Abstract
The idiotypic structure of the monoclonal immunoglobulin (Ig) in multiple myeloma (MM) might be regarded as a tumor-specific antigen. The present study was designed to identify T-cell epitopes of the variable region of the Ig heavy chain (VH) in MM (n = 5) using bioinformatics and analyze the presence of naturally occurring T cells against idiotype-derived peptides. A large number of human-leukocyte-antigen (HLA)–binding (class I and II) peptides were identified. The frequency of predicted epitopes depended on the database used: 245 in bioinformatics and molecular analysis section (BIMAS) and 601 in SYFPEITHI. Most of the peptides displayed a binding half-life or score in the low or intermediate affinity range. The majority of the predicted peptides were complementarity-determining region (CDR)–rather than framework region (FR)–derived (52%-60% vs 40%-48%, respectively). Most of the predicted peptides were confined to the CDR2-FR3-CDR3 “geographic” region of the Ig-VH region (70%), and significantly fewer peptides were found within the flanking (FR1-CDR1-FR2 and FR4) regions (P < .01). There were 8– to 10–amino acid (aa) long peptides corresponding to the CDRs and fitting to the actual HLA-A/B haplotypes that spontaneously recognized, albeit with a low magnitude, type I T cells (interferon γ), indicating an ongoing major histocompatibility complex (MHC) class I–restricted T-cell response. Most of those peptides had a low binding half-life (BIMAS) and a low/intermediate score (SYFPEITHI). Furthermore, 15- to 20-aa long CDR1-3–derived peptides also spontaneously recognized type I T cells, indicating the presence of MHC class II–restricted T cells as well. This study demonstrates that a large number of HLA-binding idiotypic peptides can be identified in patients with MM. Such peptides may spontaneously induce a type I MHC class I– as well as class II–restricted memory T-cell response.
Introduction
Multiple myeloma (MM) is a lymphoproliferative disorder characterized by clonal proliferation of B cells producing a monoclonal immunoglobulin (Ig) that can be detected in serum and/or urine. The idiotypic Ig is a unique tumor antigen of the myeloma cell clone presented as complete Ig molecules on the tumor cell surface1 but also as peptides in the peptide-binding cleft of major histocompatibility complex (MHC) molecules.2,3
Naturally occurring T cells reactive with the idiotype of the autologous monoclonal Ig have been described in murine plasmo-cytoma4 and in human monoclonal gammopathies (MM and monoclonal gammopathy of undetermined significance).5,6 Cytotoxic activity of idiotype-specific T cells against autologous human myeloma plasma cells has also been reported.7,8 Preliminary data in patients with B-cell malignancies indicate that the T-cell reactivity may be directed against unique parts of the idiotype (ie, somatically mutated complementarity-determining regions [CDRs]), especially sequences deduced from the heavy-chain variable diversity-joining part (VH-D-JH) CDR3 region.9-11 However, peptides derived from the framework regions (FRs) may also express cytotoxic T-cell epitopes.12
This study was designed to predict T-cell epitopes of the variable region of the Ig heavy chain from patients with MM using bioinformatics and to analyze functional properties of such peptides, with the aim to identify regions of interest for targeting that might form a basis for production of subunit idiotype (Id) vaccines. Active immunization using the autologous idiotype has been shown to induce an idiotype-specific cellular response, and tumor regression has been observed in non-Hodgkin lymphoma (NHL)13 and MM.14
Patients, materials, and methods
Patients
Included in the study were 5 patients with MM of IgG type. This study was approved by the Ethical Committee of Karolinska Institutet. Informed consent was provided according to the Declaration of Helsinki. The median age was 64 years (range, 47-74 years). Clinical characteristics are shown in Table 1. Patients nos. 1, 4, and 5 were previously untreated. Patient no. 2 was in partial remission and a stable plateau phase for 3 years following chemotherapy. Patient no. 3 had received only local radiotherapy 12 months prior to testing and was in an unmaintained plateau phase. The human-leukocyte-antigen (HLA)–ABC (class I) and HLA-DR (class II) genotype was determined at the Tissue Typing Laboratory, Department of Clinical Immunology, Huddinge University Hospital, Stockholm, Sweden.
Clinical characteristics and HLA-A, HLA-B, HLA-C, and HLA-DR genotypes of the patients participating in this study
Patient no. . | Ig subtype . | Clinical stage† . | Disease phase . | HLA-ABC . | HLA-DR . |
|---|---|---|---|---|---|
| 1 | IgG1κ | IIA | Untreated | A1, A2, B12(44), B15(62) | DR4 |
| 2 | IgG1λ | IIA | Stable PR‡ | A1, A28, B8, B12(44), Cw5, Cw7 | DR3(17), DR4 |
| 3 | IgG1κ | IIA | Stable PR‡ | A02, A24, B07, B15, C07, C12 | DRB1*11, DRB1*15 |
| 4 | IgG1λ | IIA | Untreated | A2, A11, B7, B35, Bw6, Cw4, Cw6 | DR3(17) |
| 5 | IgGκ | IA | Untreated | A3, A19(32), B7, B40(60), Bw6, Cw3, Cw7 | DR2(15), DR4 |
Patient no. . | Ig subtype . | Clinical stage† . | Disease phase . | HLA-ABC . | HLA-DR . |
|---|---|---|---|---|---|
| 1 | IgG1κ | IIA | Untreated | A1, A2, B12(44), B15(62) | DR4 |
| 2 | IgG1λ | IIA | Stable PR‡ | A1, A28, B8, B12(44), Cw5, Cw7 | DR3(17), DR4 |
| 3 | IgG1κ | IIA | Stable PR‡ | A02, A24, B07, B15, C07, C12 | DRB1*11, DRB1*15 |
| 4 | IgG1λ | IIA | Untreated | A2, A11, B7, B35, Bw6, Cw4, Cw6 | DR3(17) |
| 5 | IgGκ | IA | Untreated | A3, A19(32), B7, B40(60), Bw6, Cw3, Cw7 | DR2(15), DR4 |
According to Durie and Salmon.15
Unmaintained partial remission and plateau phase for 1 to 3 years.
Preparation of monoclonal IgG and F(ab′)2fragments
The procedure has been described in detail elsewhere.16 Briefly, serum was applied to a sterile MabTrapG column (Pharmacia, Uppsala, Sweden). IgG was eluted with 0.1 m glycine-HCl (pH, 2.7). Isoelectric focusing (Pharmacia Phast system) estimated that more than 90% of the IgG was monoclonal. The monoclonal IgG was dialyzed against sterile NaCl followed by filtration through a Millipore filter (0.20 μm; Bedford, MA). F(ab′)2 fragments were prepared by pepsin digestion.16
Amplification of the variable heavy chain gene
Bone marrow samples were obtained and total RNA was isolated using RNAzol reagent (Tel-Test, Friendswood, TX). First-strand cDNA synthesis was performed in a 20-μL reaction mixture containing 3 μg RNA in 10 μL volume, 4 μL 5 × buffer (GIBCO BRL Life Technologies, Paisley, United Kingdom), 1.5 μL dithiothreitol (DTT, 100 mM), 2 μL deoxynucleoside triphosphates (5 mM each; Amersham Pharmacia Biotech, Piscataway, NJ), 1.0 μL random hexamer primers (100 pmol/μL; Amersham Pharmacia Biotech), 0.5 μL RNAsin (20 U/μL; Promega, Madison, WI), and 1.0 μL Maloney murine leukemia virus reverse transcriptase (200 U/μL; GIBCO BRL Life Technologies). The reaction mixture was incubated at 42°C for 45 minutes followed by 5 minutes at 95°C to inactivate reverse transcriptase and then stored at – 20°C. Polymerase chain reaction (PCR) was performed using the consensus VH primers from all VH family genes (VH1, VH2, VH3, VH4a, VH4b, VH5, and VH6) as sense primer and a Cγ primer as antisense.17,18 All PCR reactions were performed as follows: 95°C 1 minute, 65°C 45 seconds, and 72°C 1 minute. In all samples, a final extension of 9 minutes at 72°C was applied. Serially diluted cDNA was used as template in the PCR reactions (1:10, 1:20, 1:50, 1:100, and 1:200 dilutions). The PCR products were electrophoresed on a 1.5% agarose gel containing ethidium bromide and visualized by ultraviolet exposure. The clonality of the VH genes used in the patients was selected based on the positive PCR amplification in the most diluted samples. All PCR products were cloned into the pGEM-T vector (Promega), and sequencing was performed using SP6 and T7 primers for cycle sequencing utilizing the ABI sequence detection system 310 (Applied Biosystems, Foster City, CA). The CDR regions were determined using the international ImMuneGeneTics database (http://imgt.cines.fr:8104/). All sequences were submitted to GenBank (accession numbers: AF459628, AF459629, AF459630, AF459631, and AF459632).
Epitope prediction analysis
The Ig-VH protein sequence of the respective 5 patients was analyzed for 9–amino acid (aa) long peptides, which could potentially bind to MHC class I molecules, using the bioinformatics and molecular analysis section (BIMAS) algorithm (program) for epitope prediction (http://bimas.dcrt.nih.gov/molbio/hla_bind). The following MHC class I molecules were included in the analysis: HLA-A1, HLA-A*0201, HLA-A3, HLA-A*1101, HLA-A24, HLA-B7, HLA-B8, HLA-B*3501, HLA-B40, HLA-B*4403, HLA-B62, HLA-Cw*0301, HLA-Cw*0401, HLA-Cw*0602, and HLA-Cw*0702. The sequences of peptides binding to MHC class II molecules cannot be predicted in BIMAS. The threshold for binding was set at a predicted half-life of one minute.12,19
We also performed epitope prediction analysis using the SYFPEITHI database (http://syfpeithi.bmi-heidelberg.com) for HLA-A1, HLA-A*0201, HLA-A3, HLA-B*0702, HLA-B*08, HLA-B*1510, HLA-DRB1*0301, HLA-DRB1*0401, and HLA-DRB1*1101. In this database, peptides binding to HLA-C cannot be predicted. To decide the limiting score for binding capacity to be used in the SYFPEITHI database, peptides predicted to bind to the same HLA types in both databases were compared with regard to disassociation half-time (BIMAS) and score for binding capacity (SYFPEITHI). Of all disassociation half-times for predicted peptides in the BIMAS database, the score for binding capacity for the same peptides in the SYFPEITHI database was 9 or higher. Therefore, the lowest score used for binding capacity in SYFPEITHI was set to 9.
Design of peptides for functional tests
Of the patients, 3 were analyzed for the presence of naturally occurring peptide-specific T cells. The present study was focused only on CDR-derived peptides. All peptides were designed before the programs BIMAS and SYFPEITHI were available, but upon availability the peptides' scores were checked in these 2 algorithms. The majority of peptides were designed to harbor more than 50% of the aa's within the CDR region and to fit each patient's HLA type. All peptides were randomly selected. MHC class I binding peptides varied between 8 and 10 aa, and MHC class II binding peptides were 15 to 20 aa. The aa sequences of the peptides are displayed in Tables 2,3.
Naturally occurring type I (IFN-γ) T cells (ELISPOT) recognizing 8- to 10-aa long peptides from the CDR regions of the tumor-derived idiotypic immunoglobulin
Patient no. . | HLA-ABC genotype . | Amino acid sequence . | CDR region . | Score† . | Binding half-life, min‡ . | Number of stimulated spots/105 PBMCs, mean ± SEM§ . |
|---|---|---|---|---|---|---|
| 1 | A*0201 | ALWGHGTLV | CDR3 | 25 | 577.0 | 19 ± 6.0 |
| A*0201 | SLKSRVSLSV | CDR2 | 24 | 2.2 | 18 ± 3.0 | |
| A1 | GLEWIGYIY | CDR2 | 27 | 90.0 | 17 ± 1.5 | |
| B*4403 | RVGRRAGWF | CDR3 | — | 0.15 | 17 ± 2.0 | |
| A*0201 | VGRRAGWFV | CDR3 | 11 | 2.4 | 16 ± 2.5 | |
| B62 | SGYSISDKY | CDR1 | — | 4.0 | 14 ± 2.5 | |
| B*4403 | LEWIGYIYY | CDR2 | — | 180.0 | < 10 | |
| 2 | B8 | SLKTRLTIS | CDR2 | 26 | 4.0 | 35 ± 14.0 |
| A1 | RIDWDDEKY | CDR2 | 27 | 25.0 | 26 ± 0.5 | |
| A1 | HRDNYYYAM | CDR3 | 11 | 0.05 | 23 ± 10.0 | |
| B8 | SLRTREMCV | CDR1 | 22 | 24.0 | 24 ± 10.5 | |
| B8 | DEKYYSTSL | CDR2 | 21 | 0.8 | < 10 | |
| B8 | STSLKTRL | CDR2 | 23 | 4.0 | < 10 | |
| A1 | CARIHRDNY | CDR3 | 16 | 0.05 | < 10 | |
| 5 | A3 | RLEWLSFHY | CDR3 | 20 | 12.0 | 19 ± 1.7 |
| B7 | CTRDHHRL | CDR3 | — | — | < 10 | |
| A3 | RLEWLSFHYY | CDR3 | 20 | 12.0 | < 10 |
Patient no. . | HLA-ABC genotype . | Amino acid sequence . | CDR region . | Score† . | Binding half-life, min‡ . | Number of stimulated spots/105 PBMCs, mean ± SEM§ . |
|---|---|---|---|---|---|---|
| 1 | A*0201 | ALWGHGTLV | CDR3 | 25 | 577.0 | 19 ± 6.0 |
| A*0201 | SLKSRVSLSV | CDR2 | 24 | 2.2 | 18 ± 3.0 | |
| A1 | GLEWIGYIY | CDR2 | 27 | 90.0 | 17 ± 1.5 | |
| B*4403 | RVGRRAGWF | CDR3 | — | 0.15 | 17 ± 2.0 | |
| A*0201 | VGRRAGWFV | CDR3 | 11 | 2.4 | 16 ± 2.5 | |
| B62 | SGYSISDKY | CDR1 | — | 4.0 | 14 ± 2.5 | |
| B*4403 | LEWIGYIYY | CDR2 | — | 180.0 | < 10 | |
| 2 | B8 | SLKTRLTIS | CDR2 | 26 | 4.0 | 35 ± 14.0 |
| A1 | RIDWDDEKY | CDR2 | 27 | 25.0 | 26 ± 0.5 | |
| A1 | HRDNYYYAM | CDR3 | 11 | 0.05 | 23 ± 10.0 | |
| B8 | SLRTREMCV | CDR1 | 22 | 24.0 | 24 ± 10.5 | |
| B8 | DEKYYSTSL | CDR2 | 21 | 0.8 | < 10 | |
| B8 | STSLKTRL | CDR2 | 23 | 4.0 | < 10 | |
| A1 | CARIHRDNY | CDR3 | 16 | 0.05 | < 10 | |
| 5 | A3 | RLEWLSFHY | CDR3 | 20 | 12.0 | 19 ± 1.7 |
| B7 | CTRDHHRL | CDR3 | — | — | < 10 | |
| A3 | RLEWLSFHYY | CDR3 | 20 | 12.0 | < 10 |
aa's within the CDR region are indicated in bold. — indicates not available.
SYFPEITHI.
BIMAS.
A specific T-cell response was defined as at least 10 spots/105 PBMCs.
Naturally occurring type I (IFN-γ) T cells (ELISPOT) recognizing 15- to 20-aa long peptides from the CDR regions of tumor-derived idiotypic immunoglobulin
Patient no. . | HLA . | Amino acid sequence . | CDR region . | Score† . | Number of stimulated spots/105 PBMCs, mean ± SEM‡ . |
|---|---|---|---|---|---|
| 1 | DRB10401 | FCARVGRRAGWFVALWGHG | CDR3 | 12 | 15 ± 2.4 |
| DRB10401 | IGYIYYTGTTHYNPSLKSRV | CDR2 | 22 | 12 ± 2.5 | |
| DRB10401 | GYSISDKYWTWIRQA | CDR1 | 9 | < 10 | |
| 2 | DRB10401 | FSLRTREMCVTWIRQ | CDR1 | 6 | 27 ± 11.9 |
| DRB10301 | YCARIHRDNYYYAMDVWGQG | CDR3 | 20 | 26 ± 14.0 | |
| DRB10401 | YCARIHRDNYYYAMDVWGQG | CDR3 | 20 | ||
| DRB10301 | LARIDWDDEKYYSTSLKTRLTI | CDR2 | 34 | 14 ± 2.1 | |
| DRB10401 | LARIDWDDEKYYSTSLKTRLTI | CDR2 | 22 | ||
| 5 | DRB10401 | TRDHHRLEWLSFHYYGMDV | CDR3 | 14 | 18 ± 2.3 |
| DRB10401 | YIRSKTFGGTTEYAASVKG | CDR2 | 16 | 17 ± 1.8 | |
| DRB10401 | TRDHHRLEWLSFHYYG | CDR3 | 6 | 13 ± 0.9 | |
| DRB10401 | SGFTFGDYVMNWVRQAPG | CDR1 | 16 | 13 ± 2.3 | |
| DRB10401 | WVGYIRSKTFGGTTEY | CDR2 | 16 | < 10 | |
| DRB10401 | GGTTEYAASVKGRFTI | CDR2 | 12 | < 10 | |
| DRB10401 | SFHYYGMDVWGQGTTV | CDR3 | 16 | < 10 | |
| DRB10401 | HYYGMDVWGQGTTVTT | CDR3 | 20 | < 10 |
Patient no. . | HLA . | Amino acid sequence . | CDR region . | Score† . | Number of stimulated spots/105 PBMCs, mean ± SEM‡ . |
|---|---|---|---|---|---|
| 1 | DRB10401 | FCARVGRRAGWFVALWGHG | CDR3 | 12 | 15 ± 2.4 |
| DRB10401 | IGYIYYTGTTHYNPSLKSRV | CDR2 | 22 | 12 ± 2.5 | |
| DRB10401 | GYSISDKYWTWIRQA | CDR1 | 9 | < 10 | |
| 2 | DRB10401 | FSLRTREMCVTWIRQ | CDR1 | 6 | 27 ± 11.9 |
| DRB10301 | YCARIHRDNYYYAMDVWGQG | CDR3 | 20 | 26 ± 14.0 | |
| DRB10401 | YCARIHRDNYYYAMDVWGQG | CDR3 | 20 | ||
| DRB10301 | LARIDWDDEKYYSTSLKTRLTI | CDR2 | 34 | 14 ± 2.1 | |
| DRB10401 | LARIDWDDEKYYSTSLKTRLTI | CDR2 | 22 | ||
| 5 | DRB10401 | TRDHHRLEWLSFHYYGMDV | CDR3 | 14 | 18 ± 2.3 |
| DRB10401 | YIRSKTFGGTTEYAASVKG | CDR2 | 16 | 17 ± 1.8 | |
| DRB10401 | TRDHHRLEWLSFHYYG | CDR3 | 6 | 13 ± 0.9 | |
| DRB10401 | SGFTFGDYVMNWVRQAPG | CDR1 | 16 | 13 ± 2.3 | |
| DRB10401 | WVGYIRSKTFGGTTEY | CDR2 | 16 | < 10 | |
| DRB10401 | GGTTEYAASVKGRFTI | CDR2 | 12 | < 10 | |
| DRB10401 | SFHYYGMDVWGQGTTV | CDR3 | 16 | < 10 | |
| DRB10401 | HYYGMDVWGQGTTVTT | CDR3 | 20 | < 10 |
aa's within the CDR region are indicated in bold.
Highest score found in the SYFPEITHI database.
A specific T-cell response was defined as at least 10 spots/105 PBMCs.
Peptide synthesis
Peptides were synthesized using the multiple peptide synthesizer SyRo (MultisynTech, Bochum, Germany) as described earlier.9 Solid-phase peptide synthesis was carried out with 9-fluorenyl-methoxycarbonyl methodology.20 The protocol and methods of Zsigo and Saneii21 were followed, but acetylation was not performed and coupling time was extended to 40 minutes.
Enzyme-linked immunospot (ELISPOT) assay
The ELISPOT assay for identification of IFN-γ–secreting T cells (type 1) was used as described.22,23 In patient no. 5, interleukin-4 (IL-4)–secreting cells (type 2) were also analyzed. Briefly, peripheral blood mononuclear cells (PBMCs; 1 × 105/well) were incubated with F(ab′)2 fragments of the autologous or isotypic (control) monoclonal IgG (1 pg/mL-1 μg/mL) or peptides (1-10 μg/mL) for 48 hours in humidified air with 5% CO2 at 37°C. Cells incubated with medium alone, purified protein derivative of tuberculin (PPD) (2.5 μg/mL), or concanavalin A (10 μg/mL) were used as controls. Spots corresponding to cytokine-secreting cells were counted blindly under a dissection microscope. Stimulated number of cells (total number of spots minus the number of spots in cultures with medium alone) was determined. The results are expressed as the mean number ± SEM of IFN-γ– or IL-4–secreting cells/105 PBMCs. Tests were run in duplicate and repeated 2 to 4 times.
To establish a cut-off level for a positive peptide response in the ELISPOT assay, irrelevant HLA-matched VH region–derived peptides and non–HLA-matched VH peptides were analyzed in 7 controls: 5 healthy donors and 2 control MM patients (patients nos. 1 and 2). The patients served as each other's controls. The characteristics of the idiotypic Ig-derived control peptides (score, HLA type, and location within the V-region) are shown in Tables 2,3. The peptides were used also in the control experiments of 5 healthy donors. Furthermore, 8 HLA-matched (HLA types A1, DR4), 10- to 15-aa long peptides derived of the extracellular domain of epithelial cell adhesion molecule were also tested in patient no. 2 and included as nonimmunoglobulin-derived control peptides (scores, 26-36; binding half-life, 6.7-1250). Irrelevant HLA-matched peptides induced 2.00 + 6.10 (mean + 2 SD) IFN-γ spots (n = 145). HLA nonmatched irrelevant peptides induced 1.70 + 6.66 (mean + 2 SD) IFN-γ spots (n = 84). There was no difference between healthy donors and patients with regard to the number of spots induced by irrelevant (HLA-matched or nonmatched) peptides. Based on these results and according to earlier studies,9,14 10 or more stimulated cells (total number of stimulated cells minus unstimulated cells) were considered a positive value.
Statistical methods
The Mann-Whitney U test was used to compare differences in the number of predicted HLA-binding peptides between groups.
Results
Frequency of HLA-A–,-B–,-C–, and -DR–binding peptides
The nucleotide sequence of the Ig heavy chain variable region of bone marrow plasma cells from 5 patients with MM was analyzed. The VH amino acid sequence was deduced and HLA-binding peptides predicted. The HLA-binding capacity of the derived peptides were predicted using the BIMAS and SYFPEITHI prediction programs (“Patients, materials, and methods”). In total, 245 and 601 HLA-binding peptides were identified within the whole VH region of the 5 patients using the BIMAS and SYFPEITHI systems, respectively. The numbers of predicted peptides for each patient in relation to the HLA genotypes are shown in Table 4 (BIMAS) and Table 5 (SYFPEITHI). A significantly higher number of predicted peptides were obtained using the SYFPEITHI database compared with BIMAS (P = .0006) when only HLA types that could be analyzed in both databases were compared. More than 4 times as many peptides were predicted for HLA-A (P < .0022). Almost twice as many HLA-A–binding peptides compared with HLA-B were predicted using SYFPEITHI compared with BIMAS, but the number of HLA-A– and HLA-B–binding peptides were not significantly different in the 2 systems. In BIMAS, the predicted frequency of HLA-C–binding peptides was higher than that for HLA-A– and HLA-B–binding peptides, although the difference was statistically not significant (Table 4). A large number of HLA-DR–binding peptides were also identified in SYFPEITHI (Table 5).
Number of predicted peptides that could bind to HLA-A, HLA-B, and HLA-C according to BIMAS
. | No. of peptides . | . | . | ||
|---|---|---|---|---|---|
| Patient no. . | HLA-A . | HLA-B . | HLA-C . | ||
| 1 | 4 (A1)† | 12 (B62 = B15) | ND | ||
| 9 (A*0201) | 5 (B*4403) | ||||
| 2 | 2 (A1) | 12 (B*4403) | 13 (Cw*0702) | ||
| 2 (B8) | |||||
| 3 | 7 (A*0201) | 12 (B7) | 14 (Cw*0702) | ||
| 15 (A24) | 11 (B62 = B15) | ||||
| 4 | 4 (A*0201) | 3 (B7) | 16 (Cw*0401) | ||
| 2 (A*1101) | 12 (B*3501) | 26 (Cw*0602) | |||
| 5 | 7 (A3) | 12 (B7) | 18 (Cw*0301) | ||
| 9 (B40) | 18 (Cw*0702) | ||||
| Total | 50 | 90 | 105 | ||
. | No. of peptides . | . | . | ||
|---|---|---|---|---|---|
| Patient no. . | HLA-A . | HLA-B . | HLA-C . | ||
| 1 | 4 (A1)† | 12 (B62 = B15) | ND | ||
| 9 (A*0201) | 5 (B*4403) | ||||
| 2 | 2 (A1) | 12 (B*4403) | 13 (Cw*0702) | ||
| 2 (B8) | |||||
| 3 | 7 (A*0201) | 12 (B7) | 14 (Cw*0702) | ||
| 15 (A24) | 11 (B62 = B15) | ||||
| 4 | 4 (A*0201) | 3 (B7) | 16 (Cw*0401) | ||
| 2 (A*1101) | 12 (B*3501) | 26 (Cw*0602) | |||
| 5 | 7 (A3) | 12 (B7) | 18 (Cw*0301) | ||
| 9 (B40) | 18 (Cw*0702) | ||||
| Total | 50 | 90 | 105 | ||
Number of predicted peptides that could bind to HLA-A, HLA-B, and HLA-DR according to SYFPEITHI
. | No. of peptides . | . | . | ||
|---|---|---|---|---|---|
| Patient no. . | HLA-A . | HLA-B . | HLA-DR . | ||
| 1 | 16 (A1) | 11 (B*1510) | 47 (DRB1*0401 = DR4Dw4) | ||
| 45 (A*0201) | |||||
| 2 | 24 (A1) | 31 (B*08) | 41 (DRB1*0301 = DR17) | ||
| 55 (DRB1*0401 = DR4Dw4) | |||||
| 3 | 48 (A*0201) | 23 (B*0702) | 29 (DRB1*1101) | ||
| 13 (B*1510) | |||||
| 4 | 34 (A*0201) | 13 (B*0702) | 33 (DRB1*0301 = DR17) | ||
| 5 | 58 (A3) | 23 (B*0702) | 57 (DRB1*0401 = DR4) | ||
| Total | 225 | 114 | 262 | ||
. | No. of peptides . | . | . | ||
|---|---|---|---|---|---|
| Patient no. . | HLA-A . | HLA-B . | HLA-DR . | ||
| 1 | 16 (A1) | 11 (B*1510) | 47 (DRB1*0401 = DR4Dw4) | ||
| 45 (A*0201) | |||||
| 2 | 24 (A1) | 31 (B*08) | 41 (DRB1*0301 = DR17) | ||
| 55 (DRB1*0401 = DR4Dw4) | |||||
| 3 | 48 (A*0201) | 23 (B*0702) | 29 (DRB1*1101) | ||
| 13 (B*1510) | |||||
| 4 | 34 (A*0201) | 13 (B*0702) | 33 (DRB1*0301 = DR17) | ||
| 5 | 58 (A3) | 23 (B*0702) | 57 (DRB1*0401 = DR4) | ||
| Total | 225 | 114 | 262 | ||
HLA types are indicated in parentheses.
Predicted peptides are defined as a binding capacity score of at least 9 (“Patients, materials, and methods”).
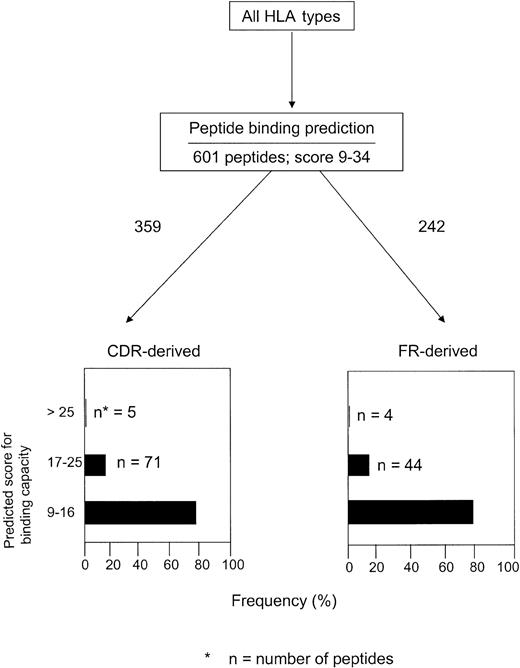
We then analyzed the frequency of HLA-binding peptides within the CDRs and FRs. A peptide was considered derived from CDR if one or more aa's were located within the CDR.12 Among the 245 peptides identified in BIMAS and 601 in SYFPEITHI, 128 (52%) and 359 (60%), respectively, were derived from CDRs. Almost all peptides (CDR- or FR-derived) had a predicted binding half-life in the range of 1 to 60 minutes (BIMAS). Only 2 of 128 CDR-derived and 6 of 117 FR-derived peptides showed a binding half-life of longer than 60 minutes (data not shown). The corresponding figures in SYFPEITHI (score, ≥ 9) are shown in Figure 1. Scoring in the lower range (9-16) were 79% of the CDR-derived peptides and 80% of the FR-derived peptides. Only 1.7% of the CDR-derived and 1.5% of the FR-derived peptides had a binding score of higher than 25. Both CDR- and FR-derived peptides had a relatively high proportion of peptides with an intermediate score (17-25) for binding (20% and 18%, respectively).
Number of HLA-binding CDR- and FR-derived peptides in relation to predicted binding score analyzed by the SYFPEITHI database in 5 patients with MM.
Number of HLA-binding CDR- and FR-derived peptides in relation to predicted binding score analyzed by the SYFPEITHI database in 5 patients with MM.
The numbers of HLA-binding peptides derived from CDR1-3 and FR1-4 are shown for the SYFPEITHI database in Table 6. Almost identical results were obtained when analyzing the proportion of HLA-binding peptides in the BIMAS database (data not shown). HLA-binding peptides were most abundant in CDR2 and FR3, followed by CDR3. Few predicted peptides were confined to FR4.
Number of predicted HLA-binding peptides within CDR1-3 and FR1-4 of the Ig-VH chain from 5 patients with MM analyzed by SYFPEITHI
. | No. of peptides . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | FR1 . | CDR1 . | FR2 . | CDR2 . | FR3 . | CDR3 . | FR4 . | ||||||
| 1 | 0 | 14 | 4 | 37 | 39 | 23 | 2 | ||||||
| 2 | 17 | 25 | 5 | 44 | 33 | 27 | 0 | ||||||
| 3 | 18 | 20 | 9 | 32 | 23 | 11 | 0 | ||||||
| 4 | 0 | 9 | 5 | 22 | 22 | 20 | 2 | ||||||
| 5 | 26 | 14 | 5 | 34 | 29 | 27 | 3 | ||||||
| Total (%) | 61 (10) | 82 (14) | 28 (5) | 169 (28) | 146 (24) | 108 (18) | 7 (1) | ||||||
. | No. of peptides . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | FR1 . | CDR1 . | FR2 . | CDR2 . | FR3 . | CDR3 . | FR4 . | ||||||
| 1 | 0 | 14 | 4 | 37 | 39 | 23 | 2 | ||||||
| 2 | 17 | 25 | 5 | 44 | 33 | 27 | 0 | ||||||
| 3 | 18 | 20 | 9 | 32 | 23 | 11 | 0 | ||||||
| 4 | 0 | 9 | 5 | 22 | 22 | 20 | 2 | ||||||
| 5 | 26 | 14 | 5 | 34 | 29 | 27 | 3 | ||||||
| Total (%) | 61 (10) | 82 (14) | 28 (5) | 169 (28) | 146 (24) | 108 (18) | 7 (1) | ||||||
A significantly higher number of HLA-binding peptides were found in a “geographic” region consisting of CDR2-FR3-CDR3 compared with the flanking (FR1-CDR1-FR2 and FR4) regions in both databases (P < .01). There was no significant difference in the frequency of predicted peptides derived from CDRs compared with FRs.
To exclude the possibility that predicted peptides were located in different Ig-VH regions for various HLA types, we made a separate analysis on the number of HLA-A*0201–binding CDRs and FRs peptides in the 3 HLA-A*0201–positive patients. Similar results were obtained for each of the 3 individual patients as well as in comparison with overall pooled data. A clustering of peptides derived from CDR2-FR3-CDR3 was observed. The same analysis was also performed for HLA-B7–, HLA-B*0702–, HLA-Cw*0702–, and HLA-DRB1*0401–positive patients and similar results were obtained (data not shown). Moreover, binding half-life and binding score of the predicted peptides in the HLA-A*0201–positive patients were comparable with those of pooled data (data not shown).
Naturally occurring peptide-specific T cells
Functional tests were performed in 3 patients (nos. 1, 2, and 5) using purified autologous idiotypic F(ab′)2 fragments and peptides derived from the VH region to stimulate PBMCs. PBMCs have previously been used to detect IFN-γ Id-reactive T cells. Id-specific T cells were found within both the CD4 and CD8 subsets.24 Anti–MHC class I and class II antibodies could specifically inhibit the response.9
All 3 patients displayed an IFN-γ T-cell response against F(ab′)2 fragments of the autologous idiotype. Found in patients nos. 1, 2, and 5 were 22, 25, and 20 stimulated spots, respectively. Isotypic (control) monoclonal IgG induced fewer than 7 spots in these patients. In patient no. 1, an IFN-γ T-cell response was noted against 6 of 7 tested 9- to 10-aa long peptides (Table 2). Of those 6 peptides, 5 were derived from CDR2 and CDR3, while 1 was derived from CDR1. In patient no. 2, specific IFN-γ–secreting T cells were detected after stimulation with 4 of 7 tested 8- to 9-aa long peptides. Also in this patient the peptides were predominantly derived from CDR2 and CDR3. In patient 5, IFN-γ–secreting T cells were detected against 1 of 3 tested 8- to 10-aa long peptides. This peptide was derived from the CDR3 region. The 2 negative peptides were also CDR3 derived. IL-4–secreting cells were analyzed in the same patient as well (no. 5), but none of the peptides induced IL-4 production (data not shown). Peptide stimulation experiments were repeated 2 to 4 times in all 3 patients, and in 85% of the tests the results were similar, but in a few cases the numbers of spots varied (Tables 2,3). The 8- to 10-aa long peptides inducing a specific T-cell response were compared with regard to half-life for binding and score for binding capacity. Of the 11 positive peptides, 7 displayed a low half-life for binding (1-60 minutes), 2 had a half-life for binding longer than 60 minutes, and 2 were under the threshold 1 minute or longer. The scores for binding capacity were evenly distributed within the low (9-16; n = 2), intermediate (17-25; n = 4), and high (> 25; n = 3) ranges.
Functional tests were also performed using 15- to 20-aa long peptides. In patient no. 1, an IFN-γ T-cell response was noted against 2 of 3 tested peptides (Table 3). These 2 peptides were derived from CDR2 and CDR3. In patient no. 2, specific IFN-γ–secreting T cells were detected after stimulation with all of the tested peptides. These peptides were derived from CDR1, CDR2, and CDR3. In patient no. 5, IFN-γ–secreting T cells were detected against 4 of 8 tested peptides; 2 of the peptides were derived from the CDR3 region and the other 2 from CDR1 and CDR2. The 4 negative peptides were derived from CDR2 and CDR3.
The 15- to 20-aa long peptides inducing a specific T-cell response were also compared with regard to score for binding capacity. From patient no. 2, 2 peptides showed different scores depending on which HLA-DR molecule they were predicted to bind. Of the 9 positive peptides, 3 had an intermediate score for binding capacity (17-25), and 2 peptides had a score lower than 9. The remaining 4 positive peptides had a score for binding capacity in the low range (9-16).
To confirm the T-cell origin of peptide-reactive cells, PBMCs of patient no. 2 were stimulated with the autologous and irrelevant idiotypic peptides and analyzed for the production of intracellular IFN-γ in phenotype-characterized CD4 and CD8 T cells. The techniques have been described previously.25 A 15-mer peptide (FSLRTREMCVTWIRQ; Table 3) was found to stimulate 1.1% of the CD4 T cells, and 2 9-aa-long peptides (SLKTRLTIS and RIDWDDEKY; Table 2) induced IFN-γ production in 0.3% and 0.6%, respectively, of the CD8 T cells (values of irrelevant control peptides have been subtracted).
Discussion
Naturally occurring idiotype-specific T cells have previously been demonstrated in patients with monoclonal gammopathies using the complete idiotypic protein for activation.5,6 Idiotype-specific type I T cells were detected, especially in patients with early indolent disease,5 whereas idiotype-specific T-cell reactivity in patients with advanced myeloma was down-regulated and/or shifted toward a type II T-cell response.26 The myeloma plasma cells may serve as antigen-presenting cells27 and might specifically be recognized and lysed by idiotype-specific T cells.7,8,28 In the blood of MM patients an expansion of activated CD57+CD8+ T cells producing perforin has been noted, which was suggested to be the result of a persistent stimulation by tumor-associated antigens.29
Somatically mutated Ig-VH CDR regions in malignant B cells, especially sequences deduced from the VH-D-JH joining region (the most unique part of Ig-VH), have been proposed to be targeted by specific T cells.9 In a B-cell lymphoma patient, T cells recognizing the CDR3 sequence of the heavy-chain variable region of the tumor-derived Ig were shown to secret IFN-γ as well as to lyse autologous PBMCs loaded with the relevant peptide.10 It has also been shown that B-cell chronic lymphocytic leukemia (CLL) patients might spontaneously exhibit T cells recognizing the tumor-derived Ig VH-CDR3 region.11
The purpose of the present study was to identify T-cell epitopes in the variable region of the Ig heavy chain derived from the tumor cells in patients with MM and to study the functional capacity of idiotypic peptides in vitro with the aim to define epitopes or geographic regions that might be preferential target structures and of potential interest for the production of subunit vaccines. Peptide fragments of the idiotype might be more immunogenic for induction of an idiotype-specific immunity than the complete intact idiotypic Ig.30 The functional parts of this study were focused on CDR-derived peptides, and FR-derived peptides were not synthesized. FR-derived peptides have been extensively studied by Trojan et al12 and shown to express cytotoxic epitopes. The magnitude of the IFN-γ response in our study was modest, which might be because idiotypes are weak antigens. The results are in line with our previous results14 as well as with those of others using other autoantigens that also showed a weak response in ELISPOT.31,32 Responses to weak antigens seem, however, to be readily detected by ELISPOT, eliminating the need for a prolonged preactivation culture period in vitro.33 The relatively low magnitude of the responses might be a limitation when interpretating the results of this as well as of other studies measuring cellular responses against “self” antigens, which have a low-to-intermediate affinity for the T-cell receptor.
In myeloma, a large number of HLA-binding peptides were predicted, which is in accordance with a previous report in B-cell malignancies.12 The number of predicted peptides was, however, related to the database, which underlines the importance of applying different databases.34 This should be kept in mind when interpreting and comparing results of various studies. Furthermore, these 2 particular databases revealed different patterns and frequencies of peptides binding to the HLA-A and -B loci, respectively.
It is not known whether CDR- and/or FR-derived peptides are important for targeting. Some authors have suggested that in particular VH-CDR3 may be an important target for lytic T cells.10,11 In the present study, predicted CDR-derived peptides were more abundant compared with FR-derived peptides. T cells spontaneously recognizing CDR1-3 were of type I, recognizing MHC class I–restricted peptides compatible with the phenotype of a cytotoxic T-cell. However in another study of B-cell malignancies,12 HLA-A*0201–binding peptides were most frequently found within the framework regions (71%) using BIMAS, applying a predicted half-life for binding of 1 minute or longer and considering a peptide as CDR derived if only a single amino acid was located within CDR. Similar results were obtained for HLA-A1, HLA-A3, HLA-A11, and HLA-B27. The FR-derived peptides expressed cytotoxic T-cell epitopes, which were shared by many patients. These results are in contrast to our findings in myeloma patients, in whom 60% of the predicted HLA-A*0201–binding peptides were CDR derived. Similar results were also found for the other HLA types in our patients. The reason for this difference is not clear.
An important aspect for the functional capacity may be the affinity of the peptide. Trojan et al12 showed that the majority of the CDR- and FR-derived peptides had a low predicted binding half-life but still expressed cytotoxic epitopes. Such peptides may have functional implications in vivo. Antigens with a high binding affinity may induce clonal deletion of T cells rather than T-cell expansion during an immune response.35 In B-cell malignancies it was also found that the CDR3 region of the Id was a key recognition element for Id-specific T cells but contributed little to the affinity of the antigen.36 In the present study, 7 of 9 functional peptides had a score of 20 or higher but none higher than 27. Only 2 of 11 functional peptides had a binding half-life of longer than 60 minutes, while the remaining peptides had a low binding half-life; 2 peptides even had a half-life of less than 1 minute. Thus, our study might support the notion that idiotypic peptides with a low and intermediate affinity may also induce a T-cell response. This is in line with a recent publication on CLL showing that CTL against low-affinity VH region–derived peptides can be identified in the blood, while modified peptides with a higher affinity (heteroclitic) increased the lytic capability (but not the frequency) of CTL, indicating the importance of the avidity of the T-cell receptor for lysis.34 Furthermore, T cells reactive with V gene segment–encoded germ-line Id peptides may be deleted in the thymus or anergized in the periphery,3 and T cells may therefore recognize only somatically mutated Id peptides. Interestingly, it was described that patients with chronic lymphocytic leukemia with somatically mutated V regions had a better prognosis than those with germ-line configuration,37 and T cells in CLL might recognize mutated VH-CDR3 regions.11
An interesting finding in our study was the clustering of HLA-binding peptides to a geographic region, CDR2-FR3-CDR-3 Ig-VH region, from which about 70% of all identified peptides originated. The number of aa's in this geographic region represented 53% to 61% of all aa's of the whole Ig-VH part. This region may represent a “hot spot” of particular interest for construction of subunit vaccines. This is further supported by our functional studies, which showed that all but 2 of 11 tested peptides—able to specifically stimulate an MHC class I peptide–restricted type I T-cell response—were derived from the CDR2 or CDR3 region. Furthermore, the experiments performed in HLA-matched controls indicated that the peptide-specific T cells were specific for the tumor and not for self.
A T-cell response against 8- to 10-aa long idiotype-derived peptides fitting to the actual HLA-A/B types is in agreement with a recent report showing that patients with myeloma exhibited naturally occurring myeloma cell–specific T cells capable of lysing the tumor cells.8 Moreover, 9-aa long CDR-derived peptides used for vaccination of NHL patients induced a CTL response.10 Vaccination with the complete Ig idiotype evoked an idiotype-specific T-cell response consisting of T cells recognizing peptides corresponding to all 3 CDR regions of the immunizing idiotype.14 However, FR regions also may express CTL epitopes.12
To induce an MHC class I–restricted T-cell response, MHC class II–restricted T cells are required.38 An IFN-γ T-cell response was also induced against 9 of 15 (15- to 20-aa long) tested idiotype-derived peptides, fitting to the actual HLA-DR types. As the myeloma cell clone consists of B cells (not only mature plasma cells) expressing MHC class I as well as class II molecules, the malignant myeloma cells might also be recognized by MHC class II–binding T cells, which may destroy the malignant cells.39 We have previously shown that the idiotypic protein recognized naturally occurring CD4 as well as CD8 T cells24 and that idiotypic peptides could stimulate T cells, whose response could be inhibited by MHC class I as well as MHC class II monoclonal antibodies, confirming the MHC restriction.9 Also in the present study, the idiotypic peptides seemed to recognize CD4 as well as CD8 T cells.
In summary, this study demonstrates the presence of many potential T-cell epitopes within the tumor-derived Ig-VH region. Such peptides may induce a spontaneous cellular immune response, consisting of MHC class I– and class II–restricted type I T cells. Further studies are warranted to define the fine functional specificity of these Id-specific T cells in myeloma patients. Such information may be of great importance for a better understanding of immune regulation of the myeloma clone. A detailed mapping of idiotype targeting structures may also be of importance to establish effective subunit vaccines.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-04-1250.
Supported by grants from the International Myeloma Foundation, Hans Edstrand Foundation, IngaBritt and Arne Lundberg's Research Foundation, the Swedish Cancer Society, the Cancer Society in Stockholm, King Gustaf V Jubilee Fund, Gunnar Nilsson Foundation, the Torsten and Ragnar Söderberg's Foundation, the Cancer and Allergy Foundation, and Karolinska Institute Foundations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are very grateful to Ms Ingrid Eriksson and Ms Barbro Näsman-Glaser for expert technical assistance; to Ms Ulla Rudén for production of peptides; to Ms Gerd Ståhlberg for secretarial help; and to Mr Adnane Achour for reviewing and giving constructive criticism of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal