Abstract
The molecular characterization of the CD4+ T-cell epitope repertoire on human tumor antigens would contribute to both clinical investigation and cancer immunotherapy. In particular, the identification of promiscuous epitopes would be beneficial to a large number of patients with neoplastic diseases regardless of their HLA-DR type. MAGE-3 is a tumor-specific antigen widely expressed in solid and hematologic malignancies; therefore, is an excellent candidate antigen. We used a major histocompatability complex (MHC) class II epitope prediction algorithm, the TEPITOPE software, to predict 11 sequence segments of MAGE-3 that could form promiscuous CD4+ T-cell epitopes. In binding assays, the synthetic peptides corresponding to the 11 predicted sequences bound at least 3 different HLA-DR alleles. Nine of the 11 peptides induced proliferation of CD4+ T cells from both healthy subjects and melanoma patients. Four immunodominant regions (residues 111-125, 146-160, 191-205, and 281-295), containing naturally processed epitopes, were recognized by most of the donors, in association with 3 to 4 different HLA-DR alleles, thus covering up to 94% of the alleles expressed in whites. On the contrary, the other promiscuous regions (residues 161-175 and 171-185) contained epitopes not naturally processed in vitro. The immunodominant epitopes identified will be useful in the design of peptide-based cancer vaccines and in the study of the functional state of tumor-specific CD4+ T cells in patients bearing tumors expressing MAGE-3.
Introduction
Evidence for a critical role of CD4+ T cells in orchestrating the antitumor response is increasing (for reviews, see Pardoll and Topalian1 and Toes et al2). CD4+ T cells provide help for induction and maintenance of antitumor CD8+ T cells and may exert effector functions, both indirectly via macrophage and eosinophil activation, and directly when inflammatory responses produce cytokines such as interferon γ (IFN-γ) that induce expression of major histocompatability complex (MHC) class II molecules on tumor cells. In addition, IFN-γ produced by antitumor CD4+ T cells may function as an antiangiogenic factor.3,4 CD4+ T cells specific for tumor antigens have been described in humans (for reviews, see Pardoll and Topalian,1 Pawelec et al,5 and Wang6).
MAGE-3 is a tumor-specific antigen widely expressed in solid tumors such as melanoma, lung carcinoma, head and neck carcinoma, and hematologic malignancies, including T-cell leukemias and myeloma but not in normal tissues (with the exception of testis).7 The MAGE gene family comprises several related genes divided into 3 clusters, named MAGE-A, -B, and -C.7 The gene previously known as MAGE-3 belongs to the MAGE-A cluster and it has received the official name MAGE-A3. We will refer to it from now on as MAGE-3, in accordance with our previous report.8 Clinical trials in patients with metastatic neoplastic disease using the HLA-A1–restricted MAGE-3 epitope yielded some clinical results.9-13 However, the immune responses were too weak and transient to eradicate tumor cells in the majority of immunized patients. The additional use of CD4 epitopes might help the induction of a memory antitumor response and improve the clinical efficacy of vaccines.
Several allele-specific CD8 and CD4 epitopes of MAGE-3 have been identified.14 Promiscuous epitopes, that is, those able to bind to different HLA-DR alleles, are crucial to increase the number of patients eligible for peptide-based cancer vaccination. Up to now, only MAGE-3146-160 has been described by us8 and others15 as a promiscuous epitope presented in association with HLA-DR11, HLA-DR4, and HLA-DR7.
In this paper, we report the results of the analysis of the MAGE-3 tumor antigen for promiscuous CD4 epitopes by a combined approach of computational identification of candidate T-cell epitopes, followed by in vitro biologic validation analysis of the predicted sequences as forming natural epitopes recognized by CD4+ T cells.
We identified 4 immunodominant regions (residues 111-125, 146-160, 191-205, and 281-295), containing naturally processed epitopes, that were recognized by most donors in association with 3 to 4 different HLA-DR alleles. These epitopes, potentially recognized by more than 90% of whites, will be useful for both clinical investigation and cancer immunotherapy.
Materials and methods
T-cell epitope prediction and peptide synthesis
We used the TEPITOPE algorithm16 that allows the identification of HLA class II ligands binding in a promiscuous or allele-specific mode and the effects of polymorphic residues on the specificity of HLA class II ligands to select 11 sequences of the MAGE-3 protein for peptide synthesis. The program permits the prediction and the parallel display of ligands for each of the 25 HLA-DR alleles incorporated in the software (examples are reported in Sturniolo et al16). We set the prediction threshold (ie, the percentage of best scoring natural peptides) at 5%, and we selected the sequences predicted to bind at least 40% of the 25 HLA-DR alleles, plus an additional sequence (residues 21-35) predicted to bind 16%. The MAGE-3 sequences comprising residues 141-155, 146-160, 156-170, 171-185, and 281-295, forming pool I, were already described.8 The MAGE-3 sequences 21-35, 111-125, 161-175, 191-205, 251-265, and 286-300, forming pool II, were synthesized by manual parallel synthesis as described.17 The peptide purity was verified by reverse-phase high-performance liquid chromatography and electron spray mass spectrometry. The synthetic peptides were lyophilized, reconstituted in dimethyl sulfoxide (DMSO) at 10 mg/mL, and diluted in RPMI 1640 (Gibco, Grand Island, NY) as needed.
Peptide-binding assays
Peptide interactions with detergent-solubilized HLA-DR molecules were measured using an enzyme-linked immunosorbent assay (ELISA)–based high-flux competition assay.18 HLA-DR molecules were affinity purified from human Epstein-Barr virus–transformed lymphoblastoid cell lines (LCLs) as described.19,20 The assays measured the ability of unlabeled peptides to compete with a biotinylated indicator peptide for binding to purified HLA-DR molecules.21 To determine relative binding affinity, the promiscuous HA307-319 peptide from influenza hemagglutinin21 was included in each competition assay.
Subjects and cells
Peripheral blood mononuclear cells (PBMCs) were obtained from 4 healthy subjects (donors 1, 2, 3, and 4) and 2 patients with melanoma (donors 5 and 6). Melanoma cell lines MD TC, OI TC, and GF TC were established from 2 cutaneous metastases and one lymph node metastasis, respectively, from 3 patients with melanoma, and line SK-Mel-24 was purchased from the American Type Culture Collection (Rockville, MD). MAGE-3 expression by patients' biopsies and melanoma cell lines was verified by reverse transcriptase–polymerase chain reaction (RT-PCR) and in melanoma cells also by intracytoplasmic staining (data not shown). The HLA-DR types of donors and melanoma cell lines were identified by molecular or serologic typing and are reported in the figures. Homozygous LCLs used were Bor (DR11) and Mun (DR3), established in our laboratory; Leis (DR4), kindly provided by F. Marincola (National Institutes of Health, Bethesda, MD); BM21 (DR11) purchased from ATCC; and Pitout (DR7), purchased from the European Collection of Cell Culture (ECCC; Salisbury, United Kingdom).
Propagation of CD4+ T cells
Synthetic peptides corresponding to pool I and pool II (Table1) were used to stimulate the PBMCs from the different donors. Briefly, 20 × 106 PBMCs were cultured for 7 days in RPMI 1640 (Gibco), supplemented with heat-inactivated human serum (10%), l-glutamine (2 mM), penicillin (100 U/mL), and streptomycin (50 μg/mL; Biowhittaker, Walkersville, MD), containing pool I or pool II (1 μg/mL of each peptide). The reactive blasts were isolated on a Percoll gradient,22 expanded in interleukin 2 (IL-2)–containing medium (10 U/mL; Lymphocult, Biotest Diagnostic, Dreieich, Germany), and restimulated at weekly intervals with the same amount of peptides plus irradiated (4000 rad) autologous PBMCs as antigen-presenting cells (APCs).
Determination of HLA-DR binding of MAGE-A3 synthetic peptides corresponding to the sequences predicted by TEPITOPE to form promiscuous epitopes
| . | Sequences . | Predicted binding alleles* . | HLA-DRβ1 alleles . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| *0101 . | *1501 . | *0301 . | *0401 . | *1101 . | *0701 . | *0801 . | |||
| Pool I residues | |||||||||
| 141-155 | GNWQYFFPVIFSKAS | 68 | 25 | 3 | >100‡ | 7 | 0.6 | 0.1 | 3.2 |
| 146-160 | FFPVIFSKASSSLQL | 66 | 10 | 0.24 | 7 | 2 | 1.8 | 0.01 | 1.5 |
| 156-170 | SSLQLVFGIELMEVD | 68 | 7 | 0.18 | 90 | 45 | 28 | 0.03 | 7 |
| 171-185 | PIGHLYIFATCLGLS | 96 | 0.3 | 0.028 | 2.8 | 0.9 | 0.9 | 0.01 | 1.5 |
| 281-295 | TSYVKVLHHMVKISG | 80 | 15 | 0.48 | 26 | 70 | 0.035 | 0.2 | 0.01 |
| Pool II residues | |||||||||
| 21-35 | EALGLVGAQAPATEE | 16 | 14 | 22 | >100‡ | >100‡ | >100‡ | 25 | >100‡ |
| 111-125 | RKVAELVHFLLLKYR | 76 | >100‡ | 0.055 | >100‡ | >100‡ | 0.7 | 55 | 7 |
| 161-175 | VFGIELMEVDPIGHL | 76 | >100‡ | 100 | 0.6 | 28 | >100‡ | 10 | 100 |
| 191-205 | GDNQIMPKAGLLIIV | 40 | >100‡ | 0.07 | >100‡ | >100‡ | 4 | 6 | 1 |
| 251-265 | VQENYLEYRQVPGSD | 48 | >100‡ | 5 | >100‡ | >100‡ | 60 | 26 | 10 |
| 286-300 | VLHHMVKISGGPHIS | 88 | 15 | 0.48 | >100‡ | >100‡ | 0.2 | 0.01 | 14 |
| HA (307-319)† | PKYVKQNTLKLAT | 84 | 0.18 | 2.4 | 6 | 0.9 | 0.7 | 0.18 | 3.2 |
| . | Sequences . | Predicted binding alleles* . | HLA-DRβ1 alleles . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| *0101 . | *1501 . | *0301 . | *0401 . | *1101 . | *0701 . | *0801 . | |||
| Pool I residues | |||||||||
| 141-155 | GNWQYFFPVIFSKAS | 68 | 25 | 3 | >100‡ | 7 | 0.6 | 0.1 | 3.2 |
| 146-160 | FFPVIFSKASSSLQL | 66 | 10 | 0.24 | 7 | 2 | 1.8 | 0.01 | 1.5 |
| 156-170 | SSLQLVFGIELMEVD | 68 | 7 | 0.18 | 90 | 45 | 28 | 0.03 | 7 |
| 171-185 | PIGHLYIFATCLGLS | 96 | 0.3 | 0.028 | 2.8 | 0.9 | 0.9 | 0.01 | 1.5 |
| 281-295 | TSYVKVLHHMVKISG | 80 | 15 | 0.48 | 26 | 70 | 0.035 | 0.2 | 0.01 |
| Pool II residues | |||||||||
| 21-35 | EALGLVGAQAPATEE | 16 | 14 | 22 | >100‡ | >100‡ | >100‡ | 25 | >100‡ |
| 111-125 | RKVAELVHFLLLKYR | 76 | >100‡ | 0.055 | >100‡ | >100‡ | 0.7 | 55 | 7 |
| 161-175 | VFGIELMEVDPIGHL | 76 | >100‡ | 100 | 0.6 | 28 | >100‡ | 10 | 100 |
| 191-205 | GDNQIMPKAGLLIIV | 40 | >100‡ | 0.07 | >100‡ | >100‡ | 4 | 6 | 1 |
| 251-265 | VQENYLEYRQVPGSD | 48 | >100‡ | 5 | >100‡ | >100‡ | 60 | 26 | 10 |
| 286-300 | VLHHMVKISGGPHIS | 88 | 15 | 0.48 | >100‡ | >100‡ | 0.2 | 0.01 | 14 |
| HA (307-319)† | PKYVKQNTLKLAT | 84 | 0.18 | 2.4 | 6 | 0.9 | 0.7 | 0.18 | 3.2 |
The binding data are expressed in terms of relative binding capacity (IC50 μM), calculated as concentration of competitor peptide required to inhibit 50% of the binding of an allele specific biotinylated peptide (indicator peptide).
Percent of alleles predicted to bind the indicated sequences, with a threshold set at 5% (100% is represented by the 25 alleles incorporated in the software). The threshold is defined as the percentage of best scoring natural peptides and comprises a range between 1% to 10%; see “Materials and methods” for details.
HA307-319 is promiscuous sequence from influenza hemagglutinin.
IC50 values higher than 100 μM are outside the sensitivity limits of the binding assay.
Flow cytometry
Cytofluorometric analyses were performed on a FACStarPlus (Becton Dickinson, Sunnyvale, CA). We used the following monoclonal antibodies (mAbs): anti-CD4–phycoerythrin (PE) and anti-CD8–fluorescein isothiocyanate (FITC; Becton Dickinson); L243 (anti–MHC class II; ATCC); 57B (originally described as an anti–MAGE-3 mAb,23 and later identified as an anti–MAGE-A4 mAb that cross-reacts with several MAGE-A proteins24) kindly provided by G. Spagnoli (Basel, Switzerland); and 20.4 (mAb that recognizes the truncated form of the human low-affinity nerve growth factor receptor, ΔNGFr; ATCC). FITC-rabbit antimouse immunoglobulin antibody (Dako, Glostrup, Denmark) was used as second-step reagent in indirect immunofluorescence stainings. For intracytoplasmic staining, melanoma cells were fixed in 3.7% paraformaldehyde, permeabilized in 0.05% NP40, and then stained as described.25
RT-PCR analysis
Total RNA was extracted by the use of RNAzolTMB (Biotech, Houston, TX), according to the manufacturer's instructions. Single-stranded cDNA was synthesized from 2 μg total RNA, by Moloney murine leukemia virus–derived reverse transcriptase (Life Technologies, Milan, Italy), in the presence of 20 U RNasin (Promega, Madison, WI). cDNA coding for MAGE-3 was detected by PCR amplification as described.26 Samples scored positive when a band of the appropriate size was visible on an agarose gel in the presence of ethidium bromide.
Proliferation assay
CD4+ T cells and autologous irradiated PBMCs or HLA-DR–matched homozygous LCLs as APCs were diluted at a 1:10 or 1:5 ratio, respectively, and used as we described previously.8Stimulants are indicated in the figure legends, where appropriate. In inhibition experiments, mAb L243 or an isotype-matched irrelevant mAb was added at the concentrations indicated in the figure legends. After 48 hours, the cultures were pulsed for 16 hours with [3H]-thymidine (TdR; 1 μCi [37 Bq], well, Amersham, Milan, Italy). The cells were collected with a Skatron Titertek multiple harvester (Skatron, Sterling, VA) and the thymidine incorporated was measured in a liquid scintillation counter.
Selection of peptide-specific CD4+ T cells by magnetic separation
To select MAGE-3161-175–specific CD4+ T cells from donor 5 from the polyclonal cell line, we first incubated the cells with the peptide in the presence of homozygous DR4-PBMCs and then we used the magnetic activating cell sorting (MACS) cytokine secretion assay-cell enrichment and detection kit (Miltenyi Biotec, Bergisch Gladback, Germany), following the manufacturer's instructions. MAGE-3161-175–specific CD4+ T cells were then restimulated weekly in the presence of the peptide (1-5 μg/mL) and DR4-PBMCs as APCs.
Recombinant viruses and infection of LCLs
The cDNA encoding Ii.MAGE-3,27 a fusion protein between the human invariant chain (Ii) and MAGE-3, was kindly provided by P. van der Bruggen (Brussels, Belgium). The retroviral vectors M3-CSM and IiM3-CSM, which encode the ΔLNGFr, and either the full-length MAGE-3 protein or the fusion protein Ii.MAGE-3, respectively, were produced as reported previously.28 LCLs were transduced by coculture with irradiated packaging cells producing the M3-CSM or the IiM3-CSM vectors, produced by Phoenix cells (GP Nolan, Stanford, CA). The percentage of infected cells was evaluated 48 hours after transduction by flow cytometry for ΔLNGFr expression with the mAb 20.4.28 The ΔLNGFr+ cells were purified by magnetic cell sorting, using rat antimouse IgG1-coated beads (Dynabeads M-450, Dynal, Oslo, Norway).
Cytotoxicity assay
CD4+ T cells were tested for specific lytic activity in a standard 4-hour 51Cr release assay as described previously.29 The following targets were used: melanoma cells MD TC, OI TC, GF TC, and SK-Mel-24, LCL and LCL engineered to express MAGE-3 (LCL-M3). In cold-target competition assays, unlabeled target cells (cold targets) were added to CD4+ T cells (effectors) and 51Cr-labeled target cells (hot targets) at serial ratios of cold-to-hot target cells. The percentage of inhibition was calculated as follows: [(% specific lysis without cold target−% specific lysis with cold target)/(% specific lysis without cold target)]× 100.
CD4+ T-cell stimulation assay
LCL-M3, LCL-IiM3 (LCLs expressing Ii.MAGE-3), or melanoma cells were tested for their ability to induce the production of IFN-γ by peptide-specific CD4+ T cells, after 24 to 48 hours of incubation, using a standard ELISA (Biosource Europe, Nivelles, Belgium), following the manufacturer's instructions.
Results
We selected 11 MAGE-3 sequences, based on prediction by TEPITOPE, and used synthetic peptides corresponding to the selected sequences to confirm their binding capability in competition assays, using 7 common HLA-DRB1 alleles (Table 1).
We grouped the 11 peptides into pool I and pool II, based on the results of the binding assays. Peptides forming pool I had the highest promiscuity, and peptides forming pool II bound well to at least 3 different HLA-DR alleles. We used pool I and pool II to propagate polyclonal T-cell lines from 6 donors. Total PBMCs were stimulated with pool I or pool II or both in independent cultures for 7 days, activated cells were enriched by a density gradient, expanded in the presence of IL-2, and restimulated weekly with irradiated peptide-pulsed autologous PBMCs. For 5 donors, after 2 cycles of stimulation, we obtained lines that comprised only CD4+ T cells. In the case of donor 1, a CD8+ T-cell depletion step was necessary to obtain more than 90% CD4+ T cells after 4 cycles of stimulation (data not shown). CD4+ T-cell lines from healthy donors 1, 2, and 4 and from donor/patient 5 could be propagated for several months. The lines from healthy subject 3 and donor/patient 6 could be propagated only for a few weeks and did not expand well in culture.
Repertoire of HLA-DR–restricted epitopes recognized by polyclonal CD4+ T cells propagated with pool I and pool II
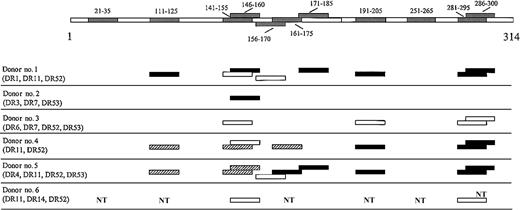
We determined the epitope repertoire of the CD4+ lines from all donors by testing, after a few cycles of stimulation, their proliferative response to each single peptide forming the pools. Figure1 summarizes the results obtained. Nine of 11 peptides tested elicited proliferation of CD4+ T cells in at least 2 donors. MAGE-3111-125, MAGE-3141-155, and the overlapping MAGE-3146-160, MAGE- 3191-205, MAGE-3281-295, and the overlapping MAGE-3286-300, were strongly recognized by most of the donors, consistent with a high degree of promiscuity. MAGE-3156-170 was weakly but significantly recognized by donor 1 and donor/patient 5, and the overlapping peptide MAGE-3161-175 was significantly and strongly recognized by donor 4 and donor/patient 5. MAGE-3171-185 was strongly recognized by donor 1 and donor/patient 5. Recognition of the MAGE-3 sequences was HLA-DR restricted as demonstrated in vitro by inhibition of the proliferation of CD4+ T cells to the peptides in the presence of an anti-DR antibody in the cultures (data not shown). MAGE-321-35 and MAGE-3251-265 were never recognized.
MAGE-3 sequence segments recognized by CD4+T cells from 6 donors and their HLA-DR type.
Polyclonal CD4+ T-cell lines propagated in culture with pool I and pool II were tested with each single peptides (10 μg/mL) forming the pools in 2-day microproliferation assays. The results are representative of several assays in the case of donors 1, 2, 4, and donor/patient 5 and, at least 2 experiments, in the case of donor 3 and donor/patient 6. Peptides that elicited a strong response are indicated in black (P < .001), peptides that elicited an intermediate response are indicated in hatched segments (.001 < P < .05), and peptides that elicited a low but still significant response are indicated in white (P < .05). Responses significantly higher than the blanks (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) were determined by unpaired, one-tailed Student t test. NT indicates not tested.
MAGE-3 sequence segments recognized by CD4+T cells from 6 donors and their HLA-DR type.
Polyclonal CD4+ T-cell lines propagated in culture with pool I and pool II were tested with each single peptides (10 μg/mL) forming the pools in 2-day microproliferation assays. The results are representative of several assays in the case of donors 1, 2, 4, and donor/patient 5 and, at least 2 experiments, in the case of donor 3 and donor/patient 6. Peptides that elicited a strong response are indicated in black (P < .001), peptides that elicited an intermediate response are indicated in hatched segments (.001 < P < .05), and peptides that elicited a low but still significant response are indicated in white (P < .05). Responses significantly higher than the blanks (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) were determined by unpaired, one-tailed Student t test. NT indicates not tested.
HLA-DR restriction molecules for the immunodominant sequences
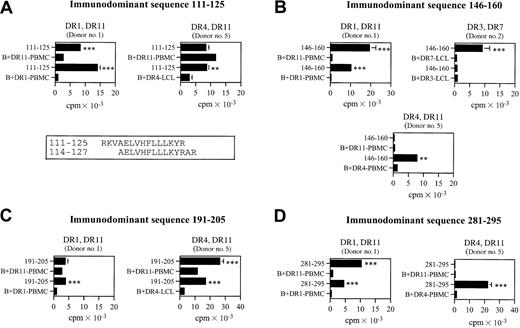
To identify the HLA-DR–restricting allele for the immunodominant sequences, CD4+ T cells from donors 1 and 2 and donor/patient 5 were challenged in microproliferation assays with APCs, homozygous for each of the 2 HLA-DRB1 alleles expressed by the donor, pulsed with individual peptides (Figure2).
HLA-DR restriction of CD4+ T cells recognizing the immunodominant epitopes.
CD4+ T cells were propagated with pool II (A,C) or pool I (B,D). The donors and their HLA-DR types are indicated. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of homozygous PBMCs or LCLs as APCs) is expressed as B + PBMC or B + LCL. Alignment of overlapping sequences 111-125 and 114-127 (presented by HLA-DR1327) is reported in the box. Responses significantly higher than the blanks are indicated: **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student ttest).
HLA-DR restriction of CD4+ T cells recognizing the immunodominant epitopes.
CD4+ T cells were propagated with pool II (A,C) or pool I (B,D). The donors and their HLA-DR types are indicated. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of homozygous PBMCs or LCLs as APCs) is expressed as B + PBMC or B + LCL. Alignment of overlapping sequences 111-125 and 114-127 (presented by HLA-DR1327) is reported in the box. Responses significantly higher than the blanks are indicated: **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student ttest).
We did not perform HLA-DR restriction on donor 4 because donor 4 is HLA-DR11 homozygous, nor on donor 3 and donor/patient 6, because CD4+ T cells did not expand in culture enough to allow further characterization. MAGE-3111-125 was recognized in association with HLA-DR1, HLA-DR4, and HLA-DR11 (Figure2A). Moreover, MAGE-3114-127, which largely overlaps MAGE-3111-125, was shown by van der Bruggen and colleagues27 to be recognized in association with HLA-DR13. MAGE-3146-160 was recognized in association with HLA-DR1, HLA-DR4, HLA-DR11, and HLA-DR7 (Figure 2B). MAGE-3191-205 was recognized in association with HLA-DR1, HLA-DR4, and HLA-DR11 alleles (Figure 2C). MAGE-3281-295was recognized in association with HLA-DR1, HLA-DR4, and HLA-DR11 (Figure 2D). The same immunodominant sequence could be recognized in different donors with either one or both of the HLA-DRB1 alleles expressed. Moreover, different donors sharing the same alleles might preferentially recognize the same epitope with different alleles, probably due to a difference in precursors' frequency.
Immunodominant sequences contain naturally processed epitopes
MAGE-3114-127, largely overlapping MAGE-3111-125, MAGE- 3146-160, and MAGE-3281-295 were already described to contain naturally processed epitopes recognized in association with HLA-DR13,27 HLA-DR4 and HLA-DR7,15 and HLA-DR11,8 respectively. MAGE-3114-127contains a natural epitope that is formed after processing through the exogenous pathway28; we could not test the natural processing pathways for MAGE-3111-125, which most likely contain the same natural epitope, because we failed to obtain CD4+ T cells recognizing exclusively that sequence. MAGE-3146-160 was formed after both endogenous and exogenous pathways.15 In the present work, we found that MAGE-3146-160 is recognized by CD4+ cytotoxic T cells (CTLs) from donor 2 (HLA-DR3, HLA-DR7) when expressed on HLA-DR7–matched melanoma cells (GF TC) and DR7-LCL engineered to express MAGE-3 (data not shown). MAGE-3281-295 was shown by us8 to contain an epitope formed after both endogenous and exogenous pathways and recognized by CD4+ CTLs.
To verify whether MAGE-3191-205 contains a natural epitope, we used polyclonal CD4+ T cells from donor 4 after 9 weeks of propagation with pool II, when the cells strongly and exclusively recognized MAGE-3191-205 (Figure3A lower panel). Recognition was HLA-DR restricted as shown in Figure 3B. DR11-LCLs were engineered to express MAGE-3 (DR11-LCL-M3) and used in cytotoxicity assays. As shown in Figure 3C, MAGE-3191-205–specific CD4+ T cells killed DR11-LCL-M3, but not wild-type DR11-LCLs. Moreover, CD4+ CTLs killed the MD TC and OI TC melanoma cells, which express MAGE-3 and HLA-DR11, but not SK-Mel 24, expressing unrelated HLA-DR (Figure 3D). In addition, we performed cold-target inhibition experiments, in which cold targets (DR11-LCLs) preincubated with peptide MAGE-3191-205 were added to the hot tumor targets. Killing of MD TC and OI TC was inhibited by addition of peptide-pulsed DR11-LCLs (up to 75% and 98%, respectively), suggesting that this peptide contains an epitope that is presented by the melanoma cells (Figure 3E-F). The percentage of inhibition of DR11-LCL (Figure 3E) or DR11-LCLs pulsed with an irrelevant peptide (Figure 3F) was comparable in the 2 cases, and up to 25% to 30%. The high cold/hot ratio (50%) required to observe a specific inhibition of MD TC killing, compared to OI TC, might be related to different binding affinity of the peptide for the 2 subtypes of DR11-LCLs (DR*1104 and DR*1101) used. In the case of MD TC, competition for MHC binding to the cold or hot targets of free peptide released from the LCLs in culture might reduce the ability of these LCLs to inhibit the killing.
Characterization of the CD4+ T-cell response to MAGE-3191-205.
CD4+ T cells were propagated from donor 4 (HLA-DR11) by weekly restimulation with pool II. (A) Proliferative responses, measured in 2-day microproliferation assays, to each single peptide forming pool II (10 μg/mL) after 4 and 9 weeks of propagation of the line. The data shown are means of triplicate determination ± SDs. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) is expressed as B + DR11-PBMC. Responses significantly higher than the blanks are indicated: **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student t test). (B) Responses to pool II (5 μg/mL), in the absence or in the presence of L243 mAb or an irrelevant control mAb (0.5 μg/mL). The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) is expressed as B + APC. (C-F) Recognition of naturally processed MAGE-3 sequences by CD4+ T cells at 9 or more weeks of propagation. (C) Cytolytic activity, measured in a51Cr release assay, against wild-type DR*1104-LCL and DR*1104-LCL engineered to express MAGE-3 (DR*1104-LCL-M3). (D) Cytolytic activity against HLA-DR–matched (MD TC and OI TC) and HLA-DR–unrelated (SK-Mel 24) melanoma cells. (E-F) Cold target inhibition experiments. (E) Cold targets (MD TC, DR*1104-LCL, and DR*1104-LCL pulsed with MAGE-3191-205) were used to inhibit the lytic activity of CD4+ cytotoxic T cells against hot MD TC (effector-target [E/T] ratio 40:1). The percentage of specific lysis against MD TC in the absence of cold target was 22 ± 1.3. (F) Cold targets (DR*1101-LCL and DR*1101-LCL pulsed with MAGE-3191-205 or MAGE-321-35) were used to inhibit the lytic activity of CD4+ cytotoxic T cells against hot OI TC (E/T ratio 40:1). The percents lytic activity against OI TC and DR*1101-LCL, as negative control, are shown in black symbols. The data presented in panels C through F are representative of at least 3 experiments and are means of triplicate determinations ± SDs.
Characterization of the CD4+ T-cell response to MAGE-3191-205.
CD4+ T cells were propagated from donor 4 (HLA-DR11) by weekly restimulation with pool II. (A) Proliferative responses, measured in 2-day microproliferation assays, to each single peptide forming pool II (10 μg/mL) after 4 and 9 weeks of propagation of the line. The data shown are means of triplicate determination ± SDs. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) is expressed as B + DR11-PBMC. Responses significantly higher than the blanks are indicated: **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student t test). (B) Responses to pool II (5 μg/mL), in the absence or in the presence of L243 mAb or an irrelevant control mAb (0.5 μg/mL). The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) is expressed as B + APC. (C-F) Recognition of naturally processed MAGE-3 sequences by CD4+ T cells at 9 or more weeks of propagation. (C) Cytolytic activity, measured in a51Cr release assay, against wild-type DR*1104-LCL and DR*1104-LCL engineered to express MAGE-3 (DR*1104-LCL-M3). (D) Cytolytic activity against HLA-DR–matched (MD TC and OI TC) and HLA-DR–unrelated (SK-Mel 24) melanoma cells. (E-F) Cold target inhibition experiments. (E) Cold targets (MD TC, DR*1104-LCL, and DR*1104-LCL pulsed with MAGE-3191-205) were used to inhibit the lytic activity of CD4+ cytotoxic T cells against hot MD TC (effector-target [E/T] ratio 40:1). The percentage of specific lysis against MD TC in the absence of cold target was 22 ± 1.3. (F) Cold targets (DR*1101-LCL and DR*1101-LCL pulsed with MAGE-3191-205 or MAGE-321-35) were used to inhibit the lytic activity of CD4+ cytotoxic T cells against hot OI TC (E/T ratio 40:1). The percents lytic activity against OI TC and DR*1101-LCL, as negative control, are shown in black symbols. The data presented in panels C through F are representative of at least 3 experiments and are means of triplicate determinations ± SDs.
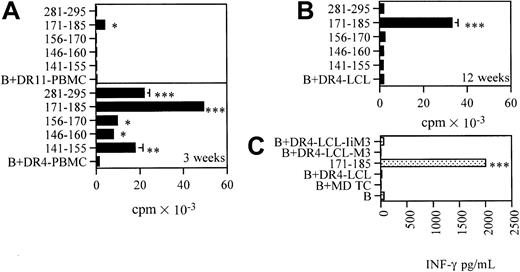
MAGE-3161-175 and MAGE-3171-185 contain epitopes not naturally processed in vitro
To verify whether MAGE-3161-175 contains a natural epitope, we used polyclonal CD4+ T cells from donor/patient 5, after selection of peptide MAGE-3161-175–specific CD4+ T cells by magnetic sorting (Figure 4B). CD4+ T cells did not secrete IFN-γ after challenge with melanoma cells or DR4-LCLs engineered to express MAGE-3 (DR4-LCL-M3), nor DR4-LCLs engineered to express Ii.MAGE-3 (DR4-LCL-IiM3), suggesting that MAGE-3161-175 does not contain a natural epitope produced after processing through both the endogenous and the exogenous pathways (Figure 4C).
Characterization of the CD4+ T-cell response to MAGE-3161-175.
CD4+ T cells from donor/patient 5 (HLA-DR4, HLA-DR11) were propagated by weekly restimulation with pool II. (A) Proliferative responses, measured in 2-day microproliferation assays, to each single peptide forming pool II (10 μg/mL) after 5 weeks of propagation of the line. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of homozygous PBMCs or LCLs as APCs) is expressed as B + DR11-PBMC or B + DR4-LCL. (B) Proliferative responses to each single peptide forming pool II (10 μg/mL) after magnetic selection of MAGE-3161-175–specific CD4+ T cells. (C) IFN-γ release of CD4+ T cells in the presence of melanoma cells, DR4-LCL, MAGE-3161-175–pulsed DR4-LCL, DR4-LCL-M3, and DR4-LCL-IiM3. The data shown in panels A to C are means of triplicate determinations ± SDs. Responses significantly higher than the blanks are indicated: **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student ttest).
Characterization of the CD4+ T-cell response to MAGE-3161-175.
CD4+ T cells from donor/patient 5 (HLA-DR4, HLA-DR11) were propagated by weekly restimulation with pool II. (A) Proliferative responses, measured in 2-day microproliferation assays, to each single peptide forming pool II (10 μg/mL) after 5 weeks of propagation of the line. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of homozygous PBMCs or LCLs as APCs) is expressed as B + DR11-PBMC or B + DR4-LCL. (B) Proliferative responses to each single peptide forming pool II (10 μg/mL) after magnetic selection of MAGE-3161-175–specific CD4+ T cells. (C) IFN-γ release of CD4+ T cells in the presence of melanoma cells, DR4-LCL, MAGE-3161-175–pulsed DR4-LCL, DR4-LCL-M3, and DR4-LCL-IiM3. The data shown in panels A to C are means of triplicate determinations ± SDs. Responses significantly higher than the blanks are indicated: **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student ttest).
To characterize the recognition of peptide MAGE-3171-185, we used polyclonal CD4+ T cells from donor/patient 5, after 12 weeks of propagation in culture, when they strongly and exclusively recognized MAGE-3171-185 (Figure5B). CD4+ T cells did not recognize melanoma cells or DR4-LCL-M3, nor DR4-LCL-IiM3, suggesting that this sequence does not contain a natural epitope as well (Figure5C). To verify that DR4-LCL-M3 and DR4-LCL-IiM3 express a sufficient amount of the MAGE-3 protein to be recognized by CD4+ T cells, we tested its expression by flow cytometry. MAGE-3 intracytoplasmic staining in transduced DR4-LCLs and DR11-LCLs recognized by CD4+ T cells specific for MAGE-3191-205 (Figure 3D) showed that they contain a comparable amount of MAGE-3 (data not shown).
Characterization of the CD4+ T-cell response to MAGE-3171-185.
CD4+ T cells from donor/patient 5 (HLA-DR4, HLA-DR11) were propagated by weekly restimulation with pool I. (A-B) Proliferative responses, measured in 2-day microproliferation assays, to each single peptide forming pool I (10 μg/mL) after 3 weeks (A) and after 12 weeks (B) of propagation of the line. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of homozygous PBMCs as APCs) is expressed as B + DR11-PBMC or B + DR4-PBMC or B + DR4-LCL. (C) IFN-γ release of CD4+ T cells in the presence of melanoma cells, DR4-LCL, MAGE-3171-185–pulsed DR4-LCL, DR4-LCL-M3, and DR4-LCL-IiM3. The data shown in panels A to C are means of triplicate determinations ± SDs. Responses significantly higher than the blanks are indicated: *P < .05, **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student t test).
Characterization of the CD4+ T-cell response to MAGE-3171-185.
CD4+ T cells from donor/patient 5 (HLA-DR4, HLA-DR11) were propagated by weekly restimulation with pool I. (A-B) Proliferative responses, measured in 2-day microproliferation assays, to each single peptide forming pool I (10 μg/mL) after 3 weeks (A) and after 12 weeks (B) of propagation of the line. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of homozygous PBMCs as APCs) is expressed as B + DR11-PBMC or B + DR4-PBMC or B + DR4-LCL. (C) IFN-γ release of CD4+ T cells in the presence of melanoma cells, DR4-LCL, MAGE-3171-185–pulsed DR4-LCL, DR4-LCL-M3, and DR4-LCL-IiM3. The data shown in panels A to C are means of triplicate determinations ± SDs. Responses significantly higher than the blanks are indicated: *P < .05, **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student t test).
Discussion
In this study, we describe a successful strategy to identify promiscuous CD4 epitopes on candidate tumor antigens. Using a combined approach of computational prediction for promiscuous MHC class II epitopes and in vitro validation by biologic assays, we identified 4 immunodominant regions of MAGE-3, which contain naturally processed epitopes presented by several different HLA-DR alleles.
For most of the predicted epitopes, we found an excellent correlation between prediction and the ability of the peptides to bind promiscuously to HLA-DR molecules and to stimulate CD4+ T cells from different donors. This was especially striking for overlapping sequences MAGE-3141-155 and MAGE-3146-160, and overlapping sequences MAGE-3281-295 and MAGE-3286-300. On the contrary, MAGE-3111-125 and MAGE-3191-205 that, as predicted, bound HLA-DR*1101 but not HLA-DR*0101 and HLA-DR*0401, could strongly stimulate proliferation of CD4+ T cells in association with all the 3 HLA-DR alleles, pointing to the importance of the in vitro studies for validation of the predicted sequences. MAGE-3251-265, which was predicted to bind 48% of the alleles, did not bind well to the HLA-DR molecules that we tested (Table 1) and was never recognized by CD4+ T cells. MAGE-3251-265 partially overlaps MAGE-3243-258, which is recognized in association with HLA-DP4 but not with HLA-DR.30 MAGE-321-35, which was predicted to bind poorly, never elicited CD4+ T-cell responses. In agreement, Kobayashi et al15 also reported that the overlapping peptide sequence MAGE-322-36 could stimulate T lymphocytes from different donors in association with HLA-DR4, but the CD4+ T cells did not expand in culture.15 All the other sequences activated CD4+ T cells from at least 2 donors and did so in association with 2 to 4 different alleles (Figure1; Table 2), confirming the ability of TEPITOPE to predict promiscuous epitopes.
Summary of CD4 immunodominant MAGE-3 epitopes recognized by CD4+ T cells and their HLA-DR presenting alleles
| HLA-DR . | Residues 111-125 . | Residues 146-160* . | Residues 191-205 . | Residues 281-295† . | HLA-DR frequency in whites, % . |
|---|---|---|---|---|---|
| DR1 | + | + | + | + | 18 |
| DR4 | + | + | + | + | 24-26 |
| DR11 | + | + | + | + | 24-27 |
| DR13 | 114-127‡ | NT | NT | NT | 19 |
| DR7 | NA | + | NA | NA | 26 |
| HLA-DR . | Residues 111-125 . | Residues 146-160* . | Residues 191-205 . | Residues 281-295† . | HLA-DR frequency in whites, % . |
|---|---|---|---|---|---|
| DR1 | + | + | + | + | 18 |
| DR4 | + | + | + | + | 24-26 |
| DR11 | + | + | + | + | 24-27 |
| DR13 | 114-127‡ | NT | NT | NT | 19 |
| DR7 | NA | + | NA | NA | 26 |
Most importantly, the immunodominant epitopes identified here are recognized in association with HLA-DR1, HLA-DR4, HLA-DR11, and HLA-DR7 that, with the addition of HLA-DR13,27 cover up to 94% of the alleles expressed in whites (Table 2). We cannot exclude that HLA-DRB3 or HLA-DRB4 molecules in addition to HLA-DRB1 can also present the identified sequences.
The immunodominant regions contain epitopes that are naturally processed from different sources of tumor antigen and by different APCs. In our present and previous studies,8 sequences MAGE-3141-160, MAGE-3191-205, and MAGE-3281-295 were recognized by cytolytic CD4+T cells after endogenous processing of MAGE-3 by LCLs and melanoma cells. Moreover, sequence MAGE-3281-295 was recognized by CD4+ T cells after processing and cross-presentation of the recombinant protein by autologous APCs.8MAGE-3141-160–reactive T-helper cells were also shown to recognize various forms of the MAGE-3 protein (tumor cell lysate, dead/apoptotic tumor cells, or recombinant protein).15MAGE- 3111-125 largely overlaps MAGE-3114-127, which is produced after processing and presentation of recombinant MAGE-3 protein by autologous dendritic cells and by melanoma cells that express an invariant chain–MAGE-3 fusion protein in the endosomal/lysosmal compartment.27 On the contrary, MAGE-3161-175–specific and MAGE-3171-185–specific CD4+ T cells did not recognize in vitro the MAGE-3 protein after processing through either the endogenous or the exogenous pathways. A possible explanation for this finding is that we failed to detect recognition of the native epitope because peptides bind to HLA molecules in a different conformation when they are loaded exogenously or when they are generated endogenously, as shown by Unanue and colleagues.31,32 In contrast with “naturally processed” epitopes that derive from processing of the whole protein antigen, “cryptic” epitopes are presented after the exposure to the antigen in peptide form, but not after processing of the whole protein antigen.33 Protease activity as well as the nature of the APCs during the processing of the protein may have a major influence on the repertoire of naturally processed or cryptic epitopes displayed for T-cell recognition.33 It is interesting that, in addition to the immunodominant epitopes, these sequences were strongly and predominantly recognized after long-term culture by CD4+ T cells from donor/patient 5, who had metastatic melanoma. We cannot completely exclude that priming of CD4+ T cells recognizing these sequences occurred in vivo as a consequence of epitope spreading during inflammatory conditions and operated by APCs not tested in our in vitro studies, leading to the formation of “cryptic” epitopes.
The use of pools of peptides for CD4+ T-cell stimulation has the advantage that competition among peptides with different binding affinity for the presenting alleles mimics in culture what occurs in vivo, where different processed peptides compete for MHC class II binding and T-cell–receptor recognition. Therefore, the finding that some peptides are predominant in our cultures may be related to their higher affinity for the MHC class II molecules, or to the presence of peptide-specific CD4+ T cells expressing T cell receptor with high affinity for the peptide/MHC class II complexes. The finding that CD4+ T cells specific for MAGE-3191-205 and MAGE-3281-295 proliferate at peptide concentrations as low as 1 to 10 ng/mL (data not shown) supports the latter possibility. The predominant recognition of different immunodominant regions in different donors may also be related to their precursor repertoires.
Animal models have shown that tumor-specific CD4+ T cells, in addition to playing a role in the induction of the antitumor immune response, have direct effector functions.1-4 The results of the present and previous studies by us and others8,33-36 strongly support a potential role of human tumor-specific CD4+ T cells as effectors of the antitumor immune response. Although the significance of the CD4+ T cells compared with CD8+ T cells in vivo remains to be clarified, lytic function of CD4+ T cells may be relevant when neoplastic cells, constitutively or as a consequence of inflammatory milieu, express MHC class II molecules. It will be interesting to verify the expansion of CD4+ CTLs ex vivo from cancer patients, as it has been shown,37 where CD4+ perforin-positive T cells were expanded in donors with chronic viral infections.
Clinical trials of vaccination with synthetic peptides corresponding to the HLA-A1–restricted MAGE-3 epitope recognized by CD8+ T cells have started in patients bearing MAGE-3+tumors.9-13 The additional use of peptides corresponding to the immunodominant CD4 epitopes identified here would increase the magnitude and life span of the antitumor immune response in these patients. Moreover, the identification of these epitopes will help in the study of the number and function of MAGE-3–specific CD4+ T cells, and the reliable monitoring of the immune responses in cancer patients before and after vaccination.
We thank Cinzia Arcelloni for high-performance liquid chromatography analyses, and Paolo Dellabona, Matteo Bellone, and Angelo Manfredi for enjoyable discussions and critical reading of the manuscript.
Prepublished online as Blood First Edition Paper, September 19, 2002; DOI 10.1182/blood-2002-03-0933.
Supported by the AIRC (Italian Association for Cancer Research), European Community (grant QLK2-CT2000-00470; M.P.P.), the MURST (Ministry of University and Scientific Research), the Ministry of Health, the National Heart, Lung, and Blood Institute (grant HL6192; B.M.C.-F.) and the National Institute of Neurological Disorders and Stroke (grant NS23919; B.M.C.-F.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Maria Pia Protti, Laboratory of Tumor Immunology, Cancer Immunotherapy and Gene Therapy Program, DIBIT, Scientific Institute H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy; e-mail: m.protti@hsr.it.

![Fig. 3. Characterization of the CD4+ T-cell response to MAGE-3191-205. / CD4+ T cells were propagated from donor 4 (HLA-DR11) by weekly restimulation with pool II. (A) Proliferative responses, measured in 2-day microproliferation assays, to each single peptide forming pool II (10 μg/mL) after 4 and 9 weeks of propagation of the line. The data shown are means of triplicate determination ± SDs. The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) is expressed as B + DR11-PBMC. Responses significantly higher than the blanks are indicated: **.001 < P < .05, ***P < .001 (determined by unpaired, one-tailed Student t test). (B) Responses to pool II (5 μg/mL), in the absence or in the presence of L243 mAb or an irrelevant control mAb (0.5 μg/mL). The blank (ie, the basal level of proliferation of CD4+ T cells in the presence of autologous PBMCs as APCs) is expressed as B + APC. (C-F) Recognition of naturally processed MAGE-3 sequences by CD4+ T cells at 9 or more weeks of propagation. (C) Cytolytic activity, measured in a51Cr release assay, against wild-type DR*1104-LCL and DR*1104-LCL engineered to express MAGE-3 (DR*1104-LCL-M3). (D) Cytolytic activity against HLA-DR–matched (MD TC and OI TC) and HLA-DR–unrelated (SK-Mel 24) melanoma cells. (E-F) Cold target inhibition experiments. (E) Cold targets (MD TC, DR*1104-LCL, and DR*1104-LCL pulsed with MAGE-3191-205) were used to inhibit the lytic activity of CD4+ cytotoxic T cells against hot MD TC (effector-target [E/T] ratio 40:1). The percentage of specific lysis against MD TC in the absence of cold target was 22 ± 1.3. (F) Cold targets (DR*1101-LCL and DR*1101-LCL pulsed with MAGE-3191-205 or MAGE-321-35) were used to inhibit the lytic activity of CD4+ cytotoxic T cells against hot OI TC (E/T ratio 40:1). The percents lytic activity against OI TC and DR*1101-LCL, as negative control, are shown in black symbols. The data presented in panels C through F are representative of at least 3 experiments and are means of triplicate determinations ± SDs.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/3/10.1182_blood-2002-03-0933/4/m_h80333786003.jpeg?Expires=1767835348&Signature=IQUBNnTHJgHzhVd99Hmfqyq9qTuYn6L9dJX-CY-bbpKQCdV~h7fp699MRAAgtQOpIFcUb~KR2MO6HiH1xT8hb4-WL8PSKeWguvZzUYeRaCOSyFv4oOWmIgjpF9fUkGEwMNvYkBGG57HJFIpGyYg5kpZlle-SSQHiCOODTifspNL~4lFQVz~Mk4fzN6b-9s4WEr6zJPD9Vnnn6GSrJoD1eAv4gQ5354~ron1D5SAew2pcPYAyQjlgbaHnYrPnxVFMgCvNs~tg0vQ4aQYUQNUnrVo4GTFWHTRj4COhRZQWFAnDRT8OnSk2qdzEYQM7TDtT0ir7A1rxGFASy-ZMTnHmJg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal