A recent article by Leung et al1 analyzed the relationship between the genetic polymorphism in exon 4 of cytochrome P450 CYP2C9 and warfarin sensitivity in Chinese patients. They used polymerase chain reaction (PCR) and direct sequencing methods to analyze the genetic changes of the CYP2C9 gene in 89 patients, and their results showed that 4 polymorphisms in exon 4 were found: heterozygosities for 608TTG>GTG (Leu208Val), 561CAG>CCG (Gln192Pro), 537CAT>CCT (His184Pro), and 527ATT>CTT (Ile181Leu) existed at frequencies 0.75, 0.20, 0.10, and 0.09, respectively. We used a similar approach to analyze the genetic polymorphisms in exon 4 of the CYP2C9 gene in Taiwanese patients. The primers used to amplify theexon 4 were 5′-AATACAGTGTTTTATATCTAAAG-3′ (GenBank accession number L16879, nucleotide [nt] 1-23) and 5′-TAAGTGGTTTCTCAGGAAGC-3′ (nt 256 to 237) as forward and reverse primers, respectively. We were unable to find these 4 polymorphisms or other new polymorphisms in the exon area in 50 healthy people (Figure 1). In order to double-check our results, we developed a PCR–restriction fragment length polymorphism (RFLP) method to analyze these 4 polymorphisms. For the detection of 608TTG>GTG, we used primer 5′-AATACAGTGTTTTATATCTAAAG-3′ (forward: nt 1 to 23) and 5′-CTGGATCCAGGGGCTGCTCG-3′ (reverse: nt 219 to 190). A mismatched base (underlined) was introduced at the 3′-end base of the reverse primer to create a TaqI restriction enzyme cutting site in combination with the base 608T of wild type after PCR and no TaqI site for the 608GTG polymorphism. For the detection of 561CAG>CCG, we used the same forward primer, and 5′-TTCCATTAAGTTAAGAAATAGC-3′ (reverse: nt 174 to 153). A mismatched base (underlined) was introduced at position 3 from the 3′ end of the reverse primer to create an AluI site in combination with the base 561A of wild type after PCR and no AluI site for the 537CCT polymorphism. For the detection of 537CAT>CCT, we used the same forward primer and 5′-CTGATCTTTATAATCAAAACGTTCA-3′ (reverse: nt 153 to 129). A mismatched base (underlined) was introduced at position 2 from the 3′ end of reverse primer to create an NlaIII site in combination with the base 536C and base 537A of wild type after PCR reaction, and no NlaIII site for the 537CCT polymorphism. For the detection of 527ATT>CTT, we used primer 5′-CCCTGCAATGTGATCTGGTC-3′ (forward: nt 97 to 116) and 5′-GCAATTCAGAGCTTGATCCATG-3′ (reverse: nt 312 to 291). A mismatched base was introduced at position 3 from 5′ end of forward primer to create an NlaIV site in combination with the base 527C of wild type after PCR, and no NlaIV site for the 527CTT polymorphism. The PCR condition was performed as described,2,3 except the annealing temperature was done at 55°C, 57°C, 56°C, and 55°C for 608, 561, 537, and 527, respectively. All the above mentioned primer sequences can be found in GenBank accession number L16879. They are specific for the CYP2C9 gene.
The representative case of direct sequencing analysis for exon 4 polymorphisms of CYP2C9 gene. No polymorphism was found in exon 4 of 50 cases after sequencing analysis.
The representative case of direct sequencing analysis for exon 4 polymorphisms of CYP2C9 gene. No polymorphism was found in exon 4 of 50 cases after sequencing analysis.
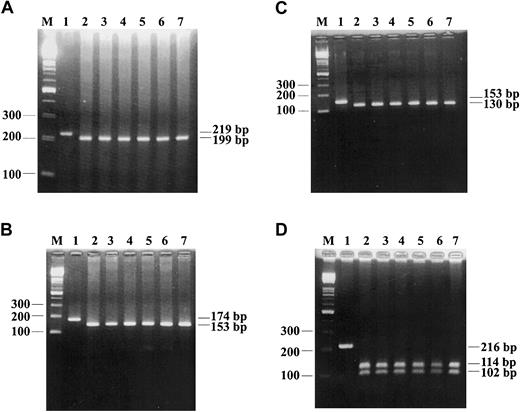
In total, 300 cases were analyzed with these 4 PCR-RFLP methods (Figure 2), and the polymorphisms found by Leung et al were not found in our analysis.
The results of PCR-RFLP analysis for 4 polymorphisms in exon 4 of CYP2C9 gene. (A) The results of detection of 608TTG>GTG are shown, the wild type (TTG) is completely digested to 199-bp fragment (lanes 2-7) by TaqI. (B) The results of detection of 561CAG>CCG are shown, the wild type (CAG) is completely digested to 153-bp fragment by AluI (lanes 2-7). (C) The results of detection of 537CAT>CCT are shown, the wild type (CAT) is completely digested to 130-bp fragment by NlaIII (lanes 2-7). (D) The results of detection of 527ATT>CTT are shown, the wild type (ATT) is completely digested to 114-bp and 102-bp fragments by NlaIV (lanes 2-7). M indicates 100-bp ladder marker; and lane 1, uncut control.
The results of PCR-RFLP analysis for 4 polymorphisms in exon 4 of CYP2C9 gene. (A) The results of detection of 608TTG>GTG are shown, the wild type (TTG) is completely digested to 199-bp fragment (lanes 2-7) by TaqI. (B) The results of detection of 561CAG>CCG are shown, the wild type (CAG) is completely digested to 153-bp fragment by AluI (lanes 2-7). (C) The results of detection of 537CAT>CCT are shown, the wild type (CAT) is completely digested to 130-bp fragment by NlaIII (lanes 2-7). (D) The results of detection of 527ATT>CTT are shown, the wild type (ATT) is completely digested to 114-bp and 102-bp fragments by NlaIV (lanes 2-7). M indicates 100-bp ladder marker; and lane 1, uncut control.
In conclusion, we demonstrated that no genetic polymorphism in exon 4 of cytochrome CYP2C9 is found in Taiwanese, which is different from the results reported by Leung et al. We suggest that further study of the Chinese population is necessary.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal