Abstract
The initial B-cell repertoire is generated by combinatorial immunoglobulin V(D)J gene segment rearrangements that occur in a preferential sequence. Because cellular proliferation occurs during the course of these rearrangement events, it has been proposed that intraclonal diversification occurs during this phase of B-cell development. An opportunity to examine this hypothesis directly was provided by the identification of a human acute lymphoblastic leukemic cell line that undergoes spontaneous differentiation from pro-B cell to the pre-B and B-cell stages with concomitant changes in the gene expression profile that normally occur during B-cell differentiation. After confirming the clonality of the progressively differentiating cells, an analysis of immunoglobulin genes and transcripts indicated that pro-B cell members marked by the same DJ rearrangement generated daughter B cells with multiple VH and VL gene segment rearrangements. These findings validate the principle of intraclonal V(D)J diversification during B-cell generation and define a manipulable model of human B-cell differentiation.
Introduction
B-cell development normally begins within specialized microenvironments in the hematopoietic fetal liver and bone marrow. Therein precursor cells undergo proliferation during the course of a series of differentiation events featuring the ordered rearrangement of variable (V), diversity (D), and joining (J) gene segments to generate a diverse antibody repertoire.1 The D→JHgene rearrangements in progenitor B (pro-B) cells are followed by V→DJH rearrangements that allow precursor B (pre-B) cells to produce μ heavy chains (HCs).2,3 The μHCs associate with the surrogate light chains (SLCs), composed of VpreB and λ5/14.1 proteins, and Igα/β heterodimers to form the pre-B cell receptor (pre-BCR; Karasuyama et al4 and Burrows et al5 and references therein). Signaling initiated via this receptor complex is essential for pre-B cell survival, proliferation, and transient down-regulation of recombination-activating genes (Rag1/2) to terminate V(D)J rearrangements in the immunoglobulin (Ig) HC locus.6,7 The subsequent extinction of SLC gene expression is temporally related to an exit from the cell cycle, Rag1/2 up-regulation, and Ig light chain (LC) V-J gene rearrangement.8-11 Immature IgM+ B cells generated by productive VL-JL rearrangements may then undergo positive or negative selection by encounter with self-antigens.12 13
B-cell clonality is usually defined operationally by the shared expression of a unique BCR specificity, although intraclonal antigen receptor diversity may be generated by several mechanisms. Antibody diversity can be generated by receptor editing, a process by which the specificity of an autoreactive BCR is changed thorough a secondary VL-JL rearrangement at an immature B-cell stage.14,15 Additional intraclonal BCR diversification can be generated by point mutations in the variable region during antigen (Ag)–driven clonal expansion in germinal centers.1,16,17B-cell malignancies, including follicular B-cell lymphoma, chronic lymphocytic leukemia, and Hodgkin disease, may exhibit this type of clonal diversification as a manifestation of their germinal center (GC) origin.18
It has been proposed that intraclonal diversity may also be generated as a consequence of the cellular proliferation that accompanies the normal stepwise V(D)J rearrangement events in pro-B and pre-B cells.19 According to this hypothesis, a pro-B cell undergoing the initial DJH rearrangement can give rise to daughter cells that select different VH genes for subsequent V-DJH rearrangement. In turn, the pre-B cell progeny would have the opportunity to select different VLand JL gene segments for VJL pairing, thereby providing additional intraclonal BCR diversity. Because a clonal model of early B-lineage differentiation would allow testing of this hypothesis, we sought a stable cell line that recapitulates this developmental process. Here, we report the characterization of a human acute lymphoblastic leukemic (ALL) cell line, EU12, that undergoes continual B-lineage differentiation from the pro-B to the pre-B and B-cell stages. Analysis of this clonal model of human B-lineage differentiation validates the principle of intraclonal V(D)J recombinatorial diversification during B-cell genesis.
Materials and methods
Cells and antibodies
The EU12 cell line was established from bone marrow cells obtained from a child with relapsed B-cell precursor ALL.20-22 The EU12 cell line was found to be negative for Epstein-Barr virus–associated nuclear antigen (EBNA) and to represent the leukemic clone by immunophenotypic analysis. The Nalm16 pro-B, OB5 pre-B, and EU12 ALL cell lines were cultured in Iscove medium supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 10% heat-inactivated fetal calf serum (FCS), and 2 mMl-glutamine. (Life Technologies, Grand Island, NY). Immunofluorescence assays used the following monoclonal antibodies (mAbs): fluorescein isothiocyanate (FITC)–conjugated CD2, CD3, CD5, CD10, CD11a, and CD34 (Becton Dickinson, Mountain View, CA); phycoerythrin (PE)–conjugated CD19, CD20, CD21, CD23, and HLA-DR (Becton Dickinson); FITC-conjugated CD45RA and antihuman λLC; PE-conjugated CD32, CD33, CD69, and VpreB8 (Pharmingen, San Diego, CA); and the CY5-labeled VpreB8 mAb as described previously.23Polyclonal antibodies included FITC-labeled anti–terminal deoxynucleotidyl transferase (TdT; Supertechs, Bethesda, MD), FITC-labeled goat anti-μHC, biotin-conjugated goat anti-μHC, and PE-labeled goat anti-IgD. Allophycocyanin (APC)–conjugated streptavidin (Southern Biotechnology, Birmingham, AL) was used as a second-step reagent for the biotin-labeled antibodies.
Immunofluorescence analysis and cell sorting
Viable cells were incubated with conjugated mAbs for immunofluorescence analysis and for fluorescence-activated cell sorting (FACS). Subpopulations of the EU12 cells were purified by 2 rounds of sorting using FACSTAR (Becton Dickinson) and MoFlow (Cytomation, Fort Collin, CO) instruments. For intracellular staining, the purified subpopulations of cells were fixed in 0.05% paraformaldehyde solution at 4°C for 1 hour, permeabilized with 0.2% Tween-20 in phosphate-buffered saline at room temperature for 20 minutes, and blocked with mouse serum for 10 minutes before incubation with fluorochrome-conjugated antibodies.
RT-PCR assays
Twice-sorted subpopulations of EU12 cells were lysed in TRIzol reagent (Gibco, Grand Island, NY), and total cellular RNA prepared following procedures recommended by the manufacturer. The synthesis of first-strand cDNA was performed as previously described.23For each cDNA preparation, a control synthesis reaction was performed without reverse transcriptase (RT) to test for genomic DNA (gDNA) contamination. The protocols for polymerase chain reaction (PCR) included denaturing at 94°C for 3 minutes amplification by 30 cycles of 94°C for 1 minute, annealing for 30 seconds at 60°C for interleukin 7 receptor (IL-7R) andmb-1; 65°C for Cκ; and 72°C forBcl-2; and extension at 72°C for 5 minutes. The primers for PCR amplification were IL-7R,5′-GTCGCTCTGTTGGTCATCTTG-3′ and 5′-TTTTG TCTTCTCTGTGCTGTG-3′;mb-1, 5′-GCTCCCCTAGAGGCAGTTAAGG GC-3′ and 5′-AGGGTAACCTCACTGTTAGGCCAGGC-3′; Cκ,5′-TGGCTGCACCATCTGTC TTCA-3′ and 5′-TTGAAGCTCTTTGTGACGGGC-3′;Bcl-2, 5′-TGCACCTGACG CCCTTCAC-3′ and 5′-AGACAGCCAGGAGAAATCAAACAG-3′. The protocols and primers used for PCR amplification of Rag1, Rag2, TdT,B29, Cμ, and β-actin have been described.11
DNA blotting
gDNA samples from sorted EU12 subpopulations and placenta (germline configuration control) were subjected to restriction enzyme digestion, electrophoresis, transfer, and hybridization with32P-labeled DNA probes for analysis of Ig HC and LC gene configuration as described.24 DNA probes, including a 3.6 kilobase (kb) BglII JH probe, aHindIII Jκ probe, and anEcoRI Cλ probe, were used as described.24
Genomic PCR assay
gDNA samples (0.5 μg) from EU12 subpopulations and the control HepG2 cells were used as templates. The protocol for PCR amplification of IgH gene segments involved denaturing at 95°C for 5 minutes, amplifying by 36 cycles of 95°C for 1 minute, annealing at 65°C for 30 seconds and at 72°C for 30 seconds, and extending at 72°C for 5 minutes. The primers for amplification, DXP′1, 5′-ATTACTATGGTTCGGGGAGTT-3′; DXP′1ext, 5′-GGTGAGGTCTGTGTCACT-3′; JH5, 5′-GTCGAACCAGTTGTCACATTG TG-3′; JH6, 5′-ACCTGAGGAGACGGTGACC-3′, and PanVHFR1, and PanVHFR3 were as described.25 The PCR products were separated by electrophoresis, transferred to a Nytran membrane (Schleicher and Schuell, Keene, NH). After standard prehybridization, the blots were hybridized at 42°C with oligonucleotide DXP′1 or JH5 probes labeled with digoxigenin (DIG) using the DIG oligonucleotide 3′-end labeling kit (Boehringer Mannheim, Germany) according to the manufacturer's instructions. After washing at 42°C, hybridization signals were revealed using the DIG luminescence detection kit for nucleic acids (Boehringer Mannheim) according to the manufacturer's instructions. To detect the VλJλ rearrangements in each of the EU12 subpopulations, the protocol of PCR amplification using a set of Vλ and Jλ primers was used as described.26
Ig gene sequencing and single-cell PCR assay
To determine μHC and λLC transcript sequences, cDNA of the test cells was synthesized for PCR amplification as previously described.23 PanVHFR1 and Cμ primers were used for amplification of the μHC cDNAs, and the set Vλand Jλ primers and PCR protocol for λLC cDNA amplification were performed as above. The PCR products of μHC or λLC transcripts were isolated and subjected to the TA cloning reaction as described by the manufacturer (Invitrogen, Carlsbad, CA). Plasmids carrying μHC or λLC transcripts were purified from randomly selected Escherichia coli colonies for sequencing reactions. For analysis of single-cell VH usage, individual μHC+ EU12 cells were deposited into tubes containing 20 μL PCR buffer with 0.25 mg/mL proteinase K. Reactions were incubated at 50°C for 1 hour and used as templates in a single-cell PCR assay. In the first-round PCR reaction, a combination of VH1,2,3-specific oligonucleotides was used as forward primer, and an oligonucleotide sequence common to JH1,2,4,5 was used as the reverse primer. In the second round of a nested PCR, 5 μL of the first PCR reaction product was used as the template, individual VH oligonucleotides were used as the forward primer, and the JH1,4,5 oligonucleotide was used as the nested reverse primer. The primer sequences have been described27 and the protocol included first-round VH gene amplification at 95°C for 5 minutes, 59°C for 4 minutes, and 72°C for 3 minutes (cycle 1); 95°C for 30 seconds, 59°C for 30 seconds, and 72°C for 80 seconds (cycles 2-36); then extension at 72°C for 5 minutes; and maintenance at 4°C. For second-round PCR, the protocol for amplification of the first round PCR product included 95°C for 3 minutes, 61°C for 1 minute, 72°C for 1 minute (cycle 1), 95°C for 1 minute, 61°C for 30 seconds, 72°C for 50 seconds (cycles 2-40), and extension at 72°C for 5 minutes, and then maintenance at 4°C. PCR products were separated by electrophoresis on a 1.5% agarose gel and purified for the sequencing reaction by using a gel extraction kit (Qiagen, Valencia, CA). Purified PCR products were sequenced directly using VH-specific primers. All sequences were analyzed with the DNAplot program (http://www.dnaplot.de/).
Results
Phenotypic and karyotypic characterization of the EU12 leukemic cell line
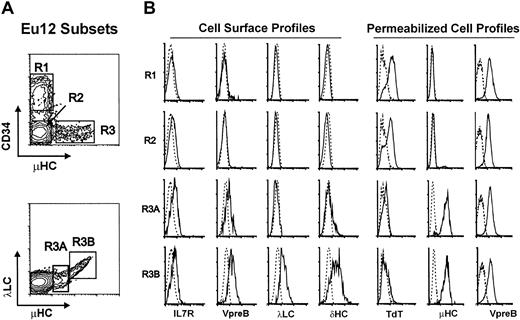
EU12 was unique among a panel of 11 ALL cell lines in that phenotypic analysis identified cells at multiple stages of B-lineage differentiation. All of the EU12 cells expressed the CD19, CD20, CD22, CD23, CD32, CD33, CD38, CD40, and HLA-DR antigens, but subpopulations of this cell line were found to be either positive or negative for CD34, CD10, and μHC (Figure 1). Differential immunofluorescence analysis indicated that the EU12 cell line is composed of 3 distinctive subpopulations: CD34+sμHC− (R1), CD34−sμHC− (R2), and CD34−sμHC+ (R3). The latter subpopulation could be subdivided into sμHClowSLC+ (R3A) and sμHChighλLC+ (R3B) subsets (Figure 1). Cells in both the R3A and R3B subsets expressed cell surface SLCs, and the cells in the R3B subset also expressed λLCs and δHCs. The μHClow R3A cells thus bear pre-BCR, whereas the μHChigh R3B cells have pre-BCR and BCR. IL-7R, barely detectable on the R1 and R2 subpopulations, was present at higher levels on the R3A and R3B subsets.
Phenotypic characterization of EU12 subpopulations.
(A) Cell surface immunofluorescence analysis of EU12 cells reveals 4 subpopulations (R1, R2, R3A, and R3B) on the basis of CD34, μHC, and λLC expression. (B) FACS-sorted EU12 cells were counterstained with anti–IL-7R, anti-VpreB, anti-λLC, or anti-δHC mAbs for cell surface profile analysis or fixed and permeabilized before analysis with anti-TdT, μHC, and VpreB Abs.
Phenotypic characterization of EU12 subpopulations.
(A) Cell surface immunofluorescence analysis of EU12 cells reveals 4 subpopulations (R1, R2, R3A, and R3B) on the basis of CD34, μHC, and λLC expression. (B) FACS-sorted EU12 cells were counterstained with anti–IL-7R, anti-VpreB, anti-λLC, or anti-δHC mAbs for cell surface profile analysis or fixed and permeabilized before analysis with anti-TdT, μHC, and VpreB Abs.
To characterize further the developmental stages of the EU12 cell line, each subpopulation was purified for intracellular immunofluorescence analysis (Figure 1). VpreB was found in permeabilized cells of all 3 subpopulations, whereas μHC was observed only in R3 cells and rarely in R2 cells (< 1%). In contrast, TdT expression was evident in most R1 and R2 cells, but not in the R3 subpopulation. The composite data from this analysis indicate that the EU12 cell line contains cells of pro-B (R1 and R2), pre-B (R3A), and B-cell (R3B) phenotypes, although the B-cell subpopulation is unusual in its IL-7R expression and simultaneous expression of surrogate and conventional λLCs.
The distinctive phenotype of EU12 cell line prompted a detailed cytogenetic analysis, and the results indicated the EU12 cell line is nearly tetraploid and has complex abnormalities involving chromosomes 2, 3, 4, 5, 6, 7, 11, 12, 17, and 21. Detailed description of its chromosome complement is as follows: 88, XXYY, +2, del(2)(q13) × 2, del(3)(q26.2),−4, der(4)t(2;4) (q13;q21), inv(5)(q15q33) × 2, del(6)(q23) × 2, t(7;21) (p13;q11) × 2,−8,−9,−11, del(11)(p13),−12, add(12)(p13), +13,−14,−14, +16, del(17)(p11) × 2, add(17)(p13), −18, +20,−21, der(21)t(7;21)(p13;q11), +mar.
EU12 cells spontaneously undergo pro-B to B-cell differentiation
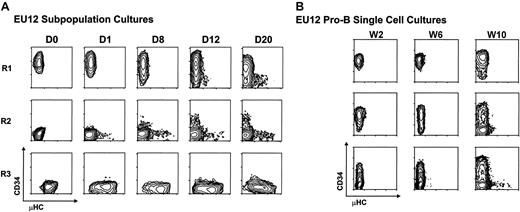
The representation of multiple stages of B-lineage differentiation among the EU12 cells suggested this cell line either is undergoing differentiation or represents a multiclonal mixture of cells. To discern between these possibilities, each subpopulation was purified by 2 rounds of FACS before reculture and phenotypic monitoring. Serial immunofluorescence analysis indicated that isolated cells of pro-B cell phenotype (CD34+μHC−) gave rise to cells of pre-B phenotype (CD34+μHClow) by day 12 and cells of B-cell phenotype (CD34−μHChigh) by 3 weeks in culture. Purified cells of the R2 subset (CD34−μHC−) likewise gave rise to cells of pre-B and B-cell phenotypes, whereas the R3 subpopulation retained cells of pre-B and B phenotypes (Figure2A). These results indicated that EU12 cells can undergo continual differentiation from pro-B cells to B cells. To verify their developmental potential at a clonal level, single R1 pro-B cells were placed in separate culture wells. Serial examination of their cellular progeny indicated that individual EU12 cells of pro-B cell phenotype are capable of giving rise to mature B-cell progeny (Figure 2B), although a longer time interval was required for this progression when the cultures are initiated with single cells, and B-cell differentiation was not seen in every subclone over the 10-week observation period. These results document the spontaneous differentiation capabilities of the EU12 cell clone and suggest variability in the progression of individual cells along the B-cell differentiation pathway.
Analysis of the differentiation potential of EU12 cells.
(A) Subpopulations of EU12 cells were purified by 2 rounds of FACS on the basis of their cell surface expression of CD34 and μHC. The purified EU12 subpopulations were cultured and their cell surface profiles reanalyzed on day 0 (D0), day 1 (D1), day 8 (D8), day 12 (D12), and day 20 (D20). (B) Single EU12 pro-B cells (CD34+μHC−) were cultured in individual wells and their progeny were characterized by immunofluorescence analysis at week 2 (W2), week 6 (W6), and week 10 (W10).
Analysis of the differentiation potential of EU12 cells.
(A) Subpopulations of EU12 cells were purified by 2 rounds of FACS on the basis of their cell surface expression of CD34 and μHC. The purified EU12 subpopulations were cultured and their cell surface profiles reanalyzed on day 0 (D0), day 1 (D1), day 8 (D8), day 12 (D12), and day 20 (D20). (B) Single EU12 pro-B cells (CD34+μHC−) were cultured in individual wells and their progeny were characterized by immunofluorescence analysis at week 2 (W2), week 6 (W6), and week 10 (W10).
Differential gene expression profiles in the EU12 subpopulations
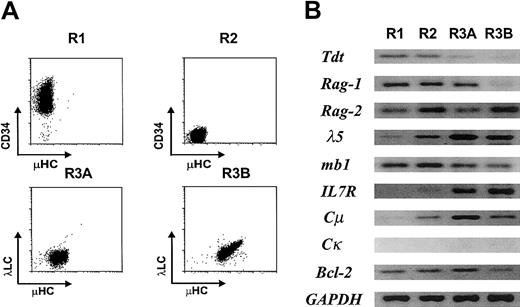
Cells representative of each definable stage in EU12 differentiation were purified by 2 rounds of FACS before RT-PCR analysis of their gene expression profile (Figure3). As anticipated from its protein expression profile (Figure 1), the TdT lymphoid cell–specific gene was down-regulated in the R3A subset and extinguished in the R3B subset. Rag1 was found to follow a similar expression pattern, whereas Rag2transcripts were expressed in all of the subpopulations, as were transcripts for the mb1, B29, λ5, and VpreB B-lineage genes. In keeping with results of the immunofluorescence analysis (Figure 1), IL-7R andCμ transcripts were not detected in the R1 pro-B cells, but were expressed in subpopulations representing later stages in differentiation. Interestingly, Bcl-2 transcription was noticeably down-regulated in the R3B B-cell subpopulation, possibly suggesting that the most mature EU12 cells are more susceptible to programmed cell death. These data support the conclusion that the EU12 cells undergo many of the changes in gene expression that characterize normal B-cell differentiation.
Expression of B lineage–specific genes in the EU12 subpopulations.
(A) Each subpopulation of EU12 cells was purified by 2 rounds of FACS. (B) RNA obtained from each subpopulation was used in RT-PCR amplification of B lineage–specific genes as described in “Material and methods.” PCR products were visualized by ethidium bromide staining of agarose gels.
Expression of B lineage–specific genes in the EU12 subpopulations.
(A) Each subpopulation of EU12 cells was purified by 2 rounds of FACS. (B) RNA obtained from each subpopulation was used in RT-PCR amplification of B lineage–specific genes as described in “Material and methods.” PCR products were visualized by ethidium bromide staining of agarose gels.
Analysis of Ig HC and LC gene configuration in the EU12 cells
Southern blot analysis was initially used to characterize the Ig gene rearrangement status at the different EU12 differentiation stages. Restriction enzyme–digested gDNA samples of each EU12 subpopulation, and of placenta as a germline control, were hybridized with JH, Cκ, and Cλ probes (Figure 4A-B; data not shown). Germline JH-containing fragments were not detectable in the unfractionated EU12 cells; instead, 2 rearranged bands were observed, a dominant 6.2-kb band and a minor 12-kb band (Figure 4A, lane 2). Of these, the R1 and R2 subpopulations contained only the 6.2-kb band (Figure 4A, lanes 3 and 4), whereas the μHC+ R3 subpopulation contained both bands of similar intensity (Figure 4A, lane 5). These findings, together with a karyotypic analysis demonstrating 2 copies of chromosome 14 (data not shown), suggest that the least differentiated R1 cells of EU12 contain 2 very similar JH rearrangements, but are incapable of producing μHCs because the rearrangements either are incomplete DJH or nonproductive VDJH. The occurrence of an additional rearrangement event on one HC allele in the R3 cells coincides with the expression of μHC.
Southern blot analysis of Ig gene configuration in the EU12 subpopulations.
gDNA from placenta and sorted EU12 subpopulations was digested withBglII before hybridization with a JH probe for IgH chain gene analysis (A), or with EcoRI before hybridization with a Cλ probe for Igλ L chain gene analysis (B). Arrows indicate HC and LC gene rearrangements in the R3 subpopulation.
Southern blot analysis of Ig gene configuration in the EU12 subpopulations.
gDNA from placenta and sorted EU12 subpopulations was digested withBglII before hybridization with a JH probe for IgH chain gene analysis (A), or with EcoRI before hybridization with a Cλ probe for Igλ L chain gene analysis (B). Arrows indicate HC and LC gene rearrangements in the R3 subpopulation.
No Jκ hybridizing bands were detected in a DNA blot analysis of the κLC locus (data not shown) in accordance with the absence of κLC transcripts in EU12 cells (Figure 3). These findings suggest that the EU12 clone has deleted the κLC locus, although chromosome 2 appeared intact at the level of resolution provided by karyotypic analysis. When EcoRI-digested DNA samples from each subpopulation were hybridized with the Cλ probe, a rearranged JλCλsegment was evident among the 7 JλCλ gene pairs in the R3 subpopulation (Figure 4B). The finding demonstrates Vλ→JλCλ gene rearrangement in the μHC+ R3 subpopulation, but not in the earlier fractions, and is consistent with the proposed developmental sequence R1→R2→R3. The cDNA sequence analysis (Figure 5) indicates that multiple rearrangements using the same JλCλ gene segments have occurred during the in vitro differentiation of the EU12 cells.
Sequence analysis of Ig HC and LC genes in μHC+ EU12 cells.
The cDNA of sorted μHC+ EU12 cells was used as template for PCR amplification at VH-Cμ (A) and VL-Cλ (C) transcripts using primer combinations described in “Materials and methods.” (B) Single μHC+ EU12 cells were sorted into PCR tubes with lysis buffer, and used for 2-round genomic PCR as described in “Materials and methods.” PCR products were purified and directly sequenced with specific VH primers.
Sequence analysis of Ig HC and LC genes in μHC+ EU12 cells.
The cDNA of sorted μHC+ EU12 cells was used as template for PCR amplification at VH-Cμ (A) and VL-Cλ (C) transcripts using primer combinations described in “Materials and methods.” (B) Single μHC+ EU12 cells were sorted into PCR tubes with lysis buffer, and used for 2-round genomic PCR as described in “Materials and methods.” PCR products were purified and directly sequenced with specific VH primers.
VH and VL gene usage in EU12 cells
To examine the extent of V(D)J recombinatorial diversification in the EU12 cells, the μHC+ R3 subpopulation of cells was purified and RNA extracted for cDNA synthesis and PCR amplification of μHC transcripts using PanVHFR3 and Cμ primers. The resultant μHC PCR products were cloned and the analysis of sequences obtained from 17 randomly selected clones identified the same DXP′1/JH4 gene segment rearrangement with no nucleotide addition in this D-JH joint region. This analysis also demonstrated the use of multiple VH gene segments by EU12 cells in that the VH3-7 gene segment was found in 12 of the 17 cDNA clones, VH1-8 in 3 cDNA clones, and VH3-11 in the other 2 cDNA clones (Figure5A). In addition, different nucleotide additions in μHC transcripts using the same VH gene provided further evidence of intraclonal V(D)J diversification.
To avoid possible transcriptional bias in this analysis of VH gene usage, we performed a single-cell genomic PCR assay of sorted μHC+ EU12 cells. The sequences derived from a panel of single-cell VH gene PCR products recaptured the 2 major VH3-7 gene rearrangements observed in the earlier analysis of μHC cDNA clones (Figure 5B). In addition, we identified a VH2-5 rearrangement with the same DXP′1/JH4 sequence observed in the cDNA clones; however, this rearrangement was nonfunctional because of a reading frame shift. The resulting transcript would be targeted for destruction by nonsense-mediated mRNA decay,28 thereby explaining why VH2-5 gene segment usage was not observed in the cDNA analysis. Consistent with the cDNA sequence analysis, 2 patterns of nucleotide additions were observed in the joint region between VH3-7 and DXP′1/JH4. The dominant joint region sequence featured a CCCC nucleotide addition, presumably added during the VH→DJH recombination process. The other additions were longer and contained a consensus CACA sequence that was also found at the 3′ end of VH2-5 gene segment.
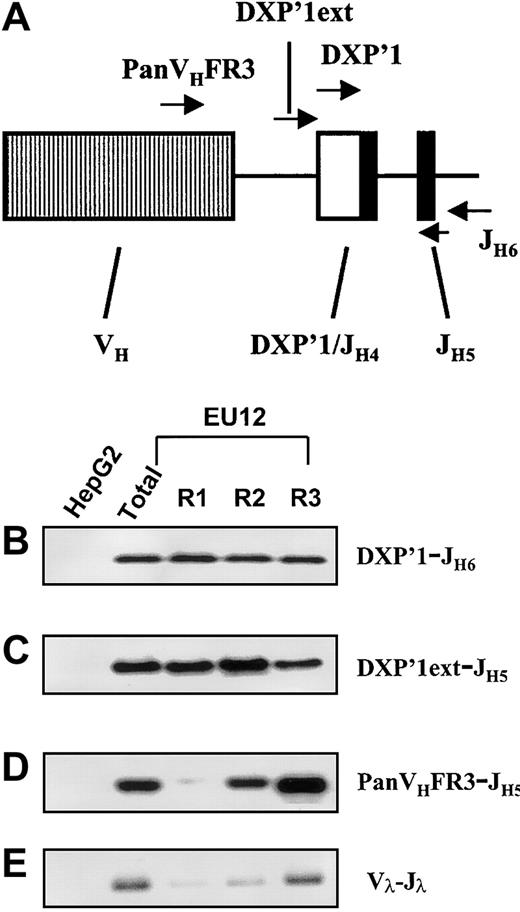
With this information in hand, genomic PCR assays were performed to determine more precisely when the V(D)J rearrangements occurred during the EU12 cell differentiation process. Primers able to discriminate between DJH and VHDJHrearrangements were used in this analysis (Figure6A). In a control experiment, the internal DXP′1 primer was used in conjunction with a downstream JH6 primer, and genomic PCR products of the expected size were detected in each EU12 subpopulation after hybridization with a JH5-specific probe (Figure 6B). This result was as expected because the primer combination amplifies both VDJH and DJH rearrangements. To assay specifically for DJH rearrangements, a primer based on the sequence upstream of the germline DXP′1 gene segment (DXP′1 ext) was used in conjunction with internal JH5 primers and the PCR products were detected by a specific internal DXP′1 probe. The EU12 cells in each purified subpopulation were found to contain a DJHsegment rearrangement, albeit at apparently reduced levels in the R3 pre-B/B cell subpopulation (Figure 6C). Using PanVHFR3 and JH5 primers in an assay that allows definition of the presence or absence of VDJH rearrangements but does not identify unique rearrangements, VDJH gene rearrangement was detected in the R2 subpopulation, and to a much greater extent in the R3 subpopulation (Figure 6D). The nonfunctional VH2-5 DXP′1/JH4 rearrangement was not detected in this assay because the PanVHFR3 primer does not recognize the VH2-5 gene segment. However, using a VH2-specific primer, a genomic PCR product was detectable in the R1, R2, and R3 subpopulations (not shown).
Genomic PCR assay of Ig HC and LC genes in EU12 subpopulations.
The diagram (A) indicates the location of each oligonucleotide used in the PCR reactions. gDNA from unfractionated EU12 cells, each subpopulation of EU12 cells, and control HepG2 cells was used as the template for PCR amplification by primers DXP′1 (internal), and JH6 (B), DXP′1 (external), and JH5 (C), and PanVHFR3, and JH5 (D) for Ig HC gene and a set of primers for Igλ LC genes (E). PCR products were subjected to electrophoresis and blotted with probes JH5 (B), and DXP′1 (internal) for panels C and D or revealed with ethidium bromide staining of agarose gels (E).
Genomic PCR assay of Ig HC and LC genes in EU12 subpopulations.
The diagram (A) indicates the location of each oligonucleotide used in the PCR reactions. gDNA from unfractionated EU12 cells, each subpopulation of EU12 cells, and control HepG2 cells was used as the template for PCR amplification by primers DXP′1 (internal), and JH6 (B), DXP′1 (external), and JH5 (C), and PanVHFR3, and JH5 (D) for Ig HC gene and a set of primers for Igλ LC genes (E). PCR products were subjected to electrophoresis and blotted with probes JH5 (B), and DXP′1 (internal) for panels C and D or revealed with ethidium bromide staining of agarose gels (E).
In an analysis of the Vλ usage, cDNA derived from the μHC+ R3 cells served as the template for PCR amplification of Vλ-Jλ transcripts using a panel of Vλ and Jλ primers.26Analysis of the sequences obtained from 8 randomly selected cDNA clones indicated the use of at least 2 Vλ gene segments each of which was recombined with the same Jλ2Cλ2 gene segment (Figure 5C), data consistent with the observation of a single rearranged band on the Southern blots (Figure 4B). Vλ3-19 gene segment usage appeared dominant, being seen in 6 of 8 clones, whereas Vλ3-10 was used in the other 2 clones. Variations in the N nucleotide additions were also observed in λLC transcripts with both Vλ gene segments, indicating the occurrence of multiple Vλ→Jλ2Cλ2 rearrangements in the R3 subpopulation. In a genomic PCR assay to detect Vλ→Jλ rearrangements, VJλrearrangements were detectable in R2 cells and to a much greater extent in the R3 cells (Figure 6E).
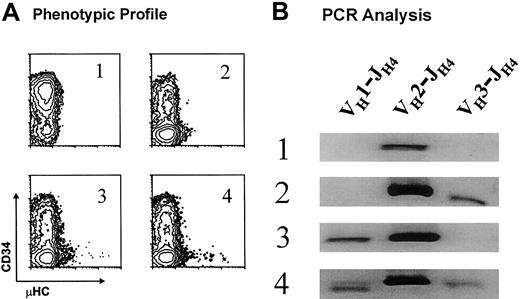
The prevalent usage of a limited number of VH gene segments and the occurrence of the same DJH rearrangements over a 2-year interval for EU12 subclones suggested a predisposition of the individual EU12 pro-B cells to undergo particular VH→DJH rearrangements. Alternatively, each pro-B cell might give rise to progeny that would use one of a limited number of VH gene segments for rearrangement. To determine whether individual EU12 pro-B cells with the DXP′1/JH4 rearrangement could generate intraclonal VH gene diversity, DNA samples of 4 EU12 subclones were isolated for genomic PCR analysis of VH gene utilization. Each of the subclones was derived from a single cell with the pro-B phenotype (R1 in Figure 2B) and the subclones were selected on the basis of their phenotypic profiles after 10 weeks in culture (Figure7A). Whereas μHC+ cells were not seen in subclone 1, subclone 2 progeny reached the pre-B cell stage, and subclones 3 and 4 contained both pre-B and B-cell subpopulations. In an assay of V(D)J rearrangements using the VH-specific primers used for the single-cell PCR analysis described, all 4 subclones were found to undergo the VH2-DJH rearrangement (Figure 7B). Subclone 2 also exhibited a VH3-DJH rearrangement, subclone 3 had a VH1-DJH rearrangement, and subclone 4 was the most diverse, manifesting all 3 VH gene segment rearrangements (Figure 7B). These results, which confirm the single-cell PCR sequence analysis, indicate that the progeny of individual EU12 pro-B cells can undergo productive VDJ rearrangements involving more than one VH gene segment. These findings formally establish that individual EU12 pro-B cells can give rise to intraclonal VH diversity during B-cell generation.
Analysis of VH gene use in EU12 subclones.
(A) 4 EU12 subclones derived from sorted single cells with pro-B phenotype were selected on the basis of their phenotypic profiles after 10 weeks in culture. (B) gDNA was isolated and used as template for PCR reactions as described for single-cell PCR analysis. The PCR products were subjected to electrophoresis and revealed with ethidium bromide staining.
Analysis of VH gene use in EU12 subclones.
(A) 4 EU12 subclones derived from sorted single cells with pro-B phenotype were selected on the basis of their phenotypic profiles after 10 weeks in culture. (B) gDNA was isolated and used as template for PCR reactions as described for single-cell PCR analysis. The PCR products were subjected to electrophoresis and revealed with ethidium bromide staining.
Discussion
These studies define a clonal model of human B-cell development in which the pro-B cell subpopulation, as well as the individual pro-B cells, can spontaneously generate pre-B and B-cell progeny. The B lineage–specific gene expression profiles for each of these subpopulations resemble those observed during normal B-cell differentiation. These characteristics define the EU12 cell line as a novel model that recapitulates many of the central features of the human B-cell differentiation pathway.
Mouse pre-B cell hybridomas,2 Abelson murine leukemia virus (AMuLV)–transformed cell lines,29 human Epstein-Barr virus–transformed B-cell precursors,24 and ALL-derived tumor cell lines30 have been used in previous studies to gain insight into the sequential nature of Ig H and L chain gene rearrangements during B-lineage differentiation. Although most of these cell lines represent clonal populations frozen in a particular stage of B-lineage differentiation, the AMuLV-transformed cell line 300-19 was found to undergo pro-B to B-cell differentiation.29 However, this proved to be an unstable phenotype because the AMuLV-transformed cell lines extinguish theirRag1/2 expression with prolonged cultivation.31 In humans, the search for an in vitro culture system or cell lines that recapitulate early B-lineage cell differentiation and self-renewal for cellular and molecular studies has been an ongoing challenge.32 One leukemic cell line, BLIN-1, was shown to undergo pre-B to B-cell differentiation at low frequency,30 and a more recently described cell line, BLIN-3, can progress from the pro-B to the pre-B cell stage in a stromal cell–dependent manner.33 The EU12 cell line is unique in maintaining 4 distinguishable subpopulations, CD34+sμHC− (R1), CD34−sμHC− (R2), CD34−sμHClow (R3A), and CD34−sμHChighsδHC+λLC+(R3B), that recapitulate sequential stages in the normal B-cell differentiation pathway. Analysis of cell surface and intracellular SLC, TdT, and μHC expression indicates the pro-B cell nature of the R1 cells, while suggesting that the R2 cells represent a transitional pro-B to pre-B cell stage. The R3A cells express pre-BCR, whereas R3B cells express BCR indicating their B-cell status.
The sorted R2 subpopulation can give rise to occasional CD34lowμHC− cells, raising the possibility that this subset may contain rare precursors at an earlier stage of development than R1 cells. Although we cannot exclude the possibility of very rare pre-R1 cells, it seems unlikely that the R2 subset contains earlier progenitors. First, cells in all of the EU12 subpopulations express high levels of the B-lineage marker CD19, indicating that all R2 cells are B lineage–committed cells. Second, although sorted R2 cells can give rise to some CD34lowμHC− cells, these cells never express levels of CD34 antigen as high as those seen on the original R1 subpopulation, possibly suggesting they are in a transition state between the R1 and R2 subsets. The CD34−μHC− cells represent a very minor proportion of B-lineage cells in normal bone marrow, yet R2 cells of this phenotype comprise a significant proportion among the EU12 cells. The explanation for this may lie in the fact that during normal B-lineage cell development, only a minor portion of the CD34+CD19+μHC− pro-B cells undergo V→DJH gene rearrangement to produce μHC and become pre-B cells. Those that fail to make productive VDJ rearrangements may undergo apoptosis in vivo and therefore are eliminated. Although the R2 subset represents a pro-B to pre-B transitional stage because that is in the process of undergoing V→DJH rearrangement, most of these cells fail to produce μHC. Nevertheless, the cells may not undergo immediate apoptosis due to the transformed characteristics and in vitro location of the EU12 cells, thereby leading to an exaggerated representation of this differentiation stage.
Another notable departure from the normal B-cell differentiation scheme was observed for the EU12 cell line. SLC expression is normally down-regulated to extinguish pre-BCR expression at the late pre-B cell stage.11 As a consequence, the receptorless pre-B cells exit the cell cycle and up-regulate their Rag1 andRag2 expression to undergo V-JL gene segment rearrangements. EU12 B cells instead were found to coexpress pre-BCR and BCR, a feature noted previously for other “transitional pre-B/B” cell lines,34-36 thus questioning why these transitional B cells fail to extinguish SLC gene expression and the possible role this failure may have in the leukemogenesis process. A complex locus control region, including the promoters and 5′ enhancer for the VpreB and λ5 genes, has been identified in mice.37-40 The promoter regions of both genes contain binding sites for Ikaros, EBF, E2A, and PAX5,41-43 and these transcription factors are expressed throughout the pre-B cell differentiation process. Ikaros-mediated transcriptional silencing44-46 may provide a mechanism for SLC gene down-regulation in normal late-stage pre-B cells. On the other hand, dominant-negative Ikaros isoforms, including Ikaros 6, have been found to be preferentially expressed in ALLs and could interfere with the usual transcriptional silencing of SLC genes.47 48 The clonal EU12 cell line may provide a suitable model to test this hypothesis.
B-cell leukemias and lymphomas are considered clonal diseases derived from a single transformed precursor, based on the analysis of their karyotypes, glucose-6-phosphate dehydrogenase (G6PD) isoenzymes, and BCR idiotypes.49-51 However, 15% to 45% of ALLs of B lineage represent oligoclonal malignancies according to the diversity of their HC gene expression.52-57 Analysis of these patients has indicated that the leukemic population may diversify by generating multiple IgH gene rearrangements during the process of tumor progression.55-57 Sequence analysis of the IgH genes has suggested that both VH→DJH and VH→VDJH recombinational events can occur during the clonal evolution of B cell leukemias.55
All of the cells in the clonal EU12 cell line, established from a childhood B-lineage ALL, apparently contain the same DXP′1/JH4 rearrangement with no N nucleotide addition in the DJH join. This suggests that the formative transformational event may have occurred during fetal life in a pro-B cell that had undergone D→JHrearrangement.58 Interestingly, overrepresentation of the DXP′1 and JH4 gene segments has been observed in other ALL samples.57,59 Sequence analysis of the VH gene rearrangements in the EU12 B cells indicated the use of 4 gene segments, VH2-5, VH3-7, VH3-11, and VH1-8, which are clustered in the DH proximal region of the HC locus. Preferential usage of the DHproximal VH gene segments has been noted previously in mice60 and in human ALL.61 The nontemplated nucleotide additions observed at the site of V-DJ joining include a dominant CCCC tetranucleotide, and other unusually long sequences that contain a consensus CACA sequence identical to the 3′ end of the VH2-5 gene segment, located just 3′ to the conserved internal cryptic RSS heptamer (TACTGTG). These results suggest that the onset of μHC expression in EU12 B cells could result from a VH→DXP′1/JH4 recombination and by VH→nonfunctional VH2-5-DXP′1/JH4 replacement events. The occurrence of VH gene replacement was initially suggested by studies of murine cell lines as a mechanism that could generate a functional μHC gene from a nonproductive VDJH rearrangement.62,63 The possible occurrence of VH gene replacement has been suggested by analysis of the VH repertoire of human B-lineage ALL55,64 and in a transgenic mouse carrying a VDJH rearrangement targeted to the endogenous IgH locus.65 Confirmation of ongoing VH gene replacement as a mechanism to generate the unique EU12 CDR3 regions would make this cell line a valuable model to define the mechanism of the VH gene replacement reaction.
The discovery of sequential rearrangement and expression of Ig HC and LC genes in the immediate precursors of mammalian B cells led to the idea that intraclonal V(D)J recombinatorial diversification might occur during B-cell genesis.19 The present studies provide direct evidence for the validity of the principle of intraclonal V(D)J diversification, in that individual EU12 pro-B cells marked with a DXP′1/JH4 gene segment give rise to a diverse repertoire during their progression from the pro-B to B-cell stage. Our analysis of the EU12 cell line model also suggests that VH→DJH recombination and VH→VDJH replacement events can occur contemporaneously in early B-lineage cells to generate intraclonal V(D)J recombinatorial diversity independent of the bone marrow microenvironment. The EU12 cell line thus provides a useful model for analysis of the progression of human B-cell development.
We thank Dr Andrew J. Carroll for performing karyotypic analysis, Dr Larry Gartland for help with flow cytometry, Charlie Mashburn for technical support, and Marsha Flurry and Ann Brookshire for help in preparing the manuscript.
Prepublished online as Blood First Edition Paper, September 19, 2002; DOI 10.1182/blood-2002-06-1828.
Supported by National Institutes of Health grants AI48098, AI42127, and AI39816. M.D.C. is a Howard Hughes Medical Institute Investigator.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Max D. Cooper, University of Alabama at Birmingham, WTI 378, 1824 6th Ave S, Birmingham, AL 35294; e-mail:max.cooper@ccc.uab.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal