Abstract
Ribosomal protein S19 (RPS19) is frequently mutated in Diamond-Blackfan anemia (DBA), a rare congenital hypoplastic anemia. Recent studies have shown that RPS19 expression decreases during terminal erythroid differentiation. Currently no information is available on the subcellular localization of normal RPS19 and the potential effects of various RPS19 mutations on cellular localization. In the present study, using wild-type and mutant RPS19 cDNA, we explored the subcellular distribution of normal and mutant proteins in a fibroblast cell line (Cos-7 cells). RPS19 was detected primarily in the nucleus, and more specifically in the nucleoli, where RPS19 colocalized with the nucleolar protein nucleolin. Using various N-terminal and C-terminal deletion constructs, we identified 2 nucleolar localization signals (NoSs) in RPS19: the first comprising amino acids Met1 to Arg16 in the NH2-terminus and the second comprising Gly120 to Asn142 in the COOH-terminus. Importantly, 2 mutations identified in DBA patients, Val15Phe and Gly127Gln, each of which localized to 1 of the 2 NoS, failed to localize RPS19 to the nucleolus. In addition to their mislocalization, there was a dramatic decrease in the expression of the 2 mutant proteins compared to the wild type. This decrease in protein expression was specific for the mutant RPS19, since expression of other proteins was normal. The present findings enable us to document the nucleolar localization signals in RPS19 and help define the phenotypic consequences of some mutations in RPS19 in DBA.
Introduction
Diamond-Blackfan anemia (DBA), a congenital hypoplastic anemia,1,2 is usually revealed early in infancy by pallor and failure to thrive. Laboratory findings include a normochromic, usually macrocytic, aregenerative anemia and erythroblastopenia with fewer than 5% of erythroid precursors in an otherwise normal bone marrow. DBA is characterized by a heterogeneous phenotype, including in 40% of affected individuals various malformations. These malformations occur mostly in the cephalic area but also in the thumbs, in the urogenital tract, and in the heart.3 A short stature, especially at birth, is considered part of the malformative syndrome.3 At diagnosis, more than 60% of patients affected by DBA respond to steroid therapy.3 Patients who either are nonresponders or require high doses of steroids with a risk of severe side effects from steroid therapy enter into a long-term transfusion therapy with iron chelation. Disease evolution in DBA patients is unpredictable. Some individuals become free of any treatment while others who initially responded to steroid therapy develop secondary steroid resistance and yet others who were initially unresponsive to steroids respond secondarily to steroid therapy. A successful bone marrow or cord blood transplantation is the only viable cure to date. A wide variety of mutations in ribosomal protein S19 (RPS19) gene has been identified in 25% of patients affected by DBA.4,5 No correlation between clinical presentation, response to treatment, and genotype has been found. The role of RPS19 in normal erythropoiesis and the impact of RPS19 mutations on disordered erythropoiesis in DBA have not been defined yet. Moreover, little information is available on nonribosomal function of RPS19 in cells. In order to obtain insights into potential function of RPS19 in erythropoiesis, we have begun a number of cell biologic studies. We recently showed that RPS19 expression decreases during terminal erythroid differentiation.6 In the present study, we explored the subcellular localization of RPS19 and the potential effects of various RPS19 mutations on cellular localization. RPS19 was detected primarily in the nucleus and, more specifically, in the nucleoli. Having identified nucleolar localization of RPS19, we characterized 2 nucleolar localization signals (NoS) in both termini regions of the protein. NoS are less well characterized than nuclear localization signals (NLS),7-9 which are usually but not always constituted by a cluster of 1 (monopartite SV40-type NLS)10,11 or 2 (bipartite nucleoplasmin-type NLS)12 small sequence(s) of positively charged amino acids such as arginine (R), histidine (H), or lysine (K).13 In marked contrast, NoS are not clustered and much less is known about nucleolar transport. The most dominant determinant appears to be the tertiary structure of the protein and the interactions between its functional domains with other nucleolar protein(s) or with rRNA.14-19 The 2 NoS that we identified in RPS19 comprised amino acids Met1 to Arg16 in the NH2-terminus and the amino acids Gly120 to Asn142 in the COOH-terminus. Importantly, 2 mutations identified in DBA patients, Val15Phe and Gly127Gln, each of which localized to 1 of the 2 NoS, failed to localize RPS19 to the nucleolus, while other mutations not localized to a NoS exhibited normal subcellular localization. In addition to their mislocalization, there was a dramatic decrease in the expression of the mutant proteins Val15Phe and Gly127Gln compared to the wild type. The present findings help define for the first time, at the cellular level, some of the phenotypic consequences of mutations in RPS19 in DBA. These findings should shed some new insights into the role of RPS19 in the pathophysiology of DBA.
Patients, materials, and methods
Description of probands
Four DBA patients from different families were studied. We summarized the data corresponding to the patients in Table 1. Variability in the mode of inheritance, in the associated malformations (including statural retardation), and in the response to therapy reflects the usual heterogeneity in DBA patients.
Description of the 4 probands affected by DBA
Proband . | Sex . | Age at diagnosis (mo) . | Inheritance . | Hb count at diagnosis (g/dL) . | Malformations . | Treatment at time of study . | RPS19 gene mutation . |
|---|---|---|---|---|---|---|---|
| 1 | F | 1 | S | 6.8 | SGA | S | Val15Phe |
| Thr55Met | |||||||
| 2 | F | 1 | S | 3.5 | SGA, deafness, hip hypoplasia | S + T | Gly127Gln |
| 3 | M | 1 | S | 6 | None | S + T | Arg56Gln |
| 4 | M | Birth | D | 10.1 | None | None | Arg62Trp |
Proband . | Sex . | Age at diagnosis (mo) . | Inheritance . | Hb count at diagnosis (g/dL) . | Malformations . | Treatment at time of study . | RPS19 gene mutation . |
|---|---|---|---|---|---|---|---|
| 1 | F | 1 | S | 6.8 | SGA | S | Val15Phe |
| Thr55Met | |||||||
| 2 | F | 1 | S | 3.5 | SGA, deafness, hip hypoplasia | S + T | Gly127Gln |
| 3 | M | 1 | S | 6 | None | S + T | Arg56Gln |
| 4 | M | Birth | D | 10.1 | None | None | Arg62Trp |
F indicates female; M, male; S, sporadic; D, dominant; SGA, small for gestational age; S, steroid; and S + T, partial response to steroid with transfusion dependence.
DNA cloning
The cDNA sequences encoding RPS19 and other proteins relevant to this study, namely nucleolin, protein 4.1R, and intercellular adhesion molecule-4 (ICAM 4), were cloned into pEGFP-C3, pEGFP-N1, or pDsRed1-C1 mammalian expression vectors (BD Biosciences-Clontech Laboratories, Palo Alto, CA), which encode a green and a red fluorescent protein, respectively. Human nucleolin cDNA (accession number XM_010858) was cloned into pDsRed1-C1 vector after polymerase chain reaction (PCR) amplification of human kidney marathon cDNA library (BD Biosciences-Clontech Laboratories) according to the manufacturer's instructions. Protein 4.1R was cloned into pEGFP-C3 using BglII and SacI restriction sites. Mouse ICAM-4 was inserted into pEGFP-N1 vector (BD Biosciences-Clontech Laboratories) using BglII and SacII restriction sites and expressed upstream of the green fluorescent protein pEGFP.
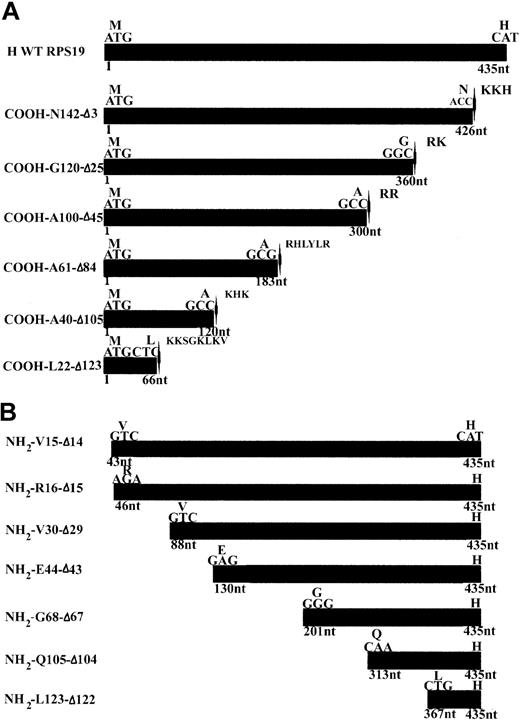
Wild-type (WT) human RPS19 cDNA and various COOH- and NH2-terminal truncated human RPS19 cDNAs, designed to map the nuclear and nucleolar localization signals (NLS and NoS) of RPS19 (Figure 1A-B), were cloned into pEGFP-C3 vector using a forward and a reverse primer containing a HindIII and a BamHI restriction site, respectively. A stop codon was inserted just before a putative NLS at the COOH-terminal end of RPS19 (Figure 1A-B).
cDNA constructs used for mapping the NoS in RPS19. (A) Human wild-type RPS19 (H WT RPS19) is shown at the top of the panel as the black box. The ATG translational start site and the last codon coding for a histidine (H) are indicated. We generated 6 COOH-terminal truncations. The mutant GFP-RPS19 fusion proteins are labeled by the last amino acid number in the new COOH-terminal and by the number of deleted amino acids (Δ). We added a stop codon (located with arrow-heads) just before a putative NLS characterized by a cluster of basic amino acids: arginine (R), histidine (H), lysine (K). (B) We also generated 7 NH2-terminal truncations in GFP-RPS19 fusion proteins. These are labeled with the amino acid number of the NH2 terminus and by the number of deleted amino acids (Δ). We used EGFP ATG as the translational start site of the GFP-RPS19 fusion proteins, meaning that RPS19 was cloned in frame with upstream EGFP coding sequence.
cDNA constructs used for mapping the NoS in RPS19. (A) Human wild-type RPS19 (H WT RPS19) is shown at the top of the panel as the black box. The ATG translational start site and the last codon coding for a histidine (H) are indicated. We generated 6 COOH-terminal truncations. The mutant GFP-RPS19 fusion proteins are labeled by the last amino acid number in the new COOH-terminal and by the number of deleted amino acids (Δ). We added a stop codon (located with arrow-heads) just before a putative NLS characterized by a cluster of basic amino acids: arginine (R), histidine (H), lysine (K). (B) We also generated 7 NH2-terminal truncations in GFP-RPS19 fusion proteins. These are labeled with the amino acid number of the NH2 terminus and by the number of deleted amino acids (Δ). We used EGFP ATG as the translational start site of the GFP-RPS19 fusion proteins, meaning that RPS19 was cloned in frame with upstream EGFP coding sequence.
Full-length human WT RPS19 or domains of RPS19 corresponding to the 15 first amino acids (aa) and to the COOH-terminal region spanning 22 aa (Gly120 to Asn142) were cloned into pcDNA3 mammalian expression vector encoding NH2-terminal c-myc epitope tagged chicken muscle pyruvate kinase (PK) (pcDNA3 c-myc PK) (generously provided by Dr G. Dreyfuss, Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia, PA), using the KpnI and NotI restriction sites.20-22 RPS19 cDNA was cloned downstream of PK, while the sequence encoding the first 15 aa of RPS19 was inserted at the NH2-terminal end of PK sequence, and the sequence corresponding to the COOH-terminal 22 aa (Gly120-Asn142) were cloned at the COOH-terminal end of PK sequence.
Mutant RPS19 cDNA carrying specific mutations identified in DBA patients were cloned downstream of either the green or the red fluorescent protein encoded by enhanced green fluorescent protein (EGFP) and DsRed1 sequences, respectively. Mutated RPS19 constructs were generated after PCR amplification of RPS19 cDNA obtained from affected DBA patients carrying the following mutations: Arg56Gln, or a double-mutant Val15Phe-Thr55Met. In addition, each of the 2 mutations in the double mutant, Val15Phe and Thr55Met, were cloned separately into pEGFP-C3 vector, and the Val15Phe mutant also was cloned into a pDsRed1-C1 vector. The pEGFP-C3-Gly127Gln and Arg62Trp mutated RPS19 constructs were generated by site-directed mutagenesis of pEGFP-C3-WT RPS19 clone using the QuickChange mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. All constructs were sequenced using the Applied Biosystems 373 DNA sequencer and ABI Big Dye Terminator sequencing kits (Perkin Elmer, Foster City, CA).23 Sequences of all the primers used in this study are available upon request.
Cell culture and transfection
Cos-7 cells were cultured in Dulbecco modified Eagle medium (Life Technologies, Gaithersburg, MD), supplemented with 1% glutamine, 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin. Cos-7 cells, seeded in either 6-well clusters or 100-mm cell culture dishes, were transfected for 16 to 40 hours with various pEGFP-C3 or pDsRed1-C1 constructs. Cells were transfected with 1.5 μg and 20 μg DNA, using 5 μL and 30 μL lipofectamine 2000 reagent and 200 μL and 1500 μL OPTI-MEM I medium (Gibco BRL, Gaithersburg, MD) per well or cell culture dish, respectively, according to the manufacturer's instructions.
Immunoblotting
Proteins were subjected to 10% or 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred as previously described.6 The membrane was blocked for 1 hour at room temperature in Blotto solution containing either 1% donkey serum (anti-RPS19) or 1% goat serum (antiactin or anti–green fluorescent protein [GFP]) as previously described.6 The membrane was then incubated overnight at 4°C with either an affinity-purified polyclonal antibody raised in chicken against His-tagged recombinant mouse RPS19 and used at a concentration of 0.05 μg/mL,6 or a mouse monoclonal antibody against chicken actin (clone C4) (ICN Biomedicals, Aurora, OH) diluted 1:20 000, or a mouse monoclonal antibody against GFP (Jackson ImmunoResearch, West Grove, PA) and used at 0.1 μg/mL. After several washes, followed by a short incubation with blocking buffer, the membranes were incubated for 1 hour at room temperature with secondary antibodies coupled to horseradish peroxidase: a donkey antichicken (Research Diagnostics, Flanders, NJ) diluted 1:5000 for detection of RPS19 or a goat antimouse (Jackson ImmnoResearch Laboratories, West Grove, PA) diluted 1:20 000 for detection of actin and GFP. After extensive washing, the membranes were probed with enhanced chemiluminescence reagent R (ECL; NEN, Life Science Products, Boston, MA).
Immunofluorescence microscopy
Confluent Cos-7 cells, either untransfected or transfected with pcDNA3-c-Myc-PK constructs, were washed twice with filtered phosphate-buffered saline (PBS) and fixed in chilled absolute methanol for 5 minutes. After fixation, cells were washed 3 times in PBS and incubated with the blocking solution (filtered PBS, 10 mg/mL bovine serum albumin [BSA], 10% donkey serum, 0.05% Tween-20) for 1 hour. Cells were then incubated for 1 hour with either the polyclonal anti-RPS19 antibody raised in chicken or a monoclonal anti–c-myc antibody (clone 9E10) raised in mouse (Santa Cruz Biotechnology, Santa Cruz, CA) diluted in the blocking solution at 15 μg/mL and 1:400, respectively. Cells were then rinsed and washed 3 times for 10 minutes with PBS + 0.1 mg/mL BSA. After blocking for 15 minutes in PBS + 10 mg/mL BSA, cells were incubated for 1 hour at room temperature with secondary antibodies coupled to fluorescein isothiocyanate (FITC), either FITC donkey antichicken or antimouse antibodies (RDI, Flanders, NJ) diluted 1:200 in PBS + 10 mg/mL BSA. Cells were rinsed and washed, and coverslips were mounted on microscope slides in a 1:1 glycerol/vectashield mixture containing 2% 1,4-diazabiclo-(2,2,2)-octan (DABCO; Merck) and sealed with nail polish. Cells were viewed using an Axiovert 132TV inverted microscope (Zeiss, Oberochen, Germany), and images were acquired using Scilimage software (Center of Pattern Recognition [CBP], Delft, the Netherlands).
Cos-7 cells, transfected with either pEGFP-C3 or pDsRed1-C1 constructs, washed and fixed as described in the previous paragraph, were mounted on microscope slides in a 1:1 glycerol/vectashield mixture containing 2% 1,4-diazabiclo-(2,2,2)-octan (DABCO), sealed with agarose, and conserved at 4°C. Cells were viewed using an Axiovert 132TV inverted microscope (Zeiss, Oberochen, Germany). We analyzed the distribution of GFP-RPS19 fusion proteins in different subcellular compartments by counting the percentage of cells exhibiting staining in the different compartments: the nucleus, the cytoplasm, and the nucleoli. Cos-7 cells cotransfected with pEGFP-C3-human WT RPS19 and pDsRed1-C1-human WT nucleolin were examined using a Zeiss LSM 410 confocal microscope (Zeiss, Thornwood, NY). Thirty slices, each 0.45-μm thick, were examined. Quantitative image processing using a Micro Imager 1400 digital camera (Xillix Technologies, Vancouver, BC, Canada) was performed, and the images derived were analyzed using Scilimage software.
Living Cos-7 cells, grown on LabTek II slides (van Waters and Rogers, Plainfield, NJ) and transfected with either pEGFP-C3 empty vector or pEGFP-C3-WT or pEGFP-C3-Val15Phe+Thr55Met or pEGFP-C3-Val15Phe or pEGFP-C3-Thr55Met RPS19 constructs, were examined without any fixation using a Nikon TE300 inverted microscope (Melville, NY) connected to a Photometrics series 300 charge coupled device camera (Photometrics, Tucson, AZ). Images were processed with V++ Imaging (Digital Optics, Browns Bay, Auckland, New Zealand) and Scilimage softwares.
Results
RPS19 is localized in the nucleoli
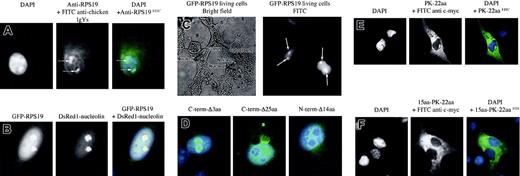
We determined the localization of endogenous RPS19 in Cos-7 cells by immunofluorescence microscopy using the antibody raised in chicken against RPS19. RPS19 localized in nuclei, where it appeared to accumulate primarily in structures corresponding to nucleoli (see arrows in Figure 2A, middle and right panels). A weaker and diffuse cytoplasmic staining also was noticed. Recombinant human WT RPS19 fused to GFP also localized in nuclei in structures corresponding likely to nucleoli (Figure 2B, left panel), while GFP alone was found exclusively in the cytoplasm (data not shown). The identical subcellular localization of native RPS19 and GFP-RPS19 implies that GFP-tagged proteins can be used as an appropriate reporter protein for nuclear and nucleolar import studies. To rule out the possibility that cell fixation might influence subcellular localization of RPS19, we determined RPS19 localization in live cells. The subcellular distribution of RPS19 in live cells (Figure 2C) and fixed cells (Figure 2B, left panel) was identical.
Subcellular localization of RPS19. (A) The use of an anti-RPS19 antibody raised in chicken shows primary localization of RPS19 in the nucleus and particularly to the nucleoli in Cos-7 cells (arrows). A weaker and diffuse cytoplasmic staining is also observed. The left panel represents 4′6-diamidino-2-phenylindole (DAPI) staining for the nucleus; the middle panel, FITC staining of RPS19; and the right panel, the merged picture of DAPI and FITC staining. (B) Nucleolar localization of RPS19 is confirmed after transfection of Cos-7 cells with the recombinant GFP–wild-type RPS19 (left panel). RPS19 colocalizes with the major nucleolar protein nucleolin (middle panel) after transfection of Cos-7 cells with GFP–wild-type RPS19 and the red fluorescent DsRed1-nucleolin fusion protein (right panel). (C) Nucleolar localization of RPS19 (arrows) is also observed in live cells after transfection of Cos-7 cells with GFP-RPS19, ruling out an artifactual effect of cell fixation on RPS19 subcellular distribution. (D) Deletion of the last 3 COOH-terminal amino acids does not impair RPS19 localization compared with wild type (left panel). In contrast, deletion of the last 25 COOH-terminal amino acids dramatically alters RPS19 nucleolar import (middle panel). Furthermore, deletion of the first 14 NH2 amino acids also impaired RPS19 nucleolar import (right panel). (E) The 22-amino-acid-long NoS identified in the last 25 amino acids fails to import pyruvate kinase into the nucleoli. (F) Addition of the first NoS to the previous pyruvate kinase construct also fails to be imported into the nucleoli of Cos-7 cells, confirming that although these 2 NoS are necessary, they are not sufficient for nucleolar import. Original magnifications: × 63 (A,B,D,E,F); × 40 (C).
Subcellular localization of RPS19. (A) The use of an anti-RPS19 antibody raised in chicken shows primary localization of RPS19 in the nucleus and particularly to the nucleoli in Cos-7 cells (arrows). A weaker and diffuse cytoplasmic staining is also observed. The left panel represents 4′6-diamidino-2-phenylindole (DAPI) staining for the nucleus; the middle panel, FITC staining of RPS19; and the right panel, the merged picture of DAPI and FITC staining. (B) Nucleolar localization of RPS19 is confirmed after transfection of Cos-7 cells with the recombinant GFP–wild-type RPS19 (left panel). RPS19 colocalizes with the major nucleolar protein nucleolin (middle panel) after transfection of Cos-7 cells with GFP–wild-type RPS19 and the red fluorescent DsRed1-nucleolin fusion protein (right panel). (C) Nucleolar localization of RPS19 (arrows) is also observed in live cells after transfection of Cos-7 cells with GFP-RPS19, ruling out an artifactual effect of cell fixation on RPS19 subcellular distribution. (D) Deletion of the last 3 COOH-terminal amino acids does not impair RPS19 localization compared with wild type (left panel). In contrast, deletion of the last 25 COOH-terminal amino acids dramatically alters RPS19 nucleolar import (middle panel). Furthermore, deletion of the first 14 NH2 amino acids also impaired RPS19 nucleolar import (right panel). (E) The 22-amino-acid-long NoS identified in the last 25 amino acids fails to import pyruvate kinase into the nucleoli. (F) Addition of the first NoS to the previous pyruvate kinase construct also fails to be imported into the nucleoli of Cos-7 cells, confirming that although these 2 NoS are necessary, they are not sufficient for nucleolar import. Original magnifications: × 63 (A,B,D,E,F); × 40 (C).
In order to further confirm the predominant nucleolar localization of RPS19, we compared the localization of RPS19 with that of nucleolin, the major nucleolar protein15,16 in Cos-7 cells cotransfected with constructs expressing RPS19 fused to GFP and nucleolin fused to red fluorescent fusion protein, DsRed1. Colocalization of RPS19 and nucleolin could be demonstrated in the transfected cells by both regular and confocal microscopy (Figure 2B, right panel). This finding confirmed the predominant nucleolar localization of RPS19.
The 15 NH2-terminal (Met1 to Val15) and 22 COOH-terminal amino acids (Gly120 to Asn142) of RPS19 are necessary but not sufficient for import of RPS19 into nucleoli
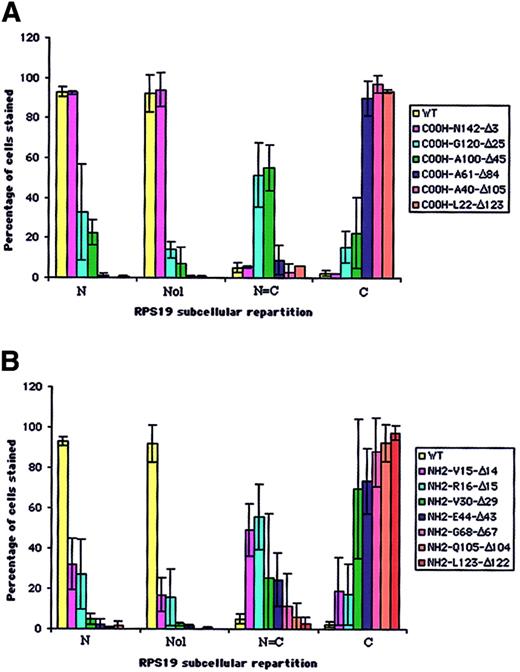
In order to map the potential NoS in RPS19 sequence, we generated a series of COOH- and NH2-terminal truncated RPS19 constructs (Figure 1A-B). These various RPS19 cDNAs cloned into pEGFP-C3 were transfected into Cos-7 cells and the subcellular distribution of various mutant proteins assessed by immunofluorescence microscopy. To obtain quantitative insights, we evaluated 200 transfected cells and determined the predominant subcellular compartment (cytoplasm or nucleus) in which the expressed protein was localized (Figure 3A-B). Nucleolar localization was evaluated separately on 200 transfected cells with the degree of staining relative to GFP-RPS19 fusion protein nucleoli staining (Figure 3A-B, second group of columns, Nol). While truncation of 3 amino acids from the COOH-terminal (COOH-N142-Δ3) did not alter nucleolar localization (Figure 2D, left panel), truncation of 25 amino acids (COOH-G120-Δ25) (Figure 2D, middle panel) significantly impaired the nucleolar import of the corresponding RPS19. Only 14% of the cells showed nucleolar staining, compared with 92% for the wild-type RPS19 (Figure 3A). Mutant proteins with further COOH-terminal deletions also failed to localize to the nucleolus (Figure 3A).
Analysis of distribution of truncated COOH- and NH2-terminal RPS19 fused to GFP in transfected Cos-7 cells. The subcellular distribution of 6 COOH-terminal (A) and 7 NH2-terminal (B) truncated RPS19 fused to GFP described in Figure 1 was compared with human WT RPS19. We evaluated among 200 transfected cells (1) the percentage of cells showing exclusive or predominant nuclear staining (N); (2) the percentage of cells showing similar nuclear and cytoplasmic staining (N = C); and (3) the percentage of cells showing exclusive or predominant cytoplasmic staining (C). Independently, we evaluated from 200 cells the percentage of cells stained in the nucleoli (Nol). The data shown are averaged from 3 independent experiments.
Analysis of distribution of truncated COOH- and NH2-terminal RPS19 fused to GFP in transfected Cos-7 cells. The subcellular distribution of 6 COOH-terminal (A) and 7 NH2-terminal (B) truncated RPS19 fused to GFP described in Figure 1 was compared with human WT RPS19. We evaluated among 200 transfected cells (1) the percentage of cells showing exclusive or predominant nuclear staining (N); (2) the percentage of cells showing similar nuclear and cytoplasmic staining (N = C); and (3) the percentage of cells showing exclusive or predominant cytoplasmic staining (C). Independently, we evaluated from 200 cells the percentage of cells stained in the nucleoli (Nol). The data shown are averaged from 3 independent experiments.
Various truncation mutants of the NH2-terminal region of RPS19 also were tested for nucleolar import. Deletion of either the first 14 (NH2-V15-Δ14) (Figure 2D, right panel) or 15 amino acids (NH2-R16-Δ15) of RPS19 resulted in significant impairment of the nucleolar localization of the corresponding truncated RPS19 proteins with only ∼16% of cells showing nucleolar staining (Figure 3B). Mutant proteins with further NH2-terminal deletions also failed to localize to the nucleolus (Figure 3B). These findings imply that 2 regions in RPS19, the first 15 NH2-terminal amino acids (Met1 to Val15), and the 22 COOH-terminal amino acids (Gly120 to Asn142) are necessary for nucleolar localization of the proteins.
To determine if either of these 2 sequences alone or in combination can act as NoS, we determined the subcellular localization of the reporter cytoplasmic protein pyruvate kinase fused to these RPS19 sequence motifs. We cloned into the pcDNA3-c-Myc-PK construct either WT RPS19 or the 2 NoS regions shown to mediate RPS19 nucleolar import. As expected in Cos-7 cells transfected with RPS19 fused to c-myc-PK, pyruvate kinase was imported into the nucleoli, while pyruvate kinase was entirely in the cytoplasm of cells transfected with c-myc-PK construct (data not shown). Interestingly, the 22 COOH-terminal sequence fused to PK failed to mediate the nucleolar import of the pyruvate kinase fusion proteins (Figure 2E). Most important, fusion of both NoS sequences motifs to PK (c-myc-first15aa-PK-22aa) also failed to facilitate the nucleolar import of the pyruvate kinase (Figure 2F). These findings imply that the 2 NoS sequences identified are necessary but not sufficient for nucleolar localization.
Two RPS19 mutations identified in DBA patients impair nucleolar localization of mutant proteins
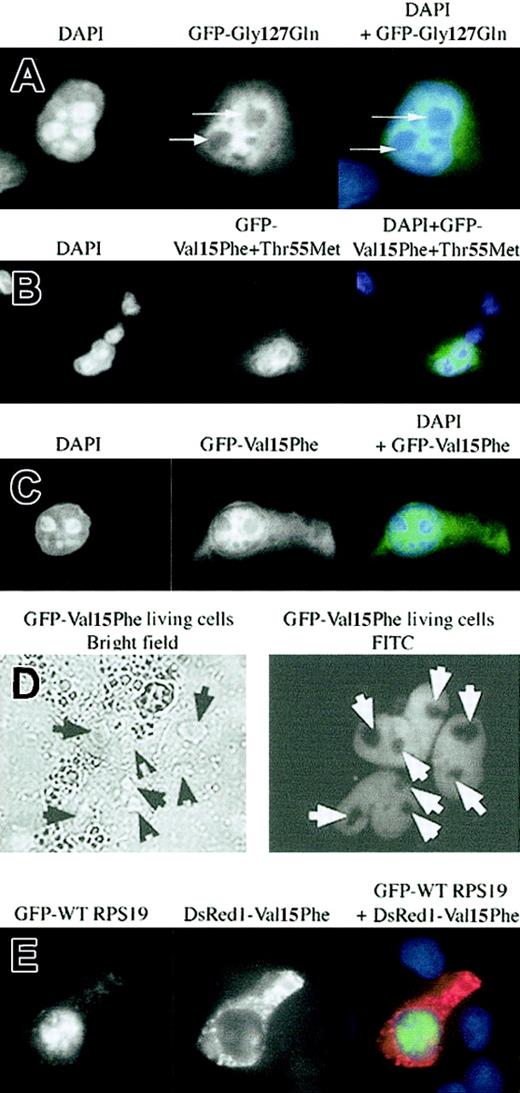
In the database of RPS19 mutations we have previously identified in DBA patients 2 mutations mapped to the 2 NoS motifs defined in the present study. The missense mutation Gly127Gln in one DBA patient was localized within the COOH-terminal NoS. When we examined the subcellular distribution pattern of mutated Gly127Gln RPS19 fused to GFP in transfected Cos-7 cells, the mutant protein failed to localize to the nucleolus (Figure 4A), implying that mutations in COOH-terminal NoS impair the appropriate subcellular localization of RPS19. In a second patient, RPS19 carried 2 mutations, Val15Phe and Thr55Met. Expression of the mutant protein in Cos-7 cells showed an impairment of nucleolar import of the protein (Figure 4B). Since it was not clear whether the Val15Phe mutation located in the N-terminal NoS or the Thr55Met located in the hot spot of DBA mutations in RPS19 was responsible for RPS19 nucleolar mislocalization, we determined the subcellular distribution of protein carrying each of the individual mutations. While the missense Val15Phe mutation reproduced the abnormal phenotype observed with the double mutant in both fixed cells (Figure 4C) and in live cells (Figure 4D), the missense Thr55Met mutation showed normal nucleolar import (data not shown). We confirmed the phenotype of the Val15Phe mutation by cotransfecting Cos-7 cells with GFP-WT RPS19 and red fluorescent DsRed1-mutated Val15Phe RPS19 constructs. As shown in Figure 4E, RPS19 as expected localized to the nucleolus (green), while Val15Phe RPS19 (red) failed to localize to the nucleolus. This finding demonstrates that in addition to Glycine127 in the COOH-terminal NoS, Valine15 in the N-terminal NoS plays a critical role in nucleolar import of RPS19. We also tested the effect of 2 other RPS19 gene mutations found in DBA, Arg56Gln or Arg62Trp, both of which are located in the hot spot of mutations found in DBA patients from codon 52 to codon 62.1 Surprisingly, neither of these 2 mutations altered the nucleolar localization of RPS19 (data not shown). These findings validate the importance of the 2 NoS sequences identified for nucleolar import of RPS19.
Altered subcellular localization of mutant RPS19 characterized in 2 DBA patients. A DBA patient carried the mutation Gly127Gln located in the C-terminal NoS, and another one carried a double missense mutation, Val15Phe + Thr55Met, in which the Val15Phe was located in the first NoS defined in this study. In each panel (A-C, E) the left field shows DAPI nuclear staining, the middle field shows FITC staining of the GFP-RPS19 fusion proteins, and the right field shows merged fields of DAPI + FITC staining. We analyzed the subcellular distribution of RPS19 after transfection of Cos-7 cells with either a GFP-mutated Gly127Gln (panel A), Val15Phe + Thr55Met (panel B), or Val15Phe (panel C) RPS19. Each DBA mutant: Gly127Gln and Val15Phe + Thr55Met impaired RPS19 nucleolar import (panels A-B). In the double mutant, the missense mutation Val15Phe is solely responsible for the impairment of RPS19 nucleolar import since Val15Phe RPS19 failed to localize to the nucleoli (panel C), while Thr55Met RPS19 exhibited normal nucleolar localization (data not shown). Arrows indicate the absence of RPS19 nucleolar staining in the Cos-7 cell shown. (D) In order to rule out a putative effect of the fixation with chilled methanol, RPS19 subcellular distribution was investigated in live transfected cells. The left panel depicts the bright field illumination, and the right panel shows the fluorescent GFP-RPS19 fusion protein. Arrows mark RPS19 nucleolar localization. (E) Green fluorescent fusion protein GFP-WT RPS19 (left panel, FITC staining) and the red fluorescent fusion protein DsRed1-mutated Val15Phe RPS19 (middle panel, Texas red staining) were coexpressed in transfected Cos-7 cells. Normal RPS19 was localized to the nucleoli, while mutant Val15Phe (red) was not imported into the nucleoli (right panel, DAPI + FITC + Texas red staining). Original magnifications: × 63 (A-C,E); × 40 (D).
Altered subcellular localization of mutant RPS19 characterized in 2 DBA patients. A DBA patient carried the mutation Gly127Gln located in the C-terminal NoS, and another one carried a double missense mutation, Val15Phe + Thr55Met, in which the Val15Phe was located in the first NoS defined in this study. In each panel (A-C, E) the left field shows DAPI nuclear staining, the middle field shows FITC staining of the GFP-RPS19 fusion proteins, and the right field shows merged fields of DAPI + FITC staining. We analyzed the subcellular distribution of RPS19 after transfection of Cos-7 cells with either a GFP-mutated Gly127Gln (panel A), Val15Phe + Thr55Met (panel B), or Val15Phe (panel C) RPS19. Each DBA mutant: Gly127Gln and Val15Phe + Thr55Met impaired RPS19 nucleolar import (panels A-B). In the double mutant, the missense mutation Val15Phe is solely responsible for the impairment of RPS19 nucleolar import since Val15Phe RPS19 failed to localize to the nucleoli (panel C), while Thr55Met RPS19 exhibited normal nucleolar localization (data not shown). Arrows indicate the absence of RPS19 nucleolar staining in the Cos-7 cell shown. (D) In order to rule out a putative effect of the fixation with chilled methanol, RPS19 subcellular distribution was investigated in live transfected cells. The left panel depicts the bright field illumination, and the right panel shows the fluorescent GFP-RPS19 fusion protein. Arrows mark RPS19 nucleolar localization. (E) Green fluorescent fusion protein GFP-WT RPS19 (left panel, FITC staining) and the red fluorescent fusion protein DsRed1-mutated Val15Phe RPS19 (middle panel, Texas red staining) were coexpressed in transfected Cos-7 cells. Normal RPS19 was localized to the nucleoli, while mutant Val15Phe (red) was not imported into the nucleoli (right panel, DAPI + FITC + Texas red staining). Original magnifications: × 63 (A-C,E); × 40 (D).
The altered nucleolar import of mutant RPS19 is associated with a dramatic decrease in protein expression
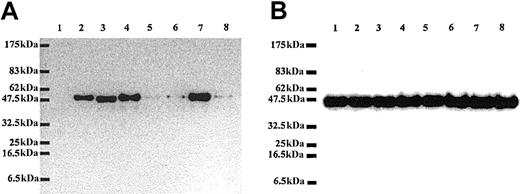
In Western blot analysis, the antibody raised in chicken against mouse recombinant RPS196 was able to recognize the GFP-RPS19 fusion protein expressed in Cos-7 cells but failed to recognize GFP alone, confirming the specificity of our antibody (Figure 5A). Strikingly, we found that the level of expression of RPS19 carrying mutations in the NoS (mutant Val15Phe and mutant Gly127Gln) was dramatically decreased (Figure 5A) compared to the expression level of normal protein. Interestingly, normal levels of expression were noted for certain RPS19 mutations that did not impair nucleolar import (Figure 5A). To rule out the possibility that these findings may be due to differences in the amount of protein loaded, we probed the same samples with a monoclonal antibody against actin. As shown in Figure 5B, similar amounts of endogenous β-actin were observed in all lanes. To assess the possibility that the decrease in expression of mutant RPS19 proteins could result from differences in transfection efficiencies or from an adverse effect of mutant RPS19 proteins on overall protein synthesis, we compared expression level of 2 different GFP fusion proteins coexpressed with either normal or various mutant RPS19. The level of expression of both GFP-4.1R and GFP–ICAM-4 was very similar in Cos-7 cells cotransfected with either normal or various RPS19 mutants (data not shown). These findings imply that neither differences in transfection efficiencies nor alteration in protein synthesis can account for the observed decreased expression in RPS19 proteins with impaired nucleolar distribution.
Dramatic differences in level of expression of normal and mutated RPS19. Lysates of Cos-7 cells transfected with different GFP fusion protein constructs including GFP alone (1), GFP-WT RPS19 (2), GFP-mutated RPS19-Arg56Gln (3), GFP-mutated RPS19-Arg62Trp (4), GFP-mutated RPS19-Val15Phe+Thr55Met (5), GFP-mutated RPS19-Val15Phe (6), GFP-mutated RPS19-Thr55Met (7), and GFP-mutated RPS19-Gly127Gln (8) were analyzed by immunoblotting. We probed the immunoblots with chicken polyclonal anti-RPS19 antibody (panel A) or mouse monoclonal antichicken actin antibody (panel B). RPS19 antibody did not cross-react with GFP (panel A, lane 1), confirming its specificity. In contrast, the antibody detected GFP-WT and mutated RPS19 fusion proteins (panel A, lanes 2-8). The Val15Phe and Gly127Gln mutations, which impaired RPS19 nucleolar import, were associated with marked decrease in RPS19 expression (panel A, lanes 5, 6, 8) compared with normal RPS19 (panel A, lane 2). In contrast, expression of RPS19 with mutations located in the hot spot of mutations found in a large number of DBA patients: Arg56Gln, Arg62Trp (panel A, lanes 3-4, respectively) was the same as normal RPS19. Expression level of endogenous β-actin detected in a duplicate blot was similar in all samples, confirming that the amount of proteins loaded in each lane was comparable (panel B, lanes 1-8). Molecular weight standards (MWs) are shown on the left side of the blots.
Dramatic differences in level of expression of normal and mutated RPS19. Lysates of Cos-7 cells transfected with different GFP fusion protein constructs including GFP alone (1), GFP-WT RPS19 (2), GFP-mutated RPS19-Arg56Gln (3), GFP-mutated RPS19-Arg62Trp (4), GFP-mutated RPS19-Val15Phe+Thr55Met (5), GFP-mutated RPS19-Val15Phe (6), GFP-mutated RPS19-Thr55Met (7), and GFP-mutated RPS19-Gly127Gln (8) were analyzed by immunoblotting. We probed the immunoblots with chicken polyclonal anti-RPS19 antibody (panel A) or mouse monoclonal antichicken actin antibody (panel B). RPS19 antibody did not cross-react with GFP (panel A, lane 1), confirming its specificity. In contrast, the antibody detected GFP-WT and mutated RPS19 fusion proteins (panel A, lanes 2-8). The Val15Phe and Gly127Gln mutations, which impaired RPS19 nucleolar import, were associated with marked decrease in RPS19 expression (panel A, lanes 5, 6, 8) compared with normal RPS19 (panel A, lane 2). In contrast, expression of RPS19 with mutations located in the hot spot of mutations found in a large number of DBA patients: Arg56Gln, Arg62Trp (panel A, lanes 3-4, respectively) was the same as normal RPS19. Expression level of endogenous β-actin detected in a duplicate blot was similar in all samples, confirming that the amount of proteins loaded in each lane was comparable (panel B, lanes 1-8). Molecular weight standards (MWs) are shown on the left side of the blots.
Discussion
DBA, a rare congenital erythroblastopenia, has been shown to be associated with mutations in the RPS19 gene in 25% of affected individuals. The mechanistic understanding of DBA pathophysiology and the role of RPS19 in erythroid cell survival, proliferation, and differentiation are yet to be defined. We have shown previously that RPS19 mRNA and protein expression decrease during terminal erythroid differentiation, an observation consistent with the finding that maturation arrest of erythroid precursors occurs at early stages of erythroid differentiation in DBA.6 In vitro erythroid progenitor cloning efficiency, which is decreased in many DBA patients carrying a mutation in the RPS19 gene, has been reported to increase after wild-type RPS19 gene transduction in CD34+ cells.24 However, there is currently no information on the intracellular localization and trafficking of RPS19 in cells.
Ribosomal protein involvement is well known in ribosome assembly and protein synthesis. Indeed, ribosome biogenesis requires important molecular trafficking between the cytoplasm and the nucleus through nuclear pore complexes (NPC).25-27 After protein synthesis has taken place in the endoplasmic reticulum, ribosomal proteins are imported into the nucleus where they assemble in the nucleolus, along with the 4 rRNA molecules, to build the complex and coordinated structure of the ribosome. They are then re-exported into the cytoplasm as ribosomal subunits to achieve mRNA translation.28-30 We expected that RPS19 would localize in the nucleolus, based on studies of other human and rat ribosomal proteins, which also localized in the nucleolus.18,19,31-35 In the present study, we established the predominant nucleolar localization of both endogenous RPS19 and recombinant GFP-WT RPS19. RPS19 nucleolar localization was confirmed by the colocalization of RPS19 with nucleolin, the major nucleolar protein.15,16,26 We also identified 2 nucleolar localization signals in RPS19, one in the NH2-terminus region and the other in the COOH-terminus of the protein. The localization of NoS in terminal regions also has been observed for other human ribosomal proteins.18,19,32 While nucleolar targeting sequences have been described for few ribosomal proteins, characteristic consensus sequences have not been identified to date.19,31,33-35 We could not identify any obvious common sequence motifs between the 2 RPS19 NoS and the NoS previously identified in other ribosomal proteins.19,31,33-35 In contrast to extensive information available on the mechanisms of nuclear import of proteins, much less is known regarding their nucleolar import.7-13
In order to characterize more precisely the functional relevance of the 2 NoS in RPS19, we analyzed the ability of the 2 NoS, either separately or in combination, to import into the nucleoli pyruvate kinase, a reporter protein known to be expressed exclusively in the cytoplasm.10,14,36 Neither of the 2 NoS, either separately or in combination, was able to target pyruvate kinase to the nucleoli. These findings imply that both sequences are necessary but not sufficient for nucleolar import of RPS19. This finding is similar to the one reported for nucleolin, for which both the glycine/arginine rich (GR) and the RNA-binding (RNP) domains were required for nucleolar targeting.15
It has been suggested that nucleolar targeting of proteins may be a 2-step process: in the first step, a nuclear localization signal mediates the initial translocation of the protein into the nucleus, where it interacts with other nucleolar proteins and/or with rRNA, and in a second step, the protein translocates into the nucleolus.14-19,33,35,37 Potential protein binding partners for the 2 nucleolar targeting sequences in RPS19 in the nucleus are still to be defined. Nucleolin, which shuttles back and forth between the nucleus and the cytoplasm,38 may be a potential binding partner for RPS19 through its RGG domain, as it has been reported for other rat and human ribosomal proteins.39 The colocalization of RPS19 with nucleolin we observed lends some support to this hypothesis.
The regulation of nucleolar import of ribosomal proteins has been shown to be quite complex. For example, RPL22 nucleolar import requires an actual interaction between the 2 NoS characterized at the NH2- and COOH-terminal ends of this ribosomal protein.18 We speculate that RPS19 nucleolar import may also be regulated by an interaction between both RPS19 NoS domains identified in this study. This hypothesis is supported by the fact that neither the NH2-terminal nor the COOH-terminal NoS in RPS19 alone can mediate nucleolar import. Another interesting hypothesis is that efficient nucleolar import of RPS19 may require oligomerization of the protein. Indeed, the second NoS in RPS19, spanning from Gly120 to Asn142, contains 2 potential sites for oligomerization that have been previously identified,40 Lys122 and Gln137.
Strikingly, 2 mutations, Val15Phe and Gly127Gln, identified in 2 DBA patients and located in each of the 2 NoS, perturbed RPS19 nucleolar localization. Each mutant RPS19 could be translocated into the nucleus but not into the nucleolus, confirming that the NLS and the NoS are distinct motifs in RPS19 sequence, as previously reported for NO3814 and for human RPS7.35 The fact that only one amino acid change in RPS19 can alter the nucleolar translocation of a ribosomal protein has precedence in that mutation of either Arg87 or Arg90 in rat ribosomal protein RPL31 was sufficient to prevent nucleolar localization of the protein.31 We hypothesize that for RPS19, Val15 and Gly127 amino acids play a key role in the protein-protein and/or protein-rRNA interactions involved in RPS19 nucleolar localization.
Another major finding was that the RPS19 protein expression level was dramatically (more than 90%) and specifically decreased when RPS19 nucleolar localization was impaired. This finding suggests that the mutant RPS19 is either not synthesized to the same extent as the normal protein or that it is more rapidly degraded because of its inability to be targeted to nucleoli. The fact that the synthesis of other proteins, such as endogenous β-actin and exogenously expressed protein 4.1R and ICAM4, is not decreased is more in favor of increased degradation of mutant RPS19. Incomplete assembly or incorrect distribution of proteins in cellular compartments can lead to the exposure of hydrophobic protein-protein interaction domains, which are recognized and destroyed by chaperones and proteases.41,42
The findings from the present study enabled us to establish for the first time a link between RPS19 mutations identified in DBA and dramatic alterations in both subcellular localization and stability of the mutant protein. These findings represent important clues in deciphering the mechanistic understanding of the role of RPS19 in DBA pathophysiology.
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-12-3878.
Supported by National Institutes of Health grant DK26263 (N.M.); The Daniella Marie Arturi Foundation; The DBA Foundation; la Direction de la Recherche Clinique Assistance Publique-Hôpitaux de Paris (CRC95183) (G.T.); and contracts INSERM/Association Franςaise contre les myopathies (AFM) (RD: 4MR09F and Association pour la recherche contre le cancer [ARC] 5636).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are very grateful to Dr Thiébaut-Noel Willig for the detailed characterization of RPS19 mutations in DBA patients, Dr Sharon Wald Krauss (Lawrence Berkeley National Laboratory [LBNL]) for providing us with the actin antibody and for helpful discussions, Michael Patterson (LBNL) and Marilyn Parra (LBNL) for performing the sequencing of the constructs, Gloria Lee (LBNL) and Dr Xiuli An (New York Blood Center [NYBC]) for providing us with the GFP-ICAM-4 and GFP-protein 4.1R constructs, respectively, and Dr Damir Sudar (LBNL) for his invaluable help in performing confocal microscopy experiments. We would like to acknowledge Dr Gydeon Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia, PA) for providing us with the pcDNA3-c-myc-PK construct and Dr Philippe Bouvet for providing the antibody against hamster nucleolin.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal