophila SL2 cells. (A) Dose-dependent repression of CD11b by ZBP-89 in stable U937 cells in whichCD11b-luciferase gene is incorporated in the genome. The various concentrations of pPacZBP-89 are indicated. Luciferase activity is normalized against β-galactosidase activity, and each histogram represents the mean ± SD of 3 independent experiments. In a typical experiment, the CD11b luciferase activity without ZBP-89 was 41 771 relative light units (RLU; normalized to 100%), compared to 22 RLU in stable cell lines lacking the exogenousCD11b promoter. (B) Histograms comparing the transrepression activity of ZBP-89 on CD11b-wt and mutantCD11b-M4a gene expression. Each transfection was performed 6 times. Luciferase activity is normalized against β-galactosidase activity, and the SD from the mean is presented with each histogram. The fold above background (conferred by the negative control plasmid pATLuc) of CD11b-wt luciferase activity without ZBP-89 is 29.32 ± 2.36 (n = 6), and this was considered as 100%. (C) Derepression of CD11b promoter activity by blocking endogenous ZBP-89 binding. U937 cells were transfected with pALbWt or pALbM4a then treated for 16 hours with PMA prior to harvesting. The level of Wt luciferase reporter gene activity, normalized against β-galactosidase activity to correct transfection efficiency, is expressed relative to that conferred by the negative control plasmid pATLuc. It represented 143.85 ± 8.5 (n = 3) fold above background and equated to 100%. (D) Transfection assays in DrosophilaSL2 cells were performed using 1 μg of the reporter construct pALbWt, 0.2 μg pPacSp1 expressing Sp1, 0.1 μg pIELacZ expressing β-galactosidase, and an increasing amount of pPacZBP-89 (expressing ZBP-89) from 0 to 1 μg. The various concentrations of pPacZBP-89 are indicated. The final DNA concentration in all transfections was adjusted to 2.6 μg by adding the empty vector pPacO. Sp1-driven promoter activity was expressed as a ratio of Sp1-driven CD11b-wt promoter activity divided by that obtained in the absence of Sp1. In the absence of ZBP-89 expression, the calculated level of Sp1-driven transcription (81.1 ± 12.2 above background, n = 3) was considered as 100%. (E) The same experiment was performed as in panel C using pALbM4a instead of pALbWt (actual values of Sp-1–driven transcription were 91 ± 10.68, n = 3, above background).
Transcriptional repression of CD11b by overexpression of ZBP-89 in differentiating U937 cells and in
Drosophila SL2 cells. (A) Dose-dependent repression of CD11b by ZBP-89 in stable U937 cells in whichCD11b-luciferase gene is incorporated in the genome. The various concentrations of pPacZBP-89 are indicated. Luciferase activity is normalized against β-galactosidase activity, and each histogram represents the mean ± SD of 3 independent experiments. In a typical experiment, the CD11b luciferase activity without ZBP-89 was 41 771 relative light units (RLU; normalized to 100%), compared to 22 RLU in stable cell lines lacking the exogenousCD11b promoter. (B) Histograms comparing the transrepression activity of ZBP-89 on CD11b-wt and mutantCD11b-M4a gene expression. Each transfection was performed 6 times. Luciferase activity is normalized against β-galactosidase activity, and the SD from the mean is presented with each histogram. The fold above background (conferred by the negative control plasmid pATLuc) of CD11b-wt luciferase activity without ZBP-89 is 29.32 ± 2.36 (n = 6), and this was considered as 100%. (C) Derepression of CD11b promoter activity by blocking endogenous ZBP-89 binding. U937 cells were transfected with pALbWt or pALbM4a then treated for 16 hours with PMA prior to harvesting. The level of Wt luciferase reporter gene activity, normalized against β-galactosidase activity to correct transfection efficiency, is expressed relative to that conferred by the negative control plasmid pATLuc. It represented 143.85 ± 8.5 (n = 3) fold above background and equated to 100%. (D) Transfection assays in DrosophilaSL2 cells were performed using 1 μg of the reporter construct pALbWt, 0.2 μg pPacSp1 expressing Sp1, 0.1 μg pIELacZ expressing β-galactosidase, and an increasing amount of pPacZBP-89 (expressing ZBP-89) from 0 to 1 μg. The various concentrations of pPacZBP-89 are indicated. The final DNA concentration in all transfections was adjusted to 2.6 μg by adding the empty vector pPacO. Sp1-driven promoter activity was expressed as a ratio of Sp1-driven CD11b-wt promoter activity divided by that obtained in the absence of Sp1. In the absence of ZBP-89 expression, the calculated level of Sp1-driven transcription (81.1 ± 12.2 above background, n = 3) was considered as 100%. (E) The same experiment was performed as in panel C using pALbM4a instead of pALbWt (actual values of Sp-1–driven transcription were 91 ± 10.68, n = 3, above background).
Loss of repression of the M4a-CD11b promoter by endogenous ZBP-89
It remains possible that exogenous ZBP-89 overexpressed in differentiating U937 cells may act indirectly in repressing theCD11b promoter. To establish whether endogenous ZBP-89 also acts as a repressor, we compared the promoter activities of the CD11b-wt and CD11b-M4a in PMA-induced U937 cells (Figure 4C). Expression of the mutant CD11b-M4a reporter in differentiating U937 increased to 160% of the wild-type reporter (Figure 4C), reflecting loss of repression by endogenous ZBP-89. Thus, the level of CD11bgene expression in differentiating U937 cells reflects the activity of endogenous ZBP-89.
Reconstitution of ZBP-89 repression in a heterologous expression system
Expression of CD11b is normally restricted to differentiating myeloid cells. The CD11b−Drosophila Schneider L2 cell line is deficient in Sp1-related proteins,24,30,31but can express a CD11b promoter-directed reporter when supplemented with Sp1.32 We found this cell line to be also lacking ZBP-89 by Western blotting as well as by EMSA (data not shown). We cotransfected Schneider L2 cells with 0.2 μg of the construct pPacSp1 (which expresses Sp1), together with increasing amounts of pPacZBP-89 expressing ZBP-89, along with either CD11b-wt or CD11b-M4a-luciferase plasmids. In the presence of Sp1, ZBP-89 repressed wild-typeCD11b promoter activity by up to 50% (Figure 4D). In contrast, CD11b-M4a promoter activity was not affected by expression of ZBP-89 (Figure 4E). Expression of ZBP-89 alone without Sp1 did not activate the CD11b promoter (data not shown). Therefore, repression of CD11b by ZBP-89 can also be achieved in a nonmyeloid cellular context and independently of the confounding effects of treatment with phorbol esters.
Expression of CD11b does not correlate with endogenous ZBP-89 protein during phorbol ester–induced differentiation of U937 cells
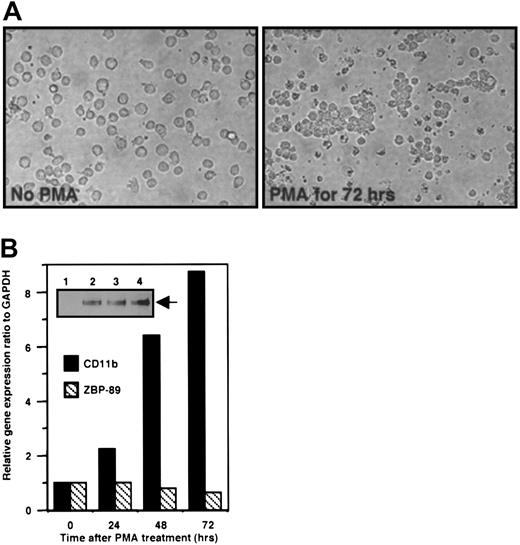
We next determined the correlation between the endogenous levels of CD11b mRNA and ZBP-89 protein in U937 cells differentiated by treatment with PMA for 72 hours, a treatment that alters homotypic adhesion in these cells (Figure 5A. Both ZBP-89 protein and CD11b mRNA increased in parallel during the differentiation of PMA-treated U937 cells into monocytelike cells (Figure 5B). Thus whereas overexpressed ZBP-89 represses theCD11b gene, we find no inverse correlation between endogenous ZBP-89 and expression of the CD11b gene in this system. These data argue against a role for endogenous ZBP-89 in repressing CD11b gene during the early stage of monocytic differentiation. Because overexpression of ZBP-89 represses the CD11b promoter-driven reporter in these cells, qualitative or quantitative changes in ZBP-89 protein levels together with the potential inaccessibility of the native CD11b promoter to ZBP-89 during early monopoiesis may account for the lack of correlation between endogenous CD11b mRNA and ZBP-89 protein at this stage.
Expression profiles of endogenous ZBP-89 protein and mRNA and CD11b mRNA during differentiation of U937.
(A) Morphology of U937 cells without (left panel) and with PMA treatment for 72 hours (right panel). The later cells form readily visible clusters (original magnification, × 40). (B) Real-time RT-PCR analysis of CD11b and ZBP-89 during U937 cell differentiation. The experiment was performed using total RNA isolated from U937 cells treated with PMA (100 ng/mL) for 0, 24, 48, and 72 hours. The ratios CD11b/GAPDH or ZBP-89/GAPDH in untreated U937 cells (0 h) were given a value of 1, and the ratios at 24, 48, and 72 hours are displayed as fold above 1. (Inset) Thirty micrograms of nuclear extract from U937 cells treated with PMA (100 ng/mL) for 0 (lane 1), 24 hours (lane 2), 48 hours (lane 3), and 72 hours (lane 4) were resolved by 6% SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and analyzed with an anti–ZBP-89 antibody followed by chemiluminescence. ZBP-89 protein is indicated with an arrow.
Expression profiles of endogenous ZBP-89 protein and mRNA and CD11b mRNA during differentiation of U937.
(A) Morphology of U937 cells without (left panel) and with PMA treatment for 72 hours (right panel). The later cells form readily visible clusters (original magnification, × 40). (B) Real-time RT-PCR analysis of CD11b and ZBP-89 during U937 cell differentiation. The experiment was performed using total RNA isolated from U937 cells treated with PMA (100 ng/mL) for 0, 24, 48, and 72 hours. The ratios CD11b/GAPDH or ZBP-89/GAPDH in untreated U937 cells (0 h) were given a value of 1, and the ratios at 24, 48, and 72 hours are displayed as fold above 1. (Inset) Thirty micrograms of nuclear extract from U937 cells treated with PMA (100 ng/mL) for 0 (lane 1), 24 hours (lane 2), 48 hours (lane 3), and 72 hours (lane 4) were resolved by 6% SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and analyzed with an anti–ZBP-89 antibody followed by chemiluminescence. ZBP-89 protein is indicated with an arrow.
Down-regulation of CD11b during monocytes to macrophage differentiation correlates with ZBP-89 protein expression
Because the level of CD11b protein is known to drop as blood monocytes differentiate into macrophages,10,13 we next assessed the impact of ZBP89 on CD11b expression in these cells. CD11b mRNA and ZBP-89 protein levels were determined during the in vitro differentiation of normal human peripheral blood monocytes into macrophages. Normal blood monocytes assumed the typical features of macrophages after 12 days in culture (Figure6A, compare left and right panels) as described.21 33 As shown in Figure 6B, CD11b mRNA dropped significantly (by ∼75%) in macrophages compared to monocytes. Importantly, this reduction coincided with an increase of protein expression of ZBP-89 (Figure 6C, compare lanes 3 and 4) that has an identical mobility to the form induced in differentiating U937 cells (Figure 6C, lanes 1 and 2; compare lanes 2 and 4). These data suggest that ZBP-89 exerts its repressor activity on CD11b during the late stages of monocytic differentiation.
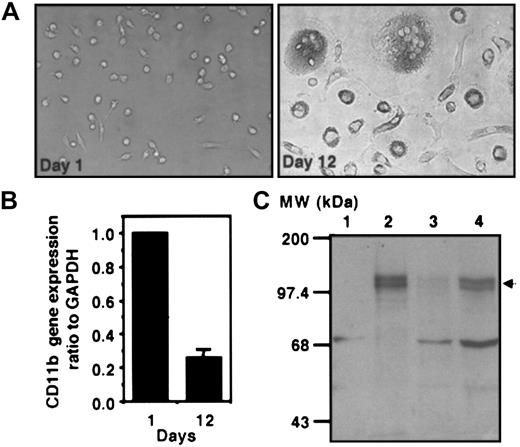
Inverse correlation between endogenous ZBP-89 protein and CD11b mRNA during differentiation of monocytes/macrophages.
(A) Morphology of human monocytes during differentiation and maturation into macrophages is shown at day 1 (left panel) and day 12 (right panel). Representative fields are shown using light microscopy (original magnification, × 40). (B) Real-time RT-PCR analysis ofCD11b during monocyte/macrophage differentiation. Total RNA isolated from monocytes/macrophages differentiated in vitro for 1 or 12 days were reverse transcribed to make cDNA. Quantitative PCR was carried out using the TaqMan system and an ABI Prism 7700 instrument as described in “Materials and methods.” The copy number of CD11b is normalized to the copy number of GAPDH and expressed as CD11b/GAPDH. The CD11b/GAPDH ratio in monocytes at day 1 was given a value of 1, and the ratio at day 12 is shown as a relative value to 1. Each histogram represents the mean ± SD of 3 independent experiments. (C) Western blot analysis showing ZBP-89 protein expression during differentiation of U937 cells and monocytes/macrophages. Five micrograms of nuclear extract from U937 cells treated with PMA for 0 hours (lane 1) or 72 hours (lane 2), and 30 μg total protein lysate from monocytes/macrophages differentiated for 1 (lane 3) or 12 days (lane 4) were resolved by 6% SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and detected with an anti–ZBP-89 antibody followed by chemiluminescence. ZBP-89 (arrow) migrates as a double band with apparent mobility of about 120 kDa, consistent with similar analyses by other investigators30,34 60 MW indicates molecular weight markers (Gibco-BRL).
Inverse correlation between endogenous ZBP-89 protein and CD11b mRNA during differentiation of monocytes/macrophages.
(A) Morphology of human monocytes during differentiation and maturation into macrophages is shown at day 1 (left panel) and day 12 (right panel). Representative fields are shown using light microscopy (original magnification, × 40). (B) Real-time RT-PCR analysis ofCD11b during monocyte/macrophage differentiation. Total RNA isolated from monocytes/macrophages differentiated in vitro for 1 or 12 days were reverse transcribed to make cDNA. Quantitative PCR was carried out using the TaqMan system and an ABI Prism 7700 instrument as described in “Materials and methods.” The copy number of CD11b is normalized to the copy number of GAPDH and expressed as CD11b/GAPDH. The CD11b/GAPDH ratio in monocytes at day 1 was given a value of 1, and the ratio at day 12 is shown as a relative value to 1. Each histogram represents the mean ± SD of 3 independent experiments. (C) Western blot analysis showing ZBP-89 protein expression during differentiation of U937 cells and monocytes/macrophages. Five micrograms of nuclear extract from U937 cells treated with PMA for 0 hours (lane 1) or 72 hours (lane 2), and 30 μg total protein lysate from monocytes/macrophages differentiated for 1 (lane 3) or 12 days (lane 4) were resolved by 6% SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and detected with an anti–ZBP-89 antibody followed by chemiluminescence. ZBP-89 (arrow) migrates as a double band with apparent mobility of about 120 kDa, consistent with similar analyses by other investigators30,34 60 MW indicates molecular weight markers (Gibco-BRL).
Discussion
The major finding in this paper is that ZBP-89 is a repressor of the CD11b gene. Repression is dose-dependent, sequence-specific, and demonstrable in nonmyeloid insect cells. In the U937 model of early-stage monocytic differentiation, no inverse correlation was observed between ZBP-89 levels andCD11b gene expression. However, in the model of late-stage monocyte-to-macrophage differentiation, such an inverse correlation was readily detected. Consequently, these findings suggest that the effect of ZBP-89 on CD11b gene expression occurs primarily at the later rather than the early stages of monocytic differentiation. This observation may underlie the observed drop in the levels of CD11b as monocytes differentiate into tissue macrophages in vivo.
ZBP-89 is a 794-amino-acid ubiquitously expressed factor,26,34 originally cloned in a truncated form, htβ, that mediates mild transactivation of the human T-cell receptor gene.27 ZBP-89 belongs to the Krüppel zinc finger family35; other members include WT1, GATA1, Sp1, and Egr1. A breakpoint in the chromosomal region housingZBP-89 is associated with hematologic malignancy23,36 and ZBP-89 is known to stabilize p53, resulting in the arrest of cell growth.37 In fact, many of the target genes of ZBP-89 are regulated by mitogens or developmental signals.26,30,34,38-40 The consensus binding site for ZBP-89 is also found in the promoters of the genes encoding PU.141 and CEBP-α,42 which play important roles in myeloid differentiation. Thus, ZBP-89 appears to be involved in regulation of cellular proliferation as well as differentiation.
The structural basis for ZBP-89–mediated repression of theCD11b gene remains to be determined. Five to 10% of zinc finger DNA-binding motifs share a conserved 120-amino-acid domain termed the POZ or BTB domain.43 Many POZ domain-containing proteins are transcriptional repressors, although some, such as the GAGA factor, can counteract repression by chromatin remodeling.44 Charged, alanine-, alanine/proline-, and alanine/glutamine-rich motifs have also been found in transcriptional repressor domains.45-47 However, homology searches reveal that ZBP-89 contains no such domains. A previous study demonstrated that a transferable region in ZBP-89 spanning amino acids 136 to 184 repressed β-enolase gene transcription during embryonic muscle cell differentiation.34 However, the expression of the truncated form of ZBP-89 called p25, which encompasses this repressor region, did not repress CD11b gene activation (data not shown). This suggests that the carboxyl-terminal domain of ZBP-89 may contribute to CD11b transcriptional repression. Because this segment contains no known repressor domains, repression may be mediated by a yet to be discovered protein domain within this region.
Functionally, 4 general mechanisms may give rise to selective transcriptional repression of genes. First, a repressor protein can mask a transcriptional activator domain. Second, the repressor may block interaction of an activator with other components of the transcription machinery.48 Third, it can displace an activator from its DNA element by competition,49 and fourth, the DNA element itself may exert allosteric effects on transcriptional regulators, such that regulators may activate transcription in the context of one gene, yet repress transcription in another.50,51 In regulating expression of theβ-enolase and gastrin genes, ZBP-89 acts as a bona fide transcriptional repressor; it inhibits basal as well as activated transcription, an effect not mediated by squelching.26,34 Recent data have shown that ZBP-89, can compete with Sp1 in regulating expression of the vimentingene.39 Sp1 is also required in CD11btranscriptional activation,32 suggesting that some of the repressor activity may be mediated through this mechanism. ZBP-89 and Sp1 do not, however, cross-compete in EMSA analyses (lanes 9 and 10 in Figure 2C), suggesting that interaction may occur at the protein-protein level. It is possible that the promoter context and cell type could also contribute to ZBP-89 repressor activity. In this situation, ZBP-89 might act through interaction with an adjacent transcriptional activator, modulating its functional state or local concentration.
The ZBP-89 element overlaps with but is distinct from that of MS-2, an ssDNA-binding factor of the CD11b gene whose identity remains to be determined18 (Figure 3). ZBP-89 may repress by competing with MS-2 for DNA binding.48 Supporting this notion is our observation that the activity of the mutant promoterCD11b-M4a is higher than that of CD11b-wt (Figure4B). This increase may also be due to the blocked binding of endogenous ZBP-89. However, the binding of adjacent proteins such as MS-2 may also affect ZBP-89 activity by modifying its association with comodulators or the basal transcription machinery. There are examples of dsDNA- and ssDNA-binding proteins binding to overlapping DNA segments and thus regulating transcription. Within the vascular smooth muscle α-actin gene, transcriptional enhancer factor 1 (TEF-1) binds to dsDNA, whereas Purα, Purβ, and MSY-1 bind to ssDNA and act as repressors.52-54 Similarly a 42-base pair regulatory element in the 5′-flanking region of the L1 transcript of the murinegrowth hormone receptor gene interacts both with MSY-1 and the dsDNA-binding protein NF-1.55 In the proximal promoter of the NMDAR1 gene, a GC-rich element contains the binding sites for the dsDNA binding proteins, Egr-1 and Sp1, and both sense and antisense strands of this element also form complexes with yet to be identified factors.56 Considering that the DNA macromolecule is a dynamic structure capable of assuming a single-stranded conformation under certain conditions, changes in the secondary structure of the CD11b promoter may be critical in controlling CD11b gene expression.
The CD11b/CD18 heterodimer is expressed on peripheral blood monocytes, neutrophilic polymorphonuclear leukocytes (PMNs), and natural killer cells, but is generally expressed at reduced amounts on tissue macrophages such as Kupffer cells, bone marrow stromal macrophages, and alveolar macrophages.10,13 57-59 The mechanisms that underlie this reduction are not known. In vitro differentiation of freshly isolated human monocytes into macrophages was accompanied by a significant drop in CD11b mRNA and induction of the ZBP-89 protein. These findings suggest that ZBP-89 acts at a later stage in monocytic differentiation as monocytes further transform into macrophages. CD11b/CD18 is known to be important not only in leukocyte recruitment into tissues but also in the many proinflammatory adhesion-dependent functions mediated by these cells in tissues. Quantitative as well as qualitative down-regulation of this receptor once monocytes transmigrate into tissues as part of their normal differentiation and maturation may be necessary to avert harmful inflammation. The data presented in this work suggest that ZBP-89 may act at the transcriptional level to elicit such a protective effect during the resolution of an inflammatory response.
We thank Dr J. L. Merchant for the anti–ZBP-89 antibody and ZBP-89 expression plasmid, Dr R. Tjian for the pPacO and pPacSp1 plasmids, Dr D. T. Scadden, Dr K. Ferenczi, Dr Boris Nikolic, and Mr D. Olson for helpful discussion and assistance with FACS analysis, Dr M. Xue for helpful suggestions on the nature of MS-2 and Miss G. Rouiffe for technical support.
Prepublished online as Blood First Edition Paper, September 19, 2002; DOI 10.1182/blood-2002-03-0680.
Supported by National Institutes of Health grants DK50305 and DK50779.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
M. Amin Arnaout, Renal Unit, Massachusetts General Hospital, 149 13th St, Charlestown, MA 02129; e-mail:arnaout@receptor.mgh.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal