Abstract
The Janus kinase Jak1 has been implicated in tumor formation by the Abelson oncogene. In this study we show that loss of Jak1 does not affect in vitro transformation by v-abl as defined by the ability to induce cytokine-independent B-cell colony formation or establishment of B-cell lines. However, Jak1-deficient, v-abl–transformed cell lines were more tumorgenic than wild-type cells when transplanted subcutaneously into severe combined immunodeficient (SCID) mice or injected intravenously into nude mice. Jak1 deficiency was associated with a loss in the ability of interferon-γ (IFN-γ)to induce growth arrest and/or apoptosis of v-abl–transformed pre-B cells or tumor growth in SCID mice. Moreover, IFN-γ mRNA could be detected in growing tumors, and tumor cells explanted from SCID mice had lost the ability to respond to IFN-γ in 9 of 20 cases, whereas the response to interferon-α (IFN-α) remained intact. Importantly, a similar increase in tumorgenicity was observed when IFN-γ–deficient cells were injected into SCID mice, identifying the tumor cell itself as the main source of IFN-γ. These findings demonstrate that Jak1, rather than promoting tumorgenesis as previously proposed, is critical in mediating an intrinsic IFN-γ–dependent tumor surveillance.
Introduction
A variety of important cellular functions, including survival, differentiation, and proliferation, are regulated by cytokines in the hematopoietic system. The Jak (Janus kinase)–Stat (signal transducer and activator of transcription) pathway is one of the important signaling pathways downstream of cytokine receptors.1,2 Following ligand binding to its cognate receptor, receptor-associated Jaks are activated and mediate the subsequent tyrosine phosphorylation and activation of Stat proteins. Activated Stat proteins form dimers, translocate to the nucleus, and bind to specific DNA elements to induce or modulate expression of target genes. Reflecting the critical role of Stats in cell growth and survival, inappropriate Stat activation has been observed in many solid malignancies3-7 as well as in acute and chronic leukemias.8-12 Among the various Stats, constitutive Stat5 activation was observed in different forms of leukemias.13,14 Recent studies15 have established a role for Stat5 activation in the induction of a myeloproliferative and lymphoproliferative disease associated with retroviral transduction of a Tel/Jak2 fusion oncogene into bone marrow cells. Because a comparable myeloproliferative disease is induced by retrovirally transduced oncostatin M (OSM), a target gene of activated Stat5, it is hypothesized that OSM may be the critical target gene of the constitutively activated Stat5 for the induction of the myeloproliferation.
The v-abl gene was originally identified as the transforming gene of the Abelson murine leukemia virus (A-MuLV), a retrovirus that causes lymphosarcomas in mice.16 The v-abl gene is a hybrid of a portion of the viral gag gene fused with a sequence derived from the endogenous c-abl gene, encoding a nonreceptor nuclear tyrosine kinase. V-abl has elevated tyrosine kinase activity and is localized in the cytoplasm. In vitro, A-MuLV transforms fibroblasts and pre–B-lymphoid cell lines, whereas in vivo the virus primarily transforms pre-B cells.17,18 V-abl–transformed primary pre-B cells are poorly tumorgenic and prone to apoptosis.19 These observations suggest that additional genetic and epigenetic changes are required to achieve full malignant transformation. Recent studies have demonstrated a role for the tumor suppressors p53 and p19ARF in progression of transformation.20,21
A critical role for the Jak-Stat pathway in v-abl transformation has been proposed. Initial studies14,22 demonstrated that Stat5 is constitutively activated in V-abl–transformed pre-B cells, leading to the suggestion that Stat5 is critical for transformation. However, using Stat5-deficient mice, we demonstrated in a recent report that Stat5 is not required for v-abl or bcr/abl transformation of pre-B cells.23 In addition, Jak1 has been implicated in transformation by the v-abl as well as by the v-src oncogene.24,25 Based on in vitro observations, it was postulated that transformation by v-abl is dependent on its ability to bind and activate Jak1. In the studies described here, we demonstrate that B-cell precursor cells lacking Jak1 can be transformed upon Abelson infection and give rise to immortal cell lines. In addition, Abelson-infected Jak1-deficient B-cell precursors induce tumor formation in severe combined immunodeficient (SCID) mice at a higher rate and with an earlier onset than Jak1 wild-type cells. Evidence is presented that the higher malignancy is due to the loss of interferon-γ (IFN-γ) sensitivity associated with the Jak1 deficiency, demonstrating that IFN-γ is a key determinant in the progression of tumorgenicity of Abelson-transformed B cells.
Materials and methods
Tissue-culture conditions and virus preparation
Transformed fetal liver cells and cell lines derived from tumor tissue were maintained in RPMI medium containing 5% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin-streptomycin, and 5 μM β-mercaptoethanol. NIH3T3 cells, T220-29 cells (NIH3T3 cells engineered to produce interleukin-7 [IL-7]), and A010 cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% heat-inactivated FCS and 100 U/mL penicillin-streptomycin. A010 cells produce an ecotropic replication-deficient form of the Abelson virus. For collection of the viral supernatant, A010 cells were plated in 100-mm dishes precoated with gelatin (1%) and grown to confluency. Supernatant was harvested every 8 hours for 40 hours, pooled, and filtered through a 0.45-μm filter.
Infection of fetal livers, in vitro transformation assays, and establishment of cell lines
For the preparation of fetal liver cells, heterozygous animals were set up for breeding, and vaginal plugs were checked daily. Sixteen days after conception the pregnant animals were killed and the fetal livers prepared. The embryo tails were used for genotyping by polymerase chain reaction (PCR).26,27 Single-cell suspensions from fetal livers were infected for 30 minutes with viral supernatant derived from A010 cells enriched with 5 ng/mL IL-7, 7 μg/mL polybrene, and 5 μM β-mercaptoethanol. The virus-infected as well as the mock-infected cells were then maintained for 15 to 20 hours on IL-7–producing feeder layers (T220-29 cells).23 Thereafter, cells were washed several times and plated in cytokine-free methylcellulose at a density of 3 × 105/mL in 35-mm dishes. An aliquot of the cells was used to determine numbers of B-cell precursors in the individual fetal livers by fluorescence-activated cell sorter (FACS) analysis. After 7 to 10 days, the cloning efficiency was evaluated by counting colonies using light microscopy. The assays were performed in triplicate. Mock-infected cells did not result in growth factor–independent colonies. The ability to form cell lines was tested by transferring an aliquot of the transduced cells (1 × 106) to growth factor–free medium. The medium was changed twice a week and the culture was observed for the outgrowth of stable clones.
Injection of tumor cells into SCID and nude mice
Ten days after infection, 1 × 106 cells were resuspended into 300 μL phosphate-buffered saline (PBS) and injected subcutaneously into SCID mice.28 At the time point of injection, the cells had been in growth factor–free medium for at least a week and consisted of CD19+/CD43+ pro-B cells. Mice were checked daily for the development of tumors. Tumors bigger than 2 cm in diameter were excised and considered terminal for analysis. The tumors were excised for further analysis. To investigate the effect of IFN-γ treatment on Abelson-induced tumor formation in SCID mice, 1 × 106 cells of 3 different Jak1(+/–) as well as 2 different Jak1(–/–) fetal liver cultures were injected into 28 SCID mice each. Fourteen mice of each group received IFN-γ treatment 4 times a week (5 × 103 units per day) at the site of tumor cell injection.
For tail-vein injections, 3 × 106 cells were resuspended in 200 μL PBS. Cells from 2 independently derived Jak1(–/–) and 2 Jak1(+/–) cell lines were injected in 3 nude mice each. At the time point of injection the cell lines were analyzed by FACS and contained of a homogenous population of CD19+ cells. Twenty-five days after injection all mice were killed, and spleen weights, white blood cell counts, and the presence of CD19+ cells in the bone marrow and blood were analyzed. Age-matched nude mice that were injected with PBS only were used as controls.
Protein analysis
Cells were lysed in a buffer containing protease and phosphatase inhibitors (50 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.5, 0.1% Tween 20, 150 mM NaCl, 1 mM EDTA [ethylenediaminetetraacetic acid], 20 mM β-glycerophosphate, 0.1 mM sodium vanadate, 1 mM sodium fluoride, 10 μg/mL each aprotinin and leupeptin, and 1 mM phenylmethylsulfonyl fluoride [PMSF]).23 Protein concentrations were determined using a BCA-kit as recommended by the manufacturer (Pierce, Rockford, IL). Stat proteins and Jak kinases were immunoprecipitated out of 600 to 800 μg of cell lysate using polyclonal antisera and probed with a monoclonal antibody directed against phosphotyrosine (4G10; UBI, Haup-page, NY). Thereafter, blots were stripped and reprobed with antisera directed against the individual Stat and Jak proteins. In addition, phospho-Stat–specific antiserum (New England Biolabs, Beverly, MA) was used to determine the activity of Stat5 proteins. To assess expression levels of the abl protein, 100 μg total protein per sample was electrophoretically resolved on a 7.5% polyacrylamide gel containing sodium dodecyl sulfate (SDS) and transferred onto Immobilon membranes. Membranes were probed with a rabbit polyclonal antiserum directed against abl (Santa Cruz Biotechnology, Santa Cruz, CA). Sites of antibody binding were detected using protein A–conjugated horseradish peroxidase (EY Laboratories, San Mateo, CA) with chemiluminescent detection (ECL detection kit, Amersham, Arlington Heights, IL).
Gel shift analysis
Electrophoretic mobility shift assays (EMSAs) were performed with whole-cell extracts according to established protocols.29,30 Whole-cell extracts (12 μg protein) from cell lines or tumor samples were incubated with γ[32P]dATP (deoxyadenosine-triphosphate)–end-labeled oligonucleotides in DNA binding buffer (10 mM Tris [tris(hydroxymethyl)aminomethane], 1 mM dithiothreitol [DTT], 0.2 mM PMSF, 0.1 mM EDTA, 5% glycerol, 50 mM NaCl, 0.1% Nonidet P-40 [NP-40], 2 μg/mL bovine serum albumin [BSA], and 0.2 mg/mL poly(dIdC)) in a final volume of 12 μL. To detect Stat5 or Stat6 DNA binding activity, a Stat response element from the bovine β-casein promotor was used, whereas the SIEm67 high-affinity c-fos promoter site was used to visualize Stat1- and Stat3-containing DNA-binding complexes. Antibodies from Santa Cruz Biotechnology were used to verify DNA binding by supershift analysis (anti-Stat5 [N20] and anti-Stat6 [M20]). Samples were analyzed on 5% polyacrylamide gels with 0.25 × Tris-borate-EDTA (TBE) buffer. Gels were finally dried and exposed directly to films.
Flow cytometric analysis (FACS)
Single-cell suspensions of cells are preincubated with αCD16/CD32 antibodies (Pharmingen, San Diego, CA) to prevent nonspecific Fc receptor–mediated binding. Thereafter, aliquots of 5 × 105 cells are stained with monoclonal antibodies conjugated with fluorescent markers and analyzed by FACS (Becton Dickinson, San Diego, CA). The antibodies used for lineage determination included the B-cell lineage markers B220, CD19, CD43; the T-cell markers CD4, CD8, Thy1.2; the myeloid markers Gr1 and Mac1; and the erythroid lineage marker Ter119 (Pharmingen). For cell cycle analysis, the cells were lysed in a buffer containing 0.05 mg/mL propidium iodide, 0.01% Triton X-100, and 0.1% sodium citrate.
Semiquantitative RT-PCR and oligohybridization
First-strand cDNA synthesis and PCR amplification were performed with the Perkin Elmer reverse transcriptase (RT)–PCR kit (GeneAmp; Roche Molecular Systems, Branchburg, NJ) according to the manufacturer's instructions. Primer sequences for the various genes were as follows: mouse IFN-γ mRNA: (forward) 5′-ATGAACGCTACACACTGCATCT-3′ and 5′-CGAATCAGCAGCGACTCCTTTT-3′ (reverse); mouse β-actin: (forward) 5′-ACTCCTATGTGGGTGACGAGG-3′ and 5′-CAGGTCCAGACGCAGGATGGC-3′ (reverse). The reactions were performed using AmpliTaq DNA Polymerase Kit (Roche Molecular Systems) under the following condition: 1.5 mM MgCl2, 200 μM deoxyribonucleoside triphosphates (dNTPs) (each), 1 μg/μL primer mix, and 0.5 units polymerase.
The amplified cDNA was visualized on a 2% agarose gel, denatured with 1.5 M NaCl/0.5 M NaOH, neutralized in 1.5 M NaCl/0.5 M Tris Cl, pH 7.0, and transferred to a membrane using 20 × SSC. Three oligonucleotide probes of IFN-γ were used for hybridization: 5′-GTGACATGAAAATCCTGCAGAGCCAGATTA-3′,5′-TGCAGCTCTTCCTCATGGCTGTTTCTGGCT-3′ (N-terminal), and 5′-ACCAGCTGTTGCCGGAATCCAGCCTC-3′ (C-terminal) and confirmed the specificity of the amplified cDNA band. Activated (IL-2 and anti-CD3) primary T cells were used as positive controls for the experiment. The probes were radiolabeled using polynucleotide kinase, and hybridization was performed using the Rapid-hyb buffer protocol (Amersham).
[3H]Thymidine incorporation
A total of 2 × 105 cells were plated in round-bottom 96-well plates and stimulated with 10 μg/mL IFN-α or IFN-γ (R&D Systems, Minneapolis, MN); 18 hours thereafter, [3H]thymidine was added and incubated for another 12 hours.
Statistical analysis
Statistics were carried out using Student t test and the χ2 test as appropriate.
Results
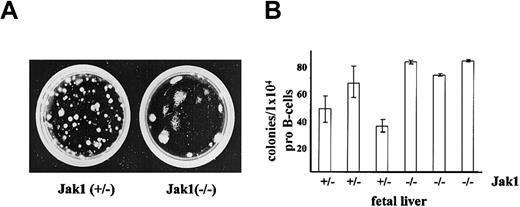
The transforming activity of Ab-MuLV includes the ability to abrogate growth factor requirements of B-cell progenitors. We therefore initially assessed the consequences of Jak1 deficiency on the ability to induce growth factor–independent growth of fetal liver cells in methylcellulose. As illustrated (Figure 1A), large colonies appeared following infection of fetal liver cells from Jak1-deficient embryos with Ab-MuLV. Immunophenotyping of the cells within the colonies identified them as CD19+/B220+ B-lineage cells (data not shown). As previously reported,26 the number of CD19+/B220+ B-cell precursor cells in Jak1-deficient fetal livers is 0.2% to 1% of wild-type fetal livers. When corrected for these differences (Figure 1B), the numbers of colonies per 104 pro-B cells were comparable between Jak1-deficient and wild-type fetal liver cell preparations.
Ab-MuLV–induced colony formation in Jak1(+/–) and Jak1(–/–) cells. (A) Fetal livers were infected with Ab-MuLV and subsequently cloned into cytokine-free methylcellulose at a density of 1 × 105 cells per milliliter. One typical colony-formation assay of each genotype is depicted. (B) Quantification of colony formation expressed as function of available B-cell precursors. B-cell precursors represent the target cell population for Ab-MuLV transformation under our conditions; numbers were determined by FACS analysis. Data represent the mean values ± standard deviation of 3 individually plated colony assays for each fetal liver.
Ab-MuLV–induced colony formation in Jak1(+/–) and Jak1(–/–) cells. (A) Fetal livers were infected with Ab-MuLV and subsequently cloned into cytokine-free methylcellulose at a density of 1 × 105 cells per milliliter. One typical colony-formation assay of each genotype is depicted. (B) Quantification of colony formation expressed as function of available B-cell precursors. B-cell precursors represent the target cell population for Ab-MuLV transformation under our conditions; numbers were determined by FACS analysis. Data represent the mean values ± standard deviation of 3 individually plated colony assays for each fetal liver.
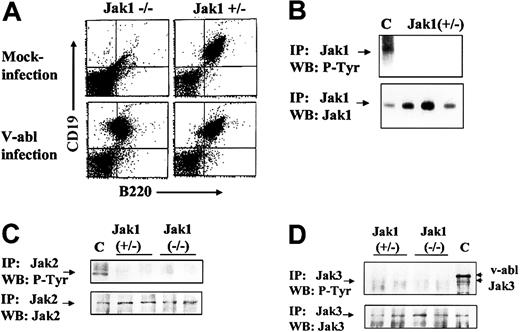
To obtain immortalized, growth factor–independent cell lines, Ab-MuLV–infected fetal liver cells were plated on IL-7–producing feeder layers for 5 days and further amplified under growth factor–free conditions. Cells were passaged twice a week. Ab-MuLV infection of Jak1-deficient fetal liver cultures resulted in the consistent establishment of cell lines (9 of 9), while infection of heterozygous fetal liver cultures gave rise to cell lines in 8 of 11 cultures (Table 1). In all cases, the cell lines obtained were CD19+/B220+ B cells (Figure 2). Cell cycle analysis did not reveal differences in cell cycle distribution between Jak1-deficient and wild-type transformed B cells (data not shown). Moreover, the doubling rates of the transformed B-cell lines were comparable (30 ± 7 hours Jak1-deficient, n = 3; 25.7 ± 2.3 hours wild-type, n = 4). We also investigated whether Jak2 or Jak3 was constitutively active in Jak1(–/–) and Jak1(+/–) cells and might compensate for the loss of Jak1. We failed to detect tyrosine phosphorylation of Jak1 in wild-type cells (Figure 2B). Also, neither Jak2 nor Jak3 was tyrosine phosphorylated in any cell line investigated (Figure 2B). Therefore, we conclude that the absence of Jak1 does not affect the ability of Ab-MuLV to transform pre-B cells or alter the growth properties of the transformed cell lines relative to wild-type cells.
Ab-MuLV induces tumorgenic cell lines in Jak1-deficient fetal liver cells
Jak1 genotype of the fetal liver . | Outgrowth of cell line . | No. of tumors per injected SCID mouse . |
|---|---|---|
| +/- | - | 0/3 |
| +/- | + | 3/3 |
| +/- | - | 0/3 |
| +/- | + | 0/4 |
| +/- | + | 0/4 |
| +/- | + | 3/4 |
| +/- | + | 4/4 |
| +/- | + | 3/4 |
| +/- | - | 0/4 |
| +/- | + | 2/4 |
| +/- | + | 2/4 |
| -/- | + | 2/3 |
| -/- | + | 2/3 |
| -/- | + | 3/3 |
| -/- | + | 4/4 |
| -/- | + | 1/4 |
| -/- | + | 4/4 |
| -/- | + | 4/4 |
| -/- | + | 4/4 |
| -/- | + | 4/4 |
Jak1 genotype of the fetal liver . | Outgrowth of cell line . | No. of tumors per injected SCID mouse . |
|---|---|---|
| +/- | - | 0/3 |
| +/- | + | 3/3 |
| +/- | - | 0/3 |
| +/- | + | 0/4 |
| +/- | + | 0/4 |
| +/- | + | 3/4 |
| +/- | + | 4/4 |
| +/- | + | 3/4 |
| +/- | - | 0/4 |
| +/- | + | 2/4 |
| +/- | + | 2/4 |
| -/- | + | 2/3 |
| -/- | + | 2/3 |
| -/- | + | 3/3 |
| -/- | + | 4/4 |
| -/- | + | 1/4 |
| -/- | + | 4/4 |
| -/- | + | 4/4 |
| -/- | + | 4/4 |
| -/- | + | 4/4 |
Ab-MuLV infection induces outgrowth of B-cell precursors in Jak1(–/–)fetal livers. (A) Fetal livers were split in half, with one half mock infected and the other half infected with Ab-MuLV, and transferred onto IL-7–producing feeder cultures (T220-29 cells). Six days thereafter, the cells were analyzed for the presence of CD19/CD43 B cells by FACS analysis. (B) Immunoprecipitation of Jak1 in wild-type cells. Three individual Jak1(+/–) cell lines were analyzed for the expression and tyrosine phosphorylation (P-Tyr) of Jak1. C indicates v-abl–transformed cells stimulated with 10 ng/mL interleukin-7. (C) Immunoprecipitation of Jak2. Two Jak1(–/–) and 2 Jak1(+/–) cell lines were used to investigate the expression and tyrosine phosphorylation of Jak2. As a control (indicated by C), v-abl–transformed Jak1(+/–) cells were stimulated with 10 ng/mL IFN-γ. (D) Immunoprecipitation of Jak3. Two Jak1(–/–) and 2 Jak1(+/–) cell lines were used to investigate the expression and tyrosine phosphorylation of Jak3. As a control (indicated by C), v-abl–transformed Jak1(+/–) cells were stimulated with 10 ng/mL interleukin-7. The upper strongly tyrosine-phosphorylated band corresponds to v-abl that associates with Jak3 upon activation.
Ab-MuLV infection induces outgrowth of B-cell precursors in Jak1(–/–)fetal livers. (A) Fetal livers were split in half, with one half mock infected and the other half infected with Ab-MuLV, and transferred onto IL-7–producing feeder cultures (T220-29 cells). Six days thereafter, the cells were analyzed for the presence of CD19/CD43 B cells by FACS analysis. (B) Immunoprecipitation of Jak1 in wild-type cells. Three individual Jak1(+/–) cell lines were analyzed for the expression and tyrosine phosphorylation (P-Tyr) of Jak1. C indicates v-abl–transformed cells stimulated with 10 ng/mL interleukin-7. (C) Immunoprecipitation of Jak2. Two Jak1(–/–) and 2 Jak1(+/–) cell lines were used to investigate the expression and tyrosine phosphorylation of Jak2. As a control (indicated by C), v-abl–transformed Jak1(+/–) cells were stimulated with 10 ng/mL IFN-γ. (D) Immunoprecipitation of Jak3. Two Jak1(–/–) and 2 Jak1(+/–) cell lines were used to investigate the expression and tyrosine phosphorylation of Jak3. As a control (indicated by C), v-abl–transformed Jak1(+/–) cells were stimulated with 10 ng/mL interleukin-7. The upper strongly tyrosine-phosphorylated band corresponds to v-abl that associates with Jak3 upon activation.
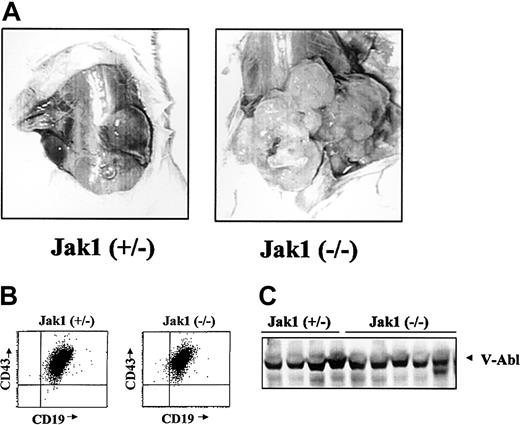
We next wanted to examine the ability of the Ab-MuLV– transformed B cells to form tumors in SCID mice as an in vivo tumor system. Cells were infected and cultured on IL-7 producer cells for 5 days and in the absence of growth factors for 5 days. Following this, the cultures contained a homogeneous population of CD43+/CD19+/B220+ cells. Injection of 1 × 106 cells resulted in the formation of tumors of at least 2 cm within 3 weeks in 17 of 41 animals injected with Jak1(+/–) fetal liver cells (Table 1). In comparison, 28 of 33 animals developed tumors of at least 2 cm within 1 to 3 weeks when Jak1-deficient, Ab-MuLV–infected fetal liver cells were used (P < .001, χ2 test). In addition to an earlier onset, transformed Jak1-deficient cells developed to visibly larger tumors (Figure 3A). Among the 7 Jak1 heterozygous and 10 Jak1-deficient tumors examined, all consisted of homogenous populations of CD43+/CD19+ B cells (Figure 3B) and expressed V-abl protein (Figure 3C).
Analysis of Abelson-transformed Jak1(+/–) and Jak1(–/–) tumors. (A) Tumor formation in SCID mice after injection with 3 × 106 transformed cells. Jak1(–/–) cells form tumors more rapidly than the Jak1(+/–) control cells. One representative tumor of each genotype is shown. (B) FACS analysis of the excised tumors. All tumors investigated express the surface markers CD19, CD43, and B220 and were therefore classified as B-cell lineage tumors. One example for each genotype is shown. (C) Analysis of v-abl expression in tumor tissue by Western blot analysis. The first 4 lanes show tumor tissue derived from Jak1(+/–)–transformed cells; the last 5 lanes show tumors derived from Jak1(–/–)–transformed cells.
Analysis of Abelson-transformed Jak1(+/–) and Jak1(–/–) tumors. (A) Tumor formation in SCID mice after injection with 3 × 106 transformed cells. Jak1(–/–) cells form tumors more rapidly than the Jak1(+/–) control cells. One representative tumor of each genotype is shown. (B) FACS analysis of the excised tumors. All tumors investigated express the surface markers CD19, CD43, and B220 and were therefore classified as B-cell lineage tumors. One example for each genotype is shown. (C) Analysis of v-abl expression in tumor tissue by Western blot analysis. The first 4 lanes show tumor tissue derived from Jak1(+/–)–transformed cells; the last 5 lanes show tumors derived from Jak1(–/–)–transformed cells.
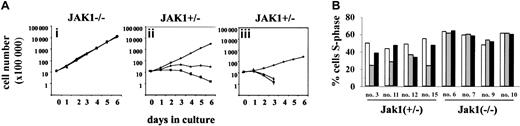
Because of the consistently increased tumorgenic nature of the Ab-MuLV–transformed Jak1-deficient cells, we reasoned that a Jak1-dependent signaling pathway may partially limit the growth of wild-type transformed cells. Potential cytokine signaling pathways associated with the inhibition of cell growth are initiated by interferons. We therefore examined the effects of IFN-α or IFN-γ on the growth of Ab-MuLV–transformed B-cell lines. As illustrated in Figure 4A, the growth of cell lines heterozygous for Jak1 deficiency was strongly inhibited by IFN-α or IFN-γ. In contrast, the growth of Jak1-deficient cell lines was unaffected. Similarly, the cell cycle distribution pattern of freshly explanted tumor cells from Jak1-deficient tumors showed no response to IFN treatment (Figure 4B). Interestingly, when we analyzed primary tumor cells derived from Jak1 heterozygous tumors, we found that 9 of 20 had lost the ability to respond to IFN-γ with growth inhibition, whereas only 1 of 20 showed an impaired ability to respond to IFN-α (P < .01, χ2 test) (Figure 4B).
Ab-MuLV–transformed Jak1(+/–) cells react with cell death or growth arrest to IFN treatment. (A) Growth curves for Jak1(–/–) cells in the absence and presence of IFN-α or IFN-γ (i). Growth curves for Jak1(+/–) cells in the absence and presence of IFN-α or IFN-γ. Cells react either with growth inhibition (ii) or cell death (iii) to IFN treatment. ⋄ indicates control; ▪, IFN-α; and ▴, IFN-γ. (B) Tumor-derived cells from Jak1(+/–) and Jak1(–/–) tumors were explanted in culture in the absence and presence of IFN-α or IFN-γ; 24 hours thereafter, the cell cycle distribution was analyzed. In 9 of 20 samples investigated, cells explanted from Jak1(+/–) tumors had at least partially lost the ability to react to IFN-γ, but only one tumor sample lost the ability to react with growth arrest to IFN-α treatment. None of the Jak1(–/–)–deficient cells reacted to IFN treatment. □ indicates control; ▦, IFN-α; and ▪, IFN-γ. Four tumor samples of each group are shown.
Ab-MuLV–transformed Jak1(+/–) cells react with cell death or growth arrest to IFN treatment. (A) Growth curves for Jak1(–/–) cells in the absence and presence of IFN-α or IFN-γ (i). Growth curves for Jak1(+/–) cells in the absence and presence of IFN-α or IFN-γ. Cells react either with growth inhibition (ii) or cell death (iii) to IFN treatment. ⋄ indicates control; ▪, IFN-α; and ▴, IFN-γ. (B) Tumor-derived cells from Jak1(+/–) and Jak1(–/–) tumors were explanted in culture in the absence and presence of IFN-α or IFN-γ; 24 hours thereafter, the cell cycle distribution was analyzed. In 9 of 20 samples investigated, cells explanted from Jak1(+/–) tumors had at least partially lost the ability to react to IFN-γ, but only one tumor sample lost the ability to react with growth arrest to IFN-α treatment. None of the Jak1(–/–)–deficient cells reacted to IFN treatment. □ indicates control; ▦, IFN-α; and ▪, IFN-γ. Four tumor samples of each group are shown.
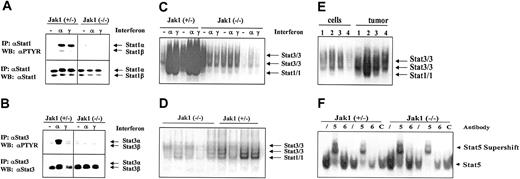
A potential mediator of IFN-induced inhibition of cell growth is Stat1. As illustrated in Figure 5A,C, IFN-α and IFN-γ strongly induced the tyrosine phosphorylation and DNA binding of Stat1 in transformed cell lines heterozygous for Jak1 while no activation was seen in Jak1-deficient cell lines. For comparison, all the cell lines contained low levels of constitutively activated Stat3 that was further activated by IFN-α stimulation in the cell lines heterozygous for Jak1 (Figure 5B-C). However, we detected constitutive Stat1 activation in tumor tissue, where it was clearly more prominent in the tumor samples heterozygous for Jak1 compared with the Jak1-deficient samples (Figure 5D). A direct comparison of cell lines and the corresponding primary tumor tissue side by side on the same gel confirmed our observation (Figure 5E). Supershift experiments with antibodies directed against Stat1 and Stat3 in wild-type and Stat1-deficient v-abl–transformed pro-B cells defined the bands corresponding to Stat1 and Stat3 activation (data not shown). The difference between cell lines and primary tumor tissue is specific for Stat1; other members of the Stat transcription factor family showed no distinct pattern of regulation. Stat5 was activated in cell lines (Figure 5F) and tumor tissue (data not shown) irrespective of whether Jak1 was expressed or not, whereas Stat6 activation was found in none of the samples investigated.
Stat activation in tumors and cell lines. (A-B) Immunoprecipitation of Stat1 and Stat3. Cell lines were either untreated (–), or stimulated with IFN-α (α), or stimulated with IFN-γ (γ) for 1 hour. Thereafter the cells were lysed and the Stat proteins immunoprecipitated. The membranes were first blotted with an antibody directed against phosphotyrosine (PTYR) and then destripped and reprobed with antisera directed against Stat1 (A) or Stat3 (B). (C) Gel shift analysis for Stat1 and Stat3 in the cell lines treated as described above for panels A and B. The specificity of the Stat1-containing complexes was proven by using Abelson-transformed B-cell lines derived from Stat1-deficient fetal livers (not shown). (D) Five tumors derived from Ab-MuLV–transformed Jak1(–/–) cells (first 5 lanes) and 4 tumors from Ab-MuLV–transformed Jak1(+/–) cells (last 4 lanes) were analyzed by gel shift for Stat1 and Stat3 activation. (E) Gel shift analysis for Stat1 and Stat3 activation in cell lines and the corresponding primary tumors. Samples 1 and 4 are deficient for Jak1; samples 2 and 3 are derived from Jak1(+/–) fetal livers. (F) Stat5 and Stat6 activation in Jak1(+/–) and Jak1(–/–) cell lines. Gel shift analysis and supershift analysis with antibodies directed against Stat5 (5) and Stat6 (6) shows activation of Stat5 but not Stat6. An unrelated antibody (C) was used as additional control. Slash indicates that no antibody was added.
Stat activation in tumors and cell lines. (A-B) Immunoprecipitation of Stat1 and Stat3. Cell lines were either untreated (–), or stimulated with IFN-α (α), or stimulated with IFN-γ (γ) for 1 hour. Thereafter the cells were lysed and the Stat proteins immunoprecipitated. The membranes were first blotted with an antibody directed against phosphotyrosine (PTYR) and then destripped and reprobed with antisera directed against Stat1 (A) or Stat3 (B). (C) Gel shift analysis for Stat1 and Stat3 in the cell lines treated as described above for panels A and B. The specificity of the Stat1-containing complexes was proven by using Abelson-transformed B-cell lines derived from Stat1-deficient fetal livers (not shown). (D) Five tumors derived from Ab-MuLV–transformed Jak1(–/–) cells (first 5 lanes) and 4 tumors from Ab-MuLV–transformed Jak1(+/–) cells (last 4 lanes) were analyzed by gel shift for Stat1 and Stat3 activation. (E) Gel shift analysis for Stat1 and Stat3 activation in cell lines and the corresponding primary tumors. Samples 1 and 4 are deficient for Jak1; samples 2 and 3 are derived from Jak1(+/–) fetal livers. (F) Stat5 and Stat6 activation in Jak1(+/–) and Jak1(–/–) cell lines. Gel shift analysis and supershift analysis with antibodies directed against Stat5 (5) and Stat6 (6) shows activation of Stat5 but not Stat6. An unrelated antibody (C) was used as additional control. Slash indicates that no antibody was added.
To examine the influence of IFN-γ on tumor growth, 1 × 106 Ab-MuLV–transformed fetal liver cells, prepared as described above, were injected into SCID mice. Concomitantly, the tumor sites were injected with IFN-γ (5 × 103 units per day; 4 days per week). Among 14 SCID mice injected with Jak1-deficient transformed B cells, 13 developed tumors after 2 to 3 weeks irrespective of whether they were treated with IFN-γ or not (Table 2). In contrast, Jak1(+/–) cells gave rise to tumors in 8 of 14 untreated animals. Here only 1 of 14 injected mice, treated with IFN-γ, developed tumors (P < .01, χ2 test).
Tumor sensitivity to IFN-γ requires Jak1
Genotype . | No. of tumors without treatment . | No. of tumors with IFN-γ treatment . |
|---|---|---|
| Jak1 (+/-) | 8 of 14 | 1 of 14 |
| Jak1 (-/-) | 13 of 14 | 14 of 14 |
Genotype . | No. of tumors without treatment . | No. of tumors with IFN-γ treatment . |
|---|---|---|
| Jak1 (+/-) | 8 of 14 | 1 of 14 |
| Jak1 (-/-) | 13 of 14 | 14 of 14 |
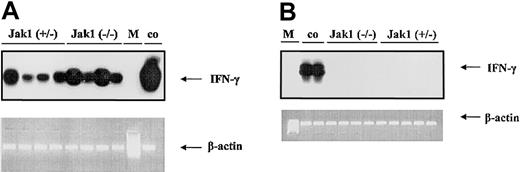
To test the hypothesis that IFN-γ is the cytokine responsible for the difference in tumor onset in nude mice, we first investigated whether IFN-γ is expressed during tumor development. We could clearly detect IFN-γ in samples derived from tumor tissues (Figure 6A), but no signal was detected in samples derived from cell lines (Figure 6B).
V-abl–transformed tumors express IFN-γ. RT-PCR and subsequent oligohybridization for IFN-γ from Jak1(+/–)– and Jak1(–/–)–derived cell lines (B) and tumor tissues (A). Representative examples of the analysis are shown in the top panels (“middle” oligo). Activated (IL-2 and anti-CD3) primary T lymphocytes were used as positive control (co); M indicates base-pair marker. Only the tumors—but not the cell lines—show a specific signal for IFN-γ. In the lower panels, an RT-PCR for β-actin is depicted to control for the prepared cDNA.
V-abl–transformed tumors express IFN-γ. RT-PCR and subsequent oligohybridization for IFN-γ from Jak1(+/–)– and Jak1(–/–)–derived cell lines (B) and tumor tissues (A). Representative examples of the analysis are shown in the top panels (“middle” oligo). Activated (IL-2 and anti-CD3) primary T lymphocytes were used as positive control (co); M indicates base-pair marker. Only the tumors—but not the cell lines—show a specific signal for IFN-γ. In the lower panels, an RT-PCR for β-actin is depicted to control for the prepared cDNA.
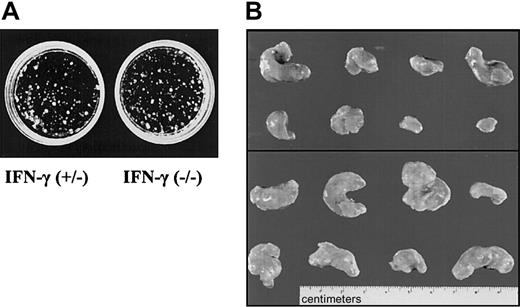
We reasoned that the most likely source for IFN-γ, in the context of tumors growing in SCID mice, would be the tumor cells themselves. Therefore, fetal liver cells from IFN-γ–deficient or control embryos were transformed with Ab-MuLV. Cloning experiments showed no differences between IFN-γ(+/–) and IFN-γ(–/–) colony-formation capability (Figure 7A). Infection gave rise to cell lines in 9 of 10 cultures when IFN-γ–deficient fetal livers cells were used. Comparably, cell lines were established from 8 of 10 cultures of fetal liver cells heterozygous for IFN-γ. Injection of the cells into SCID mice resulted in tumors with an average size of 0.8 ± 0.4 g in 7 of 12 mice when cells heterozygous for IFN-γ were used (Table 3, Figure 7B). In contrast, 11 of 12 mice developed tumors with an average size of 1.7 ± 0.8 g when IFN-γ–deficient cells were injected. This difference meets the criteria of being a statistically significant result (P < .05). In addition, when newborn mice that are deficient for IFN-γ and wild-type control mice were injected with replication-incompetentAb-MuLV, significant differences were observed between the 2 groups (B.K., unpublished data, 2002). However, this experiment does not allow us to distinguish between the impact of IFN-γ in the immune system and the impact of IFN-γ produced by the tumor cells themselves.
IFN-γ deficiency reflects the phenotype of Jak1 deficiency. (A) Colony formation of Ab-MuLV–transformed fetal liver cells from IFN-γ(+/–) and IFN-γ(–/–) embryos. No differences were observed. One representative experiment of each genotype is shown. (B) Tumor formation in SCID mice. A total of 1 × 106 transformed cells were injected. The top panel shows the 8 tumors that developed upon injection of IFN-γ(+/–) cells. The bottom panel depicts 8 of the 13 tumors that developed out of IFN-γ(–/–) cells.
IFN-γ deficiency reflects the phenotype of Jak1 deficiency. (A) Colony formation of Ab-MuLV–transformed fetal liver cells from IFN-γ(+/–) and IFN-γ(–/–) embryos. No differences were observed. One representative experiment of each genotype is shown. (B) Tumor formation in SCID mice. A total of 1 × 106 transformed cells were injected. The top panel shows the 8 tumors that developed upon injection of IFN-γ(+/–) cells. The bottom panel depicts 8 of the 13 tumors that developed out of IFN-γ(–/–) cells.
Absence of IFN-γ potentiates growth of Ab-MuLV–induced tumors
Genotype . | No. of tumors . | Average tumor weight . |
|---|---|---|
| IFN-γ (+/-) | 7 of 12 | 0.8 ± 0.4 g |
| IFN-γ (-/-) | 11 of 12 | 1.7 ± 0.8 g |
Genotype . | No. of tumors . | Average tumor weight . |
|---|---|---|
| IFN-γ (+/-) | 7 of 12 | 0.8 ± 0.4 g |
| IFN-γ (-/-) | 11 of 12 | 1.7 ± 0.8 g |
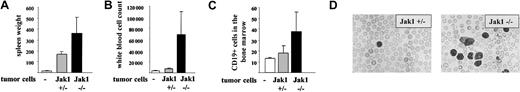
Another experimental approach is the injection of tumor cells in the tail vein of nude mice. This approach addresses the impact of the IFN-γ signaling cascade for the tumor cells and minimizes any interference with the immune system. Two individually developed cell lines of each genotype [Jak1(–/–) versus (+/–)] were injected. The experiment was terminated after 25 days, when the mice started to develop signs of disease. At that time, 5 of the 6 mice that had received Jak1(–/–) cells had developed multiple enlarged lymph nodes that exceeded 5 mm in diameter. In contrast, only one of the animals that had received Jak1(+/–) cells showed enlarged lymph nodes. The spleen was enlarged about 10-fold in animals that received Jak1(+/–) tumor cells and about 25-fold in animals that had obtained Jak1(–/–) tumor cells (Figure 8A). Similarly, the infiltration of the bone marrow with Jak1(–/–) tumor cells was doubled compared with the infiltration with Jak1(+/–) cells (Figure 8B). The most dramatic difference was observed in white blood cell counts. A 5- to 6-fold elevation in white blood cell counts was found when Jak1(–/–) tumor cells were injected, but only a 2-fold increase was found in Jak1(+/–)–injected animals (Figure 8C-D). All comparisons meet the criteria of being statistically significant (P < .05). Taken together these data clearly show that the tumor-promoting effect of Jak1 deficiency is not limited to subcutaneous tumor formation but extends to the development of leukemia.
Tail-vein injection of Jak1(–/–) cells results in a more severe form of leukemia than injection of Jak1(+/–) cells. A total of 3 × 106 tumor cells were injected into nude mice via the tail vein, and the mice were analyzed 25 days thereafter. Four nude mice that received PBS only were used as controls. Data show mean values ± SD (n = 6 for tumor cell injections and n = 4 for control mice). (A) Spleen weight of the mice at the time point of analysis. (B) White blood cell count and (C) number of CD19+ cells in the bone marrow at the time point of analysis. (D) Blood smear of one Jak1(–/–)– and one Jak1(+/–)–injected nude mouse. Hematoxylin and eosin stain, original magnification, × 100.
Tail-vein injection of Jak1(–/–) cells results in a more severe form of leukemia than injection of Jak1(+/–) cells. A total of 3 × 106 tumor cells were injected into nude mice via the tail vein, and the mice were analyzed 25 days thereafter. Four nude mice that received PBS only were used as controls. Data show mean values ± SD (n = 6 for tumor cell injections and n = 4 for control mice). (A) Spleen weight of the mice at the time point of analysis. (B) White blood cell count and (C) number of CD19+ cells in the bone marrow at the time point of analysis. (D) Blood smear of one Jak1(–/–)– and one Jak1(+/–)–injected nude mouse. Hematoxylin and eosin stain, original magnification, × 100.
Discussion
Our studies were initially predicated on recent studies24 that demonstrated an in vitro interaction of Jak1 and a carboxyl region of V-abl. Mutations within the Jak1 interacting domain of V-abl that disrupt the interaction fail to activate Jak1/Stat proteins. Cells expressing this mutant showed an extended latency and decreased ability to form tumors in nude mice. The potential direct involvement of Jak1 in v-abl transformation was suggested by the observation that a kinase-inactive, dominant-negative form of Jak1 could block v-abl–induced proliferation of BAF/3 cells. Collectively, the experiments led the authors to conclude that Jak1 may be important for the efficient transformation of immature B cells by the v-abl oncogene.
Another approach to assess the role of Jak1 in v-abl transformation is to assess the ability of v-abl to transform cells lacking Jak1. As illustrated here, the lack of Jak1 does not decrease v-abl–induced colony formation or affect the outgrowth of cytokine-independent cell lines. Therefore, v-abl–induced transformation of pre-B cells is not dependent on the presence of Jak1.
In contrast to a requirement for Jak1 for v-abl transformation, a deficiency in Jak1 facilitated tumor formation and resulted in a higher malignancy upon injection into SCID mice independently of whether the cells were injected subcutaneously or in the bloodstream of the animals. Because Jak1-deficient pre-B cells have unaltered cell cycle profiles and proliferation behavior compared with Jak1 heterozygous cells, an accelerated cell growth cannot account for the increased tumor mass or early onset of tumor formation following injection into SCID mice. Hence, we investigated the possibility that there was an altered responsiveness to cytokines that might suppress cell growth through a signaling pathway requiring Jak1. One candidate was IFN-γ because binding of IFN-γ to the IFN-γ receptor (IFN-γR) induces activation of Jak1, and fibroblasts lacking Jak1 are completely deficient in IFN-γ responses.26,31,32
IFN-γ is a strong inhibitor of pro-B cell proliferation and viability.33 Treatment of Jak1(+/–)–transformed cell lines with IFN-γ led to a complete growth arrest or induced apoptosis in all Jak1(+/–) cells within 3 days, whereas it had no effect on Jak1(–/–) cell lines. This finding is in line with the observation that overexpression of IFN-γ in mice leads to a severe B-cell lineage reduction.34 Moreover, even low doses of IFN-γ led to a nearly complete suppression of tumor growth in SCID mice but did not alter tumor initiation or progression by Jak1-deficient cells. Because SCID mice lack functional T-lymphoid cells, the major effect of IFN-γ is a direct growth inhibitory or proapoptotic effect on the tumor cells.28
Most interestingly, IFN-γ–deficient tumor cells reflect the phenotype observed within Jak1-deficient cells. Hence, we conclude that the source of IFN-γ within the growing tumor is the tumor cell itself, thereby limiting its own proliferation. Several reports show that B-lymphoid cells can express IFN-γ under certain conditions in vivo.35-38 Loss of the ability to produce IFN-γ resulted in an increased rate of tumors with an accelerated growth upon injection into SCID mice identical to Jak1 deficiency. It is currently unclear which stimuli induce IFN-γ production in the growing tumor.
IFN-γ activates the transcription factor Stat1; IFN-α activates Stat1, Stat2, as well as Stat3.39 The finding that Stat1 was not constitutively activated in the cell lines growing in tissue culture, but at various degrees in the primary Jak1(+/–) tumors, is consistent with the permanent presence of IFN-γ within the growing tumor. Recent evidence suggests that Stat3 is a transforming agent or is an essential contributor for tumors of various origins.3,40 In Abelson-induced malignancies, Stat3 is strongly activated by IFN-α treatment and is speculated to be linked to a growth-suppressing activity.
Nevertheless, IFN-γ but not IFN-α is the cytokine involved in endogenous tumor surveillance. We find a predominant activation of Stat1 but not Stat3 in primary tumors. Furthermore, 9 of 20 primary Jak1(+/–)–derived tumors had either completely or partially lost the ability to react with growth arrest or cell death to treatment with IFN-γ. In contrast, 19 of 20 tumors retained the ability to respond to IFN-α. This finding indicates the selective advantage for tumor cell clones that lose responsiveness to IFN-γ and argues for a tumor surveillance effect of IFN-γ but not IFN-α within the growing tumor. Further analysis is needed to investigate the mechanisms by which the tumor cells escape the growth inhibitory effects by IFN-γ.
A recent study in which p53-deficient mice were crossed to an IFN-γ–deficient background initially revealed the importance of IFN-γ for tumor surveillance. Mice lacking both p53 and IFN-γ suffered from premature tumor formation with a shifting of the tumor spectrum toward teratomas, sarcomas, and hemangiomas as compared with the p53(–/–) knock-outs.41 The same authors analyzed human tumor cell lines and found a loss of IFN-γ responsiveness in about 33% of adenocarcinoma cell lines. The central role for IFN-γ in tumor surveillance against carcinogen-induced sarcomas and spontaneous epithelial carcinomas has also recently be pinpointed by observations made in mice deficient for RAG2 and/or IFN-γ receptor.42
Here we show that IFN-γ is also significant for the development of leukemia and lymphoma. Growth advantages may be conferred by deficiencies in the IFN-γ receptor–Jak-Stat signaling cascade as well as by the production of IFN-γ itself. A recent report investigating acute myeloid leukemia samples supports our finding. The authors have shown that in 10 of 25 samples deletion of Jak kinases could be detected.43
Taken together, we conclude that IFN-γ produced by the tumor cells themselves exerts an important role for tumor surveillance by a direct growth inhibitory effect. This effect is mediated via activation of Jak1 that exerts a tumor-suppressing function in Abelson-transformed pro-B cells. More detailed studies of human leukemia cells will be necessary to elucidate the diagnostic and prognostic value of alterations in the IFN-γ signaling cascade.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2001-11-0142.
Supported by grant P15033 of the FWF (Fonds zur Förderung der Wissenschaftlichen Forschung) and grant 2032 of the Vienna Research Council to V.S. and by Cancer Center CORE grant (CA 21765) and ALSAC (American Lebanese Syrian Associated Charities) to J.N.I.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Udo Losert and the staff of the Biomedical Research Institute, University of Vienna, for taking care of our mice. We also thank Naomi Rosenberg, Martine Roussel, and David Levy for the generous gift of A010 cells, T220-29 cells, and Stat1-deficient mice, respectively. We are grateful to John Cleveland, Martine F. Roussel, Michael Freissmuth, and Christian Sillaber for helpful discussions in the course of this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal