Abstract
The present work investigated the mechanism for down-regulation of thrombomodulin (TM), an anticoagulant glycoprotein, on cultured umbilical vein endothelial cells (HUVECs) exposed to lipid extracts from oxidized low-density lipoprotein (ox-LDL). HUVECs exposed to phospholipid extracts, but not to free cholesterol, triglyceride, or cholesterol ester, isolated from ox-LDL reduced TM mRNA levels to nearly the same extent as native ox-LDL. Oxidized 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphocholine (ox-PAPC), but not native PAPC or a reduced form of ox-PAPC, markedly decreased TM mRNA levels. The apparent half-life (t 1/2 = 2.7 hours) of TM mRNA in control cells was not significantly different from that in cells exposed to ox-LDL or ox-PAPC. TM mRNA levels were regulated by transcriptional activation via a retinoid receptor β (RARβ). The binding activities of nuclear proteins from HUVECs treated with ox-LDL or ox-PAPC to the DR4 or stimulatory protein 1 (Sp1) sequence in the TM promoter were significantly reduced with decreased expression of RARβ, retinoid X receptor α (RXRα), Sp1, and Sp3 in the nuclei. The promoter activity in HUVECs transfected with a reporter plasmid expressing the TM promoter with targeted deletions in the DR4 and Sp1 binding elements was decreased to about 20% of that with the wild-type construct. Treatment of the cells with ox-PAPC had no additional effect on the promoter activity. These results suggest that oxidized phospholipids in ox-LDL inhibit transcription of the TM gene in HUVECs by inhibiting the binding of RARβ-RXRα heterodimer and Sp, including Sp1 and Sp3, to the DR4 element and Sp1 binding element, respectively, in the TM promoter with reduced expression of RARβ, RXRα, and Sp1 and Sp3 in the nuclei.
Introduction
Vascular endothelial cells actively participate in the regulation of hemostasis. Under normal circumstances, these cells are thrombo-resistant, a state maintained by antithrombotic mechanisms, as well as control of expression of procoagulant factors, such as tissue factor. Thrombomodulin (TM) is a high-affinity receptor for thrombin on the endothelial cells, which acts not only as a direct inhibitor for the procoagulant activity of thrombin toward fibrin formation and platelet activation, but also as a key cofactor for the TM–thrombin–protein C pathway, which is a major physiologic antithrombotic property of endothelial cells.1,2 Protein C, which circulates in the plasma as a protease zymogen, is activated by the TM-thrombin complex, and the activated protein C proteolytically inactivates coagulation factors Va and VIIIa, which are indispensable to blood coagulation.1,2 Therefore, thrombin formation in the coagulation cascade is reduced by activation of the TM–thrombin–protein C pathway. The physiologic relevance of this pathway for antithrombotic properties of endothelial cells is emphasized by the thrombosis disorders frequently observed in patients with protein C–resistant factor V3,4 or protein C deficiency,5 and it has been postulated that TM is an important regulator in maintaining the fluidity of circulating blood and lymphatic fluid. The cofactor activity of TM isolated from human placenta is markedly enhanced by reconstitution into vesicles consisting of phosphatidylcholine and acidic phospholipids, such as phosphatidylserine and phosphatidylethanolamine, so that optimal antithrombotic TM activity depends on the local cell membrane status.6,7
The cDNA sequence of human TM encodes for 575 amino acids.8,9 The 5′-flanking region of the human TM gene contains a TATA box, a CAAT box, and several conserved transcription factor binding elements such as a retinoic acid receptor responsive element (RARE) and stimulatory protein 1 (Sp1) binding element.10-12 The RARE in the TM gene consists of direct repeats of RARE separated by 4 base (DR4) sequences, and it binds a heterodimer of retinoic acid receptors (RARα, RARβ, and RARγ) and retinoid X receptor α (RXRα).10-12 We previously reported that TM expression on cultured human umbilical vein endothelial cells (HUVECs) was up-regulated by treatment of the cells with retinol or retinoids containing retinoic acid,13 increases mediated by the interactions of the RARβ-RXRα heterodimer to the DR4 sequence and nuclear Sp1 to the Sp1 binding site.12 Up-regulation of the TM cofactor activity and antigen levels in HUVECs was also observed when the cells were stimulated with histamine14 and cAMP.15 In contrast, down-regulation of TM antigen levels in HUVECs was observed in response to bacterial endotoxin16 or inflammatory cytokines such as interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α).17-20 TM down-regulation in cells exposed to the inflammatory cytokines appears to require internalization and degradation of surface TM21 and/or inhibition of TM transcription.20,22,23 The mechanism of the down-regulation of TM transcription is, however, obscure.
Oxidized low-density lipoprotein (ox-LDL) plays a pivotal role in the pathogenesis of atherosclerosis and alters various physiologic circumstances, including vascular tone, endothelial permeability, and coagulation and fibrinolysis controls within the micro-environments of atherosclerotic lesions.24 A study on the mechanisms of altered coagulation and fibrinolysis control in endothelial cells exposed to ox-LDL would contribute to understanding the pathophysiology of thrombus formation in atherosclerotic lesions. We previously demonstrated that HUVECs exposed to ox-LDL, but not to native or acetylated LDL, reduced TM expression on the cells. The decreased TM mRNA levels were mediated by the lipid components of ox-LDL through internalization of ox-LDL.25 Subsequently, down-regulation of TM expression on endothelial cells was found in lesions of coronary atherosclerosis.26 The present work was undertaken to identify an active lipid species from ox-LDL important for down-regulation of TM mRNA levels and to elucidate the molecular basis for down-regulation of TM transcription in endothelial cells treated with ox-LDL.
Materials and methods
Materials
Reagents were purchased from Wako Pure Chemical Industries (Osaka, Japan), unless otherwise indicated. Bovine serum albumin (BSA), human thrombin (4000 NIH units/mg), 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphocholine (PAPC), lysophosphatidylcholine (lysoPC) from egg yolk, 7β-hydroxyl-cholesterol, 25-hydroxycholesterol, and 7-oxocholesterol were purchased from Sigma Chemical (St Louis, MO); 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), from Calbiochem (San Diego, CA); and poly(dI-dC) and PD-10 column, from Pharmacia (Piscataway, NJ). A kit for assay of TM antigen was donated from Mitsubishi Gas Company (Tokyo, Japan). Rabbit anti–human RARα, RARβ, RARγ, RXRα, Sp3, or Sp4 immunoglobulin G (IgG) and mouse monoclonal anti–human Sp1 IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase (HRP)–conjugated sheep anti–rabbit IgG and HRP-conjugated sheep anti–mouse IgG were purchased from Southern Biotechnology Associate (Birmingham, AL). Dulbecco modified Eagle medium (DMEM) and Dulbecco phosphate-buffered saline (PBS) were purchased from Nissui Pharmaceuticals (Tokyo, Japan). [α32P]–deoxycytosine triphosphosphate (dCTP) and [γ32P]–adenosine triphosphate (ATP) were purchased from NEN Research Products (Tokyo, Japan). Oligonucleotides were synthesized by Amersham Pharmacia Biotech (Tokyo, Japan).
Cell culture
HUVECs were cultured on collagen-coated 96- or 6-well plates (Corning; Iwaki Glass, Tokyo, Japan) as described previously.15 Confluent monolayer cells were washed in serum-free medium and then incubated in DMEM supplemented with 20% fetal calf serum (FCS) containing LDL, lipid extracts from LDL, or PAPC and/or various compounds. After incubation for the indicated periods, cell viability was determined by the MTT assay as described previously.27
Isolation and modification of LDL
LDL (density range, 1.019-1.063 g/mL) was isolated from fresh human plasma of healthy subjects and ox-LDL (2 mg protein/mL) was prepared by ultraviolet (UV) irradiation (254 nm, 0.4 mW/cm2) of LDL for 12 hours at 4°C as described previously.25 Oxidation of LDL was estimated as the formation of thiobarbituric acid reactive substances (TBARS) and expressed as malondialdehyde (MDA) equivalents per milligram of protein using 1,1,3,3-tetraethoxypropane, a precursor of MDA, as a standard.28 Phospholipid and triglyceride content in the LDL were determined using assay kits Phospholipid C-test Wako and Triglyceride G-test Wako, respectively. Total cholesterol was determined using the Cholesterol CII-test Wako kit with cholesterol esterase and cholesterol oxidase. Free cholesterol was determined using the Cholesterol CII-test Wako with only cholesterol oxidase. Cholesterol ester content was determined as the total cholesterol content minus the free cholesterol content. Protein content was determined by the method of Bradford29 using BSA as a standard.
Lipid extraction from LDL and fractionation of lipid extracts
Crude lipid extracts from LDL (≈400 μg protein) in 0.2 mL PBS were obtained by the method of Bligh and Dyer30 as described previously.25 The crude lipid extract in the organic phase was evaporated and dispersed in DMEM containing 20% FCS by sonication. The lipid suspension was used in experiments as a crude lipid from native LDL or ox-LDL. For additional fractionation of lipids, crude lipid extracts in the organic phase were dried under a stream of nitrogen and then dissolved in 0.5 mL hexane–2-propanol (9:1, vol/vol). The dissolved lipids (0.5 mL) were applied onto a 2.0-mL Sep-Pak silica cartridge (Millipore Corporation, Milford, MA) equilibrated with hexane–2-propanol (9:1, vol/vol), and each lipid was separated with a modification of the method described by Hamilton and Comai.31 Free cholesterol, triglyceride, and cholesterol ester were eluted isocratically with 5 mL of hexane–2-propanol (9:1, vol/vol) and dried under a stream of nitrogen and then dissolved in hexane–ether–formic acid (90:10:0.2, vol/vol/vol). Phospholipids eluted with 24.0 mL methyltertiarybutylether–methanol–ammonium acetate (5:8:2, vol/vol/vol) were dried and dissolved in chloroform. The fraction containing free cholesterol, triglyceride, and cholesterol ester was further applied onto Sep-Pak silica cartridge column equilibrated with hexane–ether–formic acid (90:10:0.2, vol/vol/vol). Triglyceride and cholesterol ester eluted isocratically with 4 mL of the same solvent were dried and dissolved in chloroform. Free cholesterol was eluted with 3 mL hexane–2-propanol (9:1, vol/vol) and dried and then dissolved in chloroform. To identify each lipid, the separated lipids were developed in parallel by silica gel thin-layer chromatography with hexane-isopropanol (9:1). Each lipid was detected under iodine vapors and identified by comparison with the mobility of standard lipids. Each lipid in chloroform was transferred to a glass test tube and evaporated under a stream of nitrogen. The lipid residue was dispersed in DMEM containing 20% FCS by sonication for about 30 seconds. These lipid suspensions were used for the experiments.
Oxidation and reduction of phospholipid
PAPC (1 mg) in chloroform was evaporated under a stream of nitrogen. The residue dissolved in 1 mL methanol was poured in 1 well of a 6-well plate and irradiated by UV light (254 nm, 0.4 mW/cm2) for 12 hours at 4°C. The ox-PAPC was dried under a stream of nitrogen and dissolved in chloroform. Reduction of ox-PAPC or oxidized phospholipids from ox-LDL was performed as described by Watson et al.32 Ox-PAPC or oxidized phospholipids in chloroform were dried under a stream of nitrogen and resuspended in 1.0 mL of 0.1 M borate buffer (pH 8.0) by sonication for 30 seconds. The suspension was instantly reduced by adding 500 μg sodium borohydride. After 5 minutes, the suspension was mixed with 3 mL chloroform-methanol (1:1) and the chloroform phase was extracted. The reduced form of ox-PAPC or oxidized phospholipids from ox-LDL was dried under a stream of nitrogen and the lipid residue was dispersed in DMEM containing 20% FCS by sonication.
Assay of TM antigen and TM mRNA
TM antigen levels in cultured HUVECs were determined by enzyme immunoassay as previously described.33 TM mRNA levels were measured by reverse transcription–polymerase chain reaction (RT-PCR) using TM primers and β-actin primers9 as described previously.34 Amplified cDNA was detected with 0.1 mg/mL ethidium bromide staining after electrophoresis through a 2% agarose gel and photographed under UV light (254 nm). The gel areas corresponding to amplified cDNA were cut out and counted for radioactivity. There was a linear relationship between the Cerenkov counts obtained for each PCR product and the amount of total RNA obtained under the experimental conditions, in the range from 0 to 1.25 μg RNA. The PCR product counts from the TM cDNA were normalized to those from the β-actin cDNA. The half-life of mRNA was determined by the method of Xie et al.35 In the experiments with DRB, a transcriptional inhibitor, postconfluent HUVECs were incubated with 65 μM DRB and/or ox-LDL or ox-PAPC in culture medium for 0, 2, 4, and 6 hours at 37°C.
Electrophoretic mobility shift assay (EMSA) and supershift assay
Cultured HUVECs (5 × 105 cells/plate) were homogenized in a Potter Elvejem–type homogenizer and centrifuged at 2000 rpm to obtain the nuclear fraction. This fraction was suspended in 800 μL of 20 mM Tris (tris(hydroxymethyl)aminomethane)–borate (TB) buffer (pH 8.0) containing 10 mM KCl, 0.1 mM EDTA [ethylenediaminetetraacetic acid], 0.1 mM EGTA [ethylene glycol tetraacetic acid], 1 mM dithiothreitol (DTT), 2 μg/mL leupeptin, 2 μg/mL aprotinin, 0.5 mg/mL benzamidine hydrochloride, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 0.6% nonidet P-40 (NP-40) and stored at –80°C as a nuclear extract. Double-stranded oligonucleotides were prepared by annealing (heating at 95°C for 5 minutes and chilling at room temperature for 60 minutes) single-strand oligonucleotides of 5′-CGTTTGGTCACTGCAGGTCAGTCCA-3′ (at position –1535 to –1511 containing underlined consensus DR4 sequences in the 5′-flanking region of the human TM gene), 5′-CTGTGTTGCACGTGCAAGCTCCGTA-3′ (scrambled DR4 sequences), or 5′-GCGAGGGCGGGCGCAAGGGCGGCCA-3′ (at position –145 to –121 for the tandem Sp1 sites containing second and first Sp1 sites in the 5′-flanking region of the human TM gene). The purity of each double-stranded oligonucleotide was confirmed as described previously.12 The EMSA reaction mixture contained 0.5 ng labeled probe (1 × 105 cpm), 1 μg poly(dI-dC), and 2 μg nuclear proteins in a final volume of 6 μL TB buffer (pH 8.0) containing 50 mM KCl, 5 mM MgCl2, 1 mM ZnCl2, 0.5 mM DTT, 0.1 mM PMSF, 5 mM spermidine, 0.008% NP-40, and 15% glycerol. The mixture was incubated for 30 minutes at 25°C and subjected to 4% polyacrylamide gel electrophoresis (PAGE) using a buffer consisting of 10 mM Tris-HCl (pH 7.9) containing 1.5 mM EDTA and 5 mM sodium acetate. After electrophoresis, the gels were dried on filter paper and subjected to autoradiography. To identify specific RARs, RXRα, Sp1, Sp3, or Sp4 proteins involved in DNA binding (the supershift assay), 5 μg nuclear proteins were incubated with 1 μg rabbit IgG directed against human RARα, RARβ, RARγ, RXRα, Sp3 or Sp4 protein, or monoclonal anti–human Sp1 IgG for 30 minutes at 25°C. After incubation, labeled DR4 or Sp1 oligonucleotide was added and reaction was allowed to proceed for 30 minutes at 25°C before EMSA was performed.
Western blot analysis
After treatment with various reagents, nuclear proteins were extracted from HUVECs (3 × 105 cells/well) according to the method described above. Nuclear protein (5 μg) was subjected to sodium dodecyl sulfate (SDS)–PAGE under nonreduced conditions as described previously.36 After SDS-PAGE, protein was transferred to a nitrocellulose membrane and incubated with 0.5 μg/mL polyclonal antibody against human RARβ, RXRα, Sp3 or Sp4, or 0.5 μg/mL monoclonal antibody against human Sp1. Detection of these proteins was carried out using HRP-conjugated sheep anti–rabbit IgG or HRP-conjugated sheep anti–mouse IgG. A chemiluminescence detection system (Super Signal, Pierce Chemical, Rockford, IL) was used.
Transient transfection, luciferase assay, and preparation of cell lysates
The promoter activity of TM was determined using pGL3-pTM reporter plasmid containing the 5′-flanking region of TM as described previously.12 The pGL3-pTM plasmid and deletion mutants were prepared by insertion of an MluI-XhoI fragment derived from pTM-1562CAT into the pGL3 basic plasmid (Promega). HUVECs were cultured in a 6-well plate with DMEM medium supplemented with 20% FCS, human epidermal growth factor (hEGF; 10 ng/mL), human fibroblast growth factor B (hFGF-B; 5 ng/mL), heparin (10 μg/mL), and hydrocortisone (1 μg/mL) and were transfected with pGL3-pTM plasmids (2 μg/well) in the presence of the pRL-TK plasmid (0.1 μg/well) by the cationic liposome method at a cell density of 70% to 80% confluency in an OPTI-MEM medium.12 After 6 hours, the culture medium was replaced. Cells were cultured for 12 hours and then exposed to 50 μg/mL of either native PAPC (n-PAPC) or ox-PAPC for 12 hours. The promoter activity of the cell lysate was measured by a dual-luciferase reporter assay system (Promega), and the transfection efficiency was corrected with pRL-TK vector-dependent renilla luciferase activity.
Results
TM mRNA levels in HUVECs exposed to lipid extracts from ox-LDL
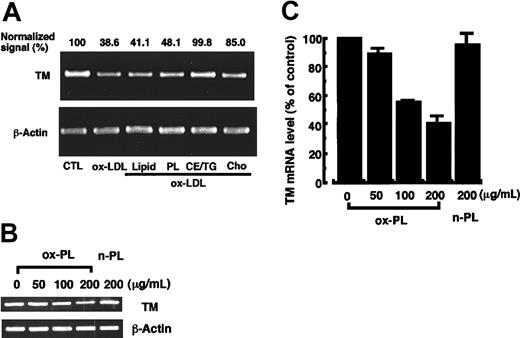
The dose- and time-dependent effects of ox-LDL or crude lipid extracts from ox-LDL on TM mRNA levels in HUVECs were investigated (Figure 1). The TBARS value of ox-LDL prepared by UV irradiation of LDL was 14.33 ± 2.66 nmol MDA/mg protein, while no TBARS was observed in native LDL. LDL containing 200 μg protein contained 200 ± 15 μg (mean ± SD) phospholipid. Cell viability as measured by the MTT assay was higher than 90% in all these experiments (data not shown). A dose- (Figure 1A-B) and time-dependent (Figure 1C-D) decrease in TM mRNA levels were observed in the cells treated with ox-LDL, and the normalized signal decreased by about 65% in the cells treated with 200 μg protein/mL ox-LDL for 12 hours (Figure 1A,C) as previously described.25 HUVECs treated with crude lipid extracts from ox-LDL also decreased TM mRNA levels in a dose- (Figure 1E-F) and time-dependent (Figure 1G-H) manner. The normalized TM mRNA signal decreased by 60% in the cells treated with crude lipid extracts containing 200 μg/mL phospholipids from ox-LDL for 12 hours (Figure 1E,G). No influence on TM mRNA levels was observed with incubations of up to 12 hours with 200 μg protein/mL native LDL or crude lipid extracts containing 200 μg/mL phospholipids from native LDL (Figure 1A,E). The results indicated that the lipid component of ox-LDL induced a decrease in HUVEC TM mRNA levels to the same extent as the ox-LDL.
Dose- and time-dependent decrease in TM mRNA levels in HUVECs treated with ox-LDL or crude lipid extracts from ox-LDL. (A-B) Cells were treated for 12 hours with or without various concentrations of ox-LDL or 200 μg protein/mL native LDL (n-LDL). (C-D) Cells were treated with 200 μg protein/mL ox-LDL for various times. (E-F) Cells were treated for 12 hours with or without crude lipids extracts (lipids) from ox-LDL or from 200 μg protein/mL n-LDL. The lipid concentrations are expressed as the equivalent concentration of phospholipids (PL) in various concentrations of ox- or n-LDL. The crude lipid extracts containing 200 μg/mL PL from LDL are of equal concentration to those in 200 μg protein/mL LDL. (G-H) Cells were treated for various times with the crude lipids containing 200 μg PL/mL extracted from ox-LDL. Total RNA was extracted from the cells, and TM mRNA levels in the cells were analyzed by quantitative RT-PCR as described in “Materials and methods.” (B,D,F,H) The autoradiograms of panels B, D, F, and H were analyzed by densitometry and the results are expressed in panels A, C, E, and G, respectively, as a percentage of control values after normalization to the β-actin mRNA signal. Panels A, C, E, and G represent the mean ± SD from 4 independent experiments, and panels B, D, F, and H show representative results.
Dose- and time-dependent decrease in TM mRNA levels in HUVECs treated with ox-LDL or crude lipid extracts from ox-LDL. (A-B) Cells were treated for 12 hours with or without various concentrations of ox-LDL or 200 μg protein/mL native LDL (n-LDL). (C-D) Cells were treated with 200 μg protein/mL ox-LDL for various times. (E-F) Cells were treated for 12 hours with or without crude lipids extracts (lipids) from ox-LDL or from 200 μg protein/mL n-LDL. The lipid concentrations are expressed as the equivalent concentration of phospholipids (PL) in various concentrations of ox- or n-LDL. The crude lipid extracts containing 200 μg/mL PL from LDL are of equal concentration to those in 200 μg protein/mL LDL. (G-H) Cells were treated for various times with the crude lipids containing 200 μg PL/mL extracted from ox-LDL. Total RNA was extracted from the cells, and TM mRNA levels in the cells were analyzed by quantitative RT-PCR as described in “Materials and methods.” (B,D,F,H) The autoradiograms of panels B, D, F, and H were analyzed by densitometry and the results are expressed in panels A, C, E, and G, respectively, as a percentage of control values after normalization to the β-actin mRNA signal. Panels A, C, E, and G represent the mean ± SD from 4 independent experiments, and panels B, D, F, and H show representative results.
TM mRNA levels in HUVECs exposed to phospholipid extracts from ox-LDL
To elucidate the active lipid species in LDL, the different lipid components of ox-LDL were separated and analyzed for their effect on TM mRNA levels in HUVECs (Figure 2A). Crude lipid extracts from ox-LDL were separated into 3 fractions, which predominantly contained phospholipid, free cholesterol, and triglyceride and cholesterol ester. On average, LDL containing 200 μg protein contained 200 μg phospholipid, 37 μg free cholesterol, 52.7 μg triglyceride, and 252 μg cholesterol ester, and these concentrations were used in the experiments. A significant decrease in TM mRNA levels was observed in the cells treated with 200 μg protein/mL ox-LDL, crude lipid extracts containing 200 μg/mL phospholipid, or 200 μg/mL phospholipid. A decrease was not observed with the cholesterol ester and triglyceride fraction. A slight decrease in TM mRNA levels was observed in the cells treated with 37 μg/mL free cholesterol isolated from the ox-LDL. However, commercially available cholesterol (40 μg/mL) irradiated with UV light for 12 hours failed to decrease in TM mRNA levels (data not shown). About a 50% decrease in TM antigen levels was observed in HUVECs treated for 12 hours with the ox-LDL phospholipid fraction (200 μg/mL) and the extent of the decrease was comparable with that induced by 200 μg protein/mL ox-LDL (data not shown). No significant decrease in TM antigen levels was observed in the cells treated with either native LDL or the 3 lipid extracts of native LDL (data not shown). A dose-dependent decrease in TM mRNA levels was observed in HUVECs treated for 12 hours with 50 to 200 μg/mL phospholipid fraction of ox-LDL (Figure 2B). The normalized TM mRNA signal decreased by 60% in the cells treated with 200 μg/mL ox-LDL phospholipid (Figure 2C). In contrast, the phospholipid (200 μg/mL) separated from native LDL did not affect TM mRNA levels (Figure 2B-C). A time-dependent decrease in TM mRNA levels also was observed in the cells treated with 200 μg/mL ox-LDL phospholipid extracts for up to 12 hours (data not shown) with results similar to that observed in Figure 1G-H. Taken together, the results indicated that oxidized phospholipids may be the primary active species of ox-LDL that contributes to reduced TM mRNA levels in HUVECs.
Phospholipids separated from ox-LDL induce a decrease in TM mRNA levels in HUVECs. The crude lipid extracts (Lipid) from ox-LDL were separated as described in “Materials and methods” into the phospholipid (PL), cholesterol ester and triglyceride (CE/TG), and free cholesterol (Cho) fractions. (A) HUVECs were treated for 12 hours with each lipid fraction at a concentration equivalent to that found in 200 μg protein/mL ox-LDL. TM mRNA levels were determined by quantitative RT-PCR. The signal is expressed as the percentage of control values (CTL) after normalization to the β-actin mRNA signal. (B) Phospholipids n-PL and ox-PL were separated from crude lipid extracts of native LDL or ox-LDL, respectively, and dispersed in DMEM containing 20% FCS. HUVECs were incubated with or without increasing concentrations of the phospholipids for 12 hours and TM mRNA levels were determined. (C) The autoradiogram of panel B was analyzed by densitometry, and the data are expressed as the percentage of control values after normalization to the β-actin mRNA signal. Data show the mean ± SD from 4 independent experiments.
Phospholipids separated from ox-LDL induce a decrease in TM mRNA levels in HUVECs. The crude lipid extracts (Lipid) from ox-LDL were separated as described in “Materials and methods” into the phospholipid (PL), cholesterol ester and triglyceride (CE/TG), and free cholesterol (Cho) fractions. (A) HUVECs were treated for 12 hours with each lipid fraction at a concentration equivalent to that found in 200 μg protein/mL ox-LDL. TM mRNA levels were determined by quantitative RT-PCR. The signal is expressed as the percentage of control values (CTL) after normalization to the β-actin mRNA signal. (B) Phospholipids n-PL and ox-PL were separated from crude lipid extracts of native LDL or ox-LDL, respectively, and dispersed in DMEM containing 20% FCS. HUVECs were incubated with or without increasing concentrations of the phospholipids for 12 hours and TM mRNA levels were determined. (C) The autoradiogram of panel B was analyzed by densitometry, and the data are expressed as the percentage of control values after normalization to the β-actin mRNA signal. Data show the mean ± SD from 4 independent experiments.
Ox-PAPC
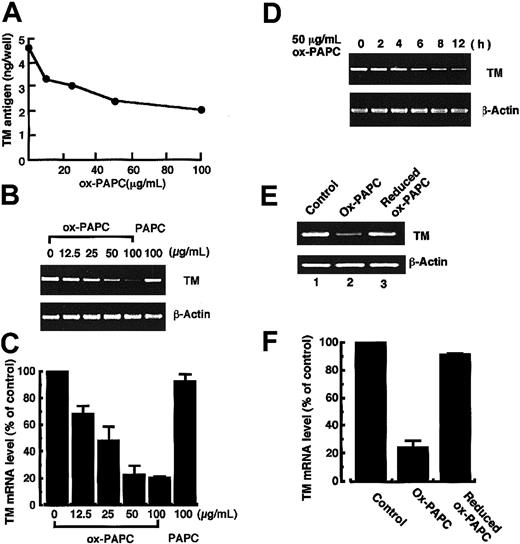
Ox-PAPC is among the oxidized phospholipids generated during oxidative modification of LDL. Ox-PAPC is a mixture of oxidized arachidonic acid–containing phospholipids that is obtained by auto-oxidation of 1-palmitoyl-2-arachidonyl-sn-glycero-3-phoshocholine and is reportedly responsible for some biologic activities of ox-LDL relevant to atherogenesis.32,37,38 Therefore, the present work investigated the effect of ox-PAPC on TM antigen and mRNA levels in HUVECs (Figure 3). Treatment of HUVECs with ox-PAPC (TBARS = 20.03 ± 0.10 nmol MDA/mg phospholipid) induced a dose-dependent decrease in TM antigen and mRNA levels (Figure 3A-C). Treatment of the cells with 50 μg/mL ox-PAPC for 12 hours induced a 54% decrease of TM antigen and 75% decrease of TM mRNA levels relative to control cells. No significant decrease in TM antigen and mRNA levels was observed in cells treated with 100 μg/mL nonoxidized native PAPC for 12 hours. A time-dependent decrease in TM mRNA levels in the cells treated with 50 μg/mL ox-PAPC was observed during the 12-hour incubation (Figure 3D). Ox-PAPC was reduced immediately by incubation with sodium borohydride for 5 minutes and its effect on TM mRNA levels was investigated (Figure 3E-F). Although down-regulation of TM mRNA levels in the cells treated with 50 μg/mL ox-PAPC for 12 hours were observed to the same extent as the above results, no decrease in TM mRNA levels was observed in the cells treated with the reduced form of ox-PAPC. These results suggest that ox-PAPC, a representative component of oxidized phospholipids in ox-LDL, may participate in the reduction of TM antigen and mRNA levels in HUVECs.
TM antigen and its mRNA levels in HUVECs treated with ox-PAPC, native PAPC, or the reduced form of ox-PAPC. (A-B) HUVECs were incubated with various concentrations of ox-PAPC or 100 μg/mL native PAPC for 12 hours. TM antigen (A) and its mRNA (B) levels in the cells were assayed by the method as described in “Materials and methods.” Data shown in panel A are the mean from 3 independent experiments. (C) The autoradiogram of panel B was analyzed by densitometry and the data are expressed as the percentage of control values after normalization to the β-actin mRNA signal. Data shown represent the mean ± SD from 4 independent experiments. (D) HUVECs were treated with 50 μg/mL ox-PAPC for various times, and TM mRNA levels were quantitated by the RT-PCR method. (E) ox-PAPC was reduced by incubation with 500 μg sodium borohydride for 5 minutes, and the reduced form of ox-PAPC was extracted and dispersed in DMEM as described in “Materials and methods.” HUVECs were treated with 50 μg/mL ox-PAPC or 50 μg/mL reduced form of ox-PAPC for 12 hours, and TM mRNA levels were assayed. (F) The autoradiogram of panel E was analyzed by densitometry, and the data are expressed as the percentage of control values after normalization to the β-actin mRNA signal. Data shown are the mean ± SD from 4 independent experiments.
TM antigen and its mRNA levels in HUVECs treated with ox-PAPC, native PAPC, or the reduced form of ox-PAPC. (A-B) HUVECs were incubated with various concentrations of ox-PAPC or 100 μg/mL native PAPC for 12 hours. TM antigen (A) and its mRNA (B) levels in the cells were assayed by the method as described in “Materials and methods.” Data shown in panel A are the mean from 3 independent experiments. (C) The autoradiogram of panel B was analyzed by densitometry and the data are expressed as the percentage of control values after normalization to the β-actin mRNA signal. Data shown represent the mean ± SD from 4 independent experiments. (D) HUVECs were treated with 50 μg/mL ox-PAPC for various times, and TM mRNA levels were quantitated by the RT-PCR method. (E) ox-PAPC was reduced by incubation with 500 μg sodium borohydride for 5 minutes, and the reduced form of ox-PAPC was extracted and dispersed in DMEM as described in “Materials and methods.” HUVECs were treated with 50 μg/mL ox-PAPC or 50 μg/mL reduced form of ox-PAPC for 12 hours, and TM mRNA levels were assayed. (F) The autoradiogram of panel E was analyzed by densitometry, and the data are expressed as the percentage of control values after normalization to the β-actin mRNA signal. Data shown are the mean ± SD from 4 independent experiments.
Half-life of TM mRNA in HUVECs exposed to ox-LDL or ox-PAPC
The half-life of TM mRNA in HUVECs treated with or without ox-LDL or ox-PAPC was measured using DRB, a transcription inhibitor, to investigate effects on TM mRNA stability (Figure 4). TM mRNA levels in the cells treated with 65 μM DRB alone decreased in a time-dependent manner (Figure 4A,C), and the apparent half-life of normalized TM mRNA was calculated as 2.7 ± 0.3 hours (Figure 4B,D). The half-life of TM mRNA in cells treated with ox-LDL plus DRB or ox-LDL alone was 3.1 ± 0.3 hours and 3.0 ± 0.3 hours, respectively, and did not significantly differ from that in the cells treated with DRB alone. The half-life of TM mRNA in cells treated with ox-PAPC plus DRB or ox-PAPC alone was 2.8 ± 0.2 hours and 3.1 ± 0.2 hours, respectively. These also did not differ significantly from that observed in the cells treated with DRB or ox-LDL alone. The results indicated that ox-LDL or ox-PAPC decreased TM mRNA levels without an apparent effect on the stability of the mRNA in HUVECs.
The half-life of TM mRNA in HUVECs treated with DRB, ox-LDL, or ox-PAPC. HUVECs were treated with 65 mM DRB alone (DRB), DRB plus 200 μg protein/mL ox-LDL (DRB + ox-LDL), ox-LDL alone (ox-LDL), DRB plus 50 μg/mL ox-PAPC (DRB + ox-PAPC), or ox-PAPC alone (ox-PAPC) for various times. Total RNA was isolated from the cells for RT-PCR analysis as described in “Materials and methods” (A,C). The autoradiogram was analyzed by densitometry and the data are expressed as the percentage of control after normalization to β-actin mRNA levels (B,D). Each symbol (▪, •, and ▴) represents 3 different experiments. The half-life (t 1/2) for each condition was calculated by linear regression analysis from semilog plots of the mRNA remaining versus incubation times and is reported as the mean.
The half-life of TM mRNA in HUVECs treated with DRB, ox-LDL, or ox-PAPC. HUVECs were treated with 65 mM DRB alone (DRB), DRB plus 200 μg protein/mL ox-LDL (DRB + ox-LDL), ox-LDL alone (ox-LDL), DRB plus 50 μg/mL ox-PAPC (DRB + ox-PAPC), or ox-PAPC alone (ox-PAPC) for various times. Total RNA was isolated from the cells for RT-PCR analysis as described in “Materials and methods” (A,C). The autoradiogram was analyzed by densitometry and the data are expressed as the percentage of control after normalization to β-actin mRNA levels (B,D). Each symbol (▪, •, and ▴) represents 3 different experiments. The half-life (t 1/2) for each condition was calculated by linear regression analysis from semilog plots of the mRNA remaining versus incubation times and is reported as the mean.
RAR, RXR, and Sp proteins in the nuclei
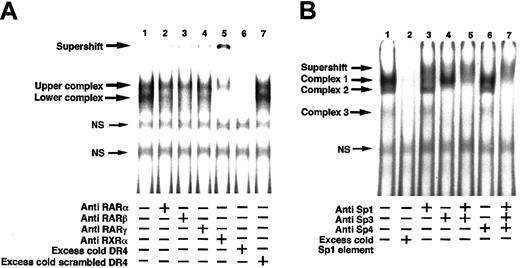
The 5′-flanking region of the TM gene contains a DR4 sequence binding for the RARs-RXRα heterodimer and an Sp1 binding sequence binding for Sp1 protein.10-12 The nuclear proteins from untreated control HUVECs that bind to the DR4 oligonucleotide sequence were investigated with an EMSA using rabbit antihuman RARα, RARβ, RARγ, or RXRα IgG (Figure 5A). There were 2 unique bands observed in the control EMSA (lane 1). A slightly reduced intensity of the lower band was observed by preincubation of the nuclear proteins with anti-RARα or RARγ IgG before assay (lanes 2 and 4, respectively), and a marked decrease in the intensity was observed by preincubation of the nuclear proteins with anti-RARβ IgG (lane 3). This indicates that RARα, RARβ, and RARγ proteins were components in the lower band, and RARβ was a major component in the RARs. Preincubation of the nuclear proteins with anti–human RXRα IgG before EMSA resulted in disappearance of the lower band, a marked decrease in intensity of the upper band and appearance of a new band at the gel top (lane 5), indicating that RXRα was an essential component in the lower band and a major component in the upper band.
Supershift assay of nuclear proteins extracted from HUVECs with the DR4 sequence and Sp1 binding sequence in the TM promoter. Nuclear proteins were prepared from cultured control HUVECs as described in “Materials and methods.” Nuclear proteins (5 μg) were incubated with or without 1 μg antibody directed against human RARα, RARβ, RARγ, or RXRα protein (A) and Sp1, Sp3, or Sp4 proteins (B) for 30 minutes at 25°C. After incubation, 0.5 ng 32P-labeled DR4 or Sp1 oligonucleotide (1 × 105 cpm) was added, and the reaction was allowed to proceed for 30 minutes at 25°C before EMSA was performed. EMSAs were performed using 4% polyacrylamide gels, and complexes bound with the oligonucleotide were detected with a Bio-Imaging Analyzer (Fuji Film, Tokyo, Japan). Binding specificity of nuclear proteins with the labeled oligonucleotides was confirmed by addition of excess cold DR4 and a scrambled DR4 (A, lanes 6-7) or excess cold Sp1 oligonucleotide (B, lane 2). NS indicates nonspecific binding.
Supershift assay of nuclear proteins extracted from HUVECs with the DR4 sequence and Sp1 binding sequence in the TM promoter. Nuclear proteins were prepared from cultured control HUVECs as described in “Materials and methods.” Nuclear proteins (5 μg) were incubated with or without 1 μg antibody directed against human RARα, RARβ, RARγ, or RXRα protein (A) and Sp1, Sp3, or Sp4 proteins (B) for 30 minutes at 25°C. After incubation, 0.5 ng 32P-labeled DR4 or Sp1 oligonucleotide (1 × 105 cpm) was added, and the reaction was allowed to proceed for 30 minutes at 25°C before EMSA was performed. EMSAs were performed using 4% polyacrylamide gels, and complexes bound with the oligonucleotide were detected with a Bio-Imaging Analyzer (Fuji Film, Tokyo, Japan). Binding specificity of nuclear proteins with the labeled oligonucleotides was confirmed by addition of excess cold DR4 and a scrambled DR4 (A, lanes 6-7) or excess cold Sp1 oligonucleotide (B, lane 2). NS indicates nonspecific binding.
The nuclear proteins from untreated control HUVECs that bind to the Sp1 oligonucleotide sequence were investigated with an EMSA using anti–human Sp1, Sp3, or Sp4 IgG (Figure 5B). There were 3 bands observed in the control EMSA (lane 1). There was a marked decrease in intensity of the complex 1 band and appearance of a weak broad band just above the complex 1 band after preincubation of the nuclear proteins with anti–Sp1 IgG (lane 3). Disappearance of the complex 2 and 3 bands was observed after preincubation of the proteins with anti–Sp3 IgG (lane 4). Preincubation of the nuclear proteins with both the anti–Sp1 and –Sp3 IgGs (lane 5) resulted in a marked decrease in the complex 1 band, appearance of a broad supershift band above the complex 1 band, and disappearance of the complex 2 and 3 bands. In contrast, preincubation of the nuclear proteins with anti–Sp4 IgG did not affect appearance of the original 3 bands (lane 6). Preincubation with anti–Sp1, –Sp3, and –Sp4 IgGs (lane 7) resulted in the same image as that observed in lane 5. These results indicated that Sp1 was a major component in the complex 1 band and Sp3 was an essential component in the complex 2 and 3 bands. The binding specificity of nuclear proteins to radiolabeled DR4 element or Sp1 element was confirmed by competition with excess unlabeled DR4 (Figure 5A, lane 6) or Sp1 (Figure 5B, lane 2) oligonucleotide.
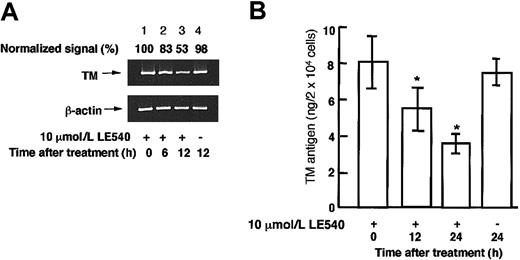
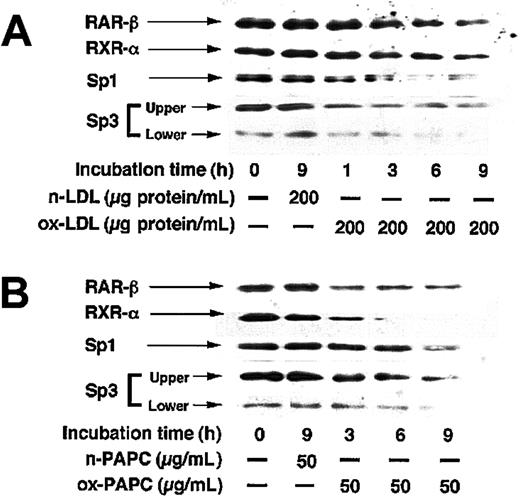
LE540 is a specific antagonist of transcriptional activation of RARβ, but not RARα, RARγ, and RXRα.39 When HUVECs were treated with 10 μM LE540 for 6 or 12 hours, TM mRNA levels decreased by about 20% and 45%, respectively (Figure 6A), and when the cells were treated with 10 μM LE540 for 12 or 24 hours, TM antigen levels decreased by 30% and 55%, respectively (Figure 6B), relative to controls. These results indicated that TM mRNA and antigen levels were controlled by transcriptional activation via RARβ. The effect of ox-LDL or ox-PAPC on RARβ, RXRα, Sp1, and Sp3 expression levels in the nuclei of HUVECs was investigated (Figure 7). Sp3 formed 2 complexes, the complex 2 and 3, with the Sp1 sequence as shown in Figure 5B. Therefore, it was observed that 2 protein bands were recognized with anti-Sp3 antibody, a clear upper band (about 100 kDa) and a weak lower band (about 60 kDa), in the Western blotting of the nuclear proteins from untreated control HUVECs. Expression levels of RARβ, RXRα, Sp1, and Sp3 in HUVECs treated with 200 μg protein/mL native LDL or 50 μg/mL native PAPC for 9 hours did not differ from those in the untreated control cells. However, a time-dependent decrease in the expression levels of RARβ, RXRα, Sp1, and Sp3 was observed in the nuclei of the cells treated with 200 μg protein/mL ox-LDL (Figure 7A) or 50 μg/mL ox-PAPC (Figure 7B) for 9 hours.
The effect of LE540, a specific antagonist for RARβ, on TM mRNA levels in the nuclei of HUVECs. HUVECs were incubated with or without 10 μM LE540 for 6, 12, or 24 hours. (A) TM mRNA levels in the cells were assayed by a quantitative RT-PCR method described in “Materials and methods.” The normalized signal is expressed as the percentage of control values (0 hours) after normalization to the β-actin mRNA signal. (B) TM antigen levels in cell lysate were determined by enzyme immunoassay. The data represent the mean ± SD from 4 independent experiments and are analyzed by the Student t test; *P < .05 when compared with control without LE540 (0 hours).
The effect of LE540, a specific antagonist for RARβ, on TM mRNA levels in the nuclei of HUVECs. HUVECs were incubated with or without 10 μM LE540 for 6, 12, or 24 hours. (A) TM mRNA levels in the cells were assayed by a quantitative RT-PCR method described in “Materials and methods.” The normalized signal is expressed as the percentage of control values (0 hours) after normalization to the β-actin mRNA signal. (B) TM antigen levels in cell lysate were determined by enzyme immunoassay. The data represent the mean ± SD from 4 independent experiments and are analyzed by the Student t test; *P < .05 when compared with control without LE540 (0 hours).
Western blots for RARβ, RXRα, Sp1, and Sp3 in nuclear proteins extracted from HUVECs treated with native LDL, ox-LDL, native PAPC, or ox-PAPC. HUVECs were incubated with or without 200 μg protein/mL native LDL (n-LDL) or ox-LDL (A), or 50 μg/mL native PAPC (n-PAPC) or ox-PAPC (B) for various times, and nuclear proteins were extracted from the cells. Nuclear proteins (5 μg) were subjected to SDS-PAGE under nonreducing conditions and transferred to a nitrocellulose membrane. The membrane was incubated with polyclonal antibody to RARβ, RXRα, or Sp3, or with monoclonal antibody to Sp1. Detection of RARβ, RXRα and Sp3, and Sp1 were carried out using HRP–conjugated sheep anti–rabbit IgG and HRP-conjugated sheep anti–mouse IgG, respectively, with detection using chemiluminescence and exposure of X-ray film.
Western blots for RARβ, RXRα, Sp1, and Sp3 in nuclear proteins extracted from HUVECs treated with native LDL, ox-LDL, native PAPC, or ox-PAPC. HUVECs were incubated with or without 200 μg protein/mL native LDL (n-LDL) or ox-LDL (A), or 50 μg/mL native PAPC (n-PAPC) or ox-PAPC (B) for various times, and nuclear proteins were extracted from the cells. Nuclear proteins (5 μg) were subjected to SDS-PAGE under nonreducing conditions and transferred to a nitrocellulose membrane. The membrane was incubated with polyclonal antibody to RARβ, RXRα, or Sp3, or with monoclonal antibody to Sp1. Detection of RARβ, RXRα and Sp3, and Sp1 were carried out using HRP–conjugated sheep anti–rabbit IgG and HRP-conjugated sheep anti–mouse IgG, respectively, with detection using chemiluminescence and exposure of X-ray film.
Binding activity of nuclear proteins from HUVECs treated with oxidized phospholipids to the TM DR4 or Sp1 sequence and deletion analysis of the DR4 and/or Sp1 sequences within the TM promoter
Interaction of the DR4 sequence or Sp1 binding sequence with the nuclear proteins from HUVECs treated with ox-PAPC or phospholipids from ox-LDL was investigated by the EMSA method (Figure 8). Binding activity of the nuclear proteins from HUVECs treated with 50 μg/mL ox-PAPC for 9 hours to the DR4 sequence apparently decreased (Figure 8A, lane 2) compared with that of the proteins from untreated control cells (lane 1). In contrast, no significant changes were observed in the EMSA between the DR4 and the nuclear proteins from HUVECs exposed to the reduced form of ox-PAPC (lane 3) or to native PAPC for 9 hours (lane 4). Similarly, binding of the Sp1 sequence with the nuclear proteins from HUVECs treated with ox-PAPC for 9 hours (Figure 8B, lane 2) also decreased in comparison with the proteins from untreated control cells (lane 1). Binding of the Sp1 sequence with nuclear proteins from the cells treated with the reduced form of ox-PAPC (Figure 8B, lane 3) or with native PAPC (lane 4) was similar to untreated control cells. Addition of excess unlabeled oligonucleotides abolished binding of the labeled oligonucleotide with the nuclear proteins from the cells (Figure 8A-B, lane 5), demonstrating specificity of the binding of oligonucleotides and nuclear proteins from the cells. Essentially identical results were obtained using phospholipids (200 μg/mL) from ox-LDL or native LDL, or the NaBH4-treated reduced form of phospholipid from ox-LDL (Figure 8C-D). These results demonstrated that treatment of HUVECs with oxidized phospholipids from ox-LDL or ox-PAPC repress the binding activity of nuclear proteins to both the DR4 and Sp1 binding elements of the 5′-flanking region of the TM gene.
EMSA of nuclear proteins from HUVECs treated with or without oxidized lipids with the DR4 and Sp1 sequences from the TM promoter. Ox-PAPC and ox-LDL were prepared by UV irradiation of PAPC and LDL, respectively, for 12 hours. Oxidized phospholipids (ox-PLs) and native phospholipids (n-PLs) were separated from crude lipid extracts prepared from their respective LDLs. Reduced forms of ox-PLs (Reduced ox-PLs) and ox-PAPC (Reduced ox-PAPC) were prepared by incubation of the ox-PLs and ox-PAPC with sodium borohydride as described in “Materials and methods.” (A-B) HUVECs were incubated for 9 hours with or without 50 μg/mL ox-PAPC, reduced ox-PAPC, or n-PAPC. (C-D) HUVECs were incubated for 9 hours with or without 200 μg/mL ox-PLs, reduced ox-PLs, or n-PLs. Nuclear proteins (2 μg) isolated from the cells were incubated with 0.5 ng 32P-labeled oligonucleotide probes (1 × 105 cpm) for DR4 (A,C) or Sp1 (B,D) for 30 minutes at 25°C. EMSAs were performed using 4% polyacrylamide gels and complexes bound with oligonucleotide were detected with a Bio-Imaging Analyzer.
EMSA of nuclear proteins from HUVECs treated with or without oxidized lipids with the DR4 and Sp1 sequences from the TM promoter. Ox-PAPC and ox-LDL were prepared by UV irradiation of PAPC and LDL, respectively, for 12 hours. Oxidized phospholipids (ox-PLs) and native phospholipids (n-PLs) were separated from crude lipid extracts prepared from their respective LDLs. Reduced forms of ox-PLs (Reduced ox-PLs) and ox-PAPC (Reduced ox-PAPC) were prepared by incubation of the ox-PLs and ox-PAPC with sodium borohydride as described in “Materials and methods.” (A-B) HUVECs were incubated for 9 hours with or without 50 μg/mL ox-PAPC, reduced ox-PAPC, or n-PAPC. (C-D) HUVECs were incubated for 9 hours with or without 200 μg/mL ox-PLs, reduced ox-PLs, or n-PLs. Nuclear proteins (2 μg) isolated from the cells were incubated with 0.5 ng 32P-labeled oligonucleotide probes (1 × 105 cpm) for DR4 (A,C) or Sp1 (B,D) for 30 minutes at 25°C. EMSAs were performed using 4% polyacrylamide gels and complexes bound with oligonucleotide were detected with a Bio-Imaging Analyzer.
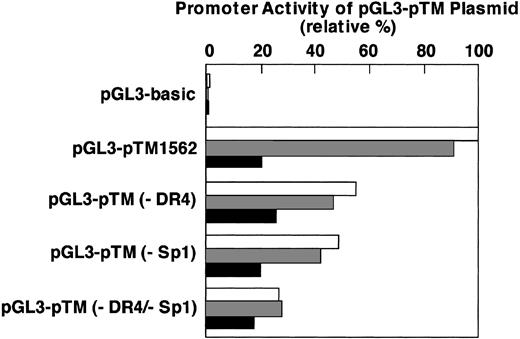
The binding activity of nuclear proteins to both the DR4 and Sp1 elements within the TM promoter with respect to oxidized phospholipid-induced down-regulation of TM expression was studied with TM promoter-luciferase fusion constructs. The wild-type construct pGL3-pTM1562, DR4 deletion mutant construct pGL3-pTM (–DR4), Sp1 deletion mutant construct pGL3-pTM (–Sp1), or DR4 and Sp1 deletion mutant construct pGL3-TM (–DR4/–Sp1) was fused to a luciferase reporter plasmid and cotransfected into HUVECs with a transfection efficiency reporter construct. The transfected cells were treated with 50 μg/mL n-PAPC or ox-PAPC for 12 hours (Figure 9). The promoter activity was determined by firefly luciferase activity in the cell extracts and expressed as a percentage of that with the wild-type construct not treated with PAPC. All activities were normalized with renilla luciferase activity. The DR4 deletion mutant (–DR4)–, Sp1 deletion mutant (–Sp1)–, and both deletion mutant (–DR4/–Sp1)–transfected cells exhibited about 45%, 50%, and 75% decreases in the promoter activity, respectively, compared with that of the cells transfected with the wild-type construct. After treatment with ox-PAPC, the cells transfected with the wild-type TM promoter exhibited only 20% to 23% of the activity observed when they were treated with medium alone or n-PAPC. The promoter activities of –DR4-transfected and –Sp1-transfected cells treated with ox-PAPC exhibited 47% and 44%, respectively, of each control. In contrast, the activity observed with –DR4/–Sp1-transfected cells treated with ox-PAPC did not decrease compared with treatment with medium alone or n-PAPC. The results provided evidence that the binding of nuclear proteins to both the DR4 element and Sp1 binding elements of the TM promoter is essential to promote TM transcription in HUVECs. Furthermore, reduction of TM promoter activity by ox-PAPC, but not n-PAPC, clearly relates to the interaction of proteins with these binding elements.
Deletion analysis of the DR4 and/or Sp1 sequences within the TM promoter. HUVECs were transfected with pGL3-pTM1562 plasmid or some deletion mutants as well as pRL-TK plasmid as described in “Materials and methods.” The culture medium was changed after 4 hours of catinoic liposome transfection, and 24 hours thereafter, the cells were treated with medium alone, or either 50 μg/mL of n-PAPC or ox-PAPC for 12 hours. The activities of firefly and renilla luciferases in the cell extracts were determined and the former activity normalized with respect to the later activity was expressed as a percentage of that with pGL3-pTM1562 plasmid treated without PAPC. □ indicates medium alone; ▦, n-PAPC treated; and ▪, ox-PAPC treated. Results were the average values of 3 independent experiments. pGL3-basic indicates plasmid not containing any TM promoter region; pGL3-pTM1562, pGL3-control plasmid containing –1562 bp from transcription site; pGL3-pTM(–DR4), pGL3-pTM1562 plasmid deleted from –1524 to –1521 in the DR4 site; pGL3-pTM(–Sp1), pGL3-pTM1562 plasmid deleted from –145 to –121 in the Sp1 site; and pGL3-pTM(–DR4/–Sp1), pGL3-pTM1562 plasmid deleted from –1524 to –1521 and from –145 to –121.
Deletion analysis of the DR4 and/or Sp1 sequences within the TM promoter. HUVECs were transfected with pGL3-pTM1562 plasmid or some deletion mutants as well as pRL-TK plasmid as described in “Materials and methods.” The culture medium was changed after 4 hours of catinoic liposome transfection, and 24 hours thereafter, the cells were treated with medium alone, or either 50 μg/mL of n-PAPC or ox-PAPC for 12 hours. The activities of firefly and renilla luciferases in the cell extracts were determined and the former activity normalized with respect to the later activity was expressed as a percentage of that with pGL3-pTM1562 plasmid treated without PAPC. □ indicates medium alone; ▦, n-PAPC treated; and ▪, ox-PAPC treated. Results were the average values of 3 independent experiments. pGL3-basic indicates plasmid not containing any TM promoter region; pGL3-pTM1562, pGL3-control plasmid containing –1562 bp from transcription site; pGL3-pTM(–DR4), pGL3-pTM1562 plasmid deleted from –1524 to –1521 in the DR4 site; pGL3-pTM(–Sp1), pGL3-pTM1562 plasmid deleted from –145 to –121 in the Sp1 site; and pGL3-pTM(–DR4/–Sp1), pGL3-pTM1562 plasmid deleted from –1524 to –1521 and from –145 to –121.
Discussion
The present work demonstrated that endothelial cells treated with phospholipid extracts, but not free cholesterol or triglyceride and cholesterol ester, separated from ox-LDL significantly reduced TM mRNA levels. Further, the degree of down-regulation was nearly the same extent as that in cells treated with intact ox-LDL and with extracts containing phospholipids at a concentration equivalent to that in ox-LDL. This result suggests that the phospholipid component of ox-LDL is a major active species that contributes to down-regulation of TM mRNA levels. This supports the previous suggestion that down-regulation of TM antigen and its mRNA levels in HUVECs exposed to ox-LDL could be mediated by the lipid components in ox-LDL.25 Among the lipid components found in ox-LDL, lysoPC (50 μg/mL), 7-oxocholesterol (40 μg/mL), 7β-hydroxyl-cholesterol (40 μg/mL), and 25-hydroxyl-cholesterol (40 μg/mL) failed to mimic the down-regulation of TM mRNA levels in HUVECs treated with ox-LDL (data not shown). In contrast, ox-PAPC produced by UV irradiation of PAPC induced a decrease in TM mRNA levels in a dose- and time-dependent manner. The oxidation was a requirement because treatment of the cells with native PAPC or the NaBH4 reduced form of ox-PAPC failed to affect TM mRNA levels. Ox-PAPC is a mixture of products obtained by auto-oxidation of PAPC, some of which were identified in mildly ox-LDL as 1-palmitoyl-2-oxovaleryl-sn-gylcero-3-phosphocholine, 1-palmitoyl-2-glutaryl-sn-gylcero-3-phosphocholine, and 1-palmitoyl-2-epoxyisoprostane-sn-gylcero-3-phosphocholine.32,37,38,40 Treatment of ox-PAPC with sodium borohydride reduces the hydroperoxides, epoxides, and carbonyls (aldehyde and ketone) in ox-PAPC to hydroxides.32 Therefore, the present results imply that a major active component of ox-LDL that influences TM mRNA levels may be oxidized phospholipids, including ox-PAPC with hydroperoxides, epoxides, and/or carbonyls moieties. Ox-PAPC is responsible for the biologic activity of mildly ox-LDL and induces expression of adhesion proteins for monocytes in endothelial cells.32,41-43 The presence of these oxidized phospholipids in fatty streak lesions from cholesterol-fed rabbits has been documented, suggesting that specific oxidized derivation of arachidonic acid–containing phospholipids may be important initiators of atherogenesis.32
When we treated HUVECs with cytochalasin B (final concentration 80 ng/mL), an endocytosis inhibitor, for 20 minutes before exposure of the cells to ox-LDL or ox-PAPC, the ox-LDL induced down-regulation of TM mRNA levels, but the effect of ox-PAPC was abolished (data not shown). This suggests that down-regulation of TM mRNA levels by ox-LDL was mediated by endocytosis of the lipoprotein into the cells, whereas the ox-PAPC–induced down-regulation was mediated by permeation of ox-PAPC through the plasma membrane without endocytosis. Our previous report suggested that down-regulation of TM expression by ox-LDL could be mediated by degradation of the lipoprotein in lysosomes after internalization of ox-LDL, because bafilomycin A1, an inhibitor of lysosome proton pumps, prevented the loss of TM antigen and mRNA by ox-LDL.25 Therefore, it is likely that ox-LDL internalized into the cell cytosol may release oxidized phospholipids through a lysosomal pathway, and the released oxidized phospholipids may affect TM mRNA levels. However, the precise mechanism for how this might occur is not known as yet. Ox-LDL is internalized after binding several specific endothelial receptors such as CD36,44 a lectinlike ox-LDL receptor-1 (LOX-1),45 and a scavenger receptor (SREC).46 We pretreated HUVECs with phosphatidylserine (200 μg/mL), a potent competitive binding inhibitor for CD36 and SREC; polyinosinic acid (200 μg/mL), a potent competitive binding inhibitor for LOX-1 and SREC; or acetyl LDL (400 μg protein/mL), a potent competitive binding inhibitor for SREC, for 20 minutes before exposure to ox-LDL to estimate the receptor involving the ox-LDL–induced down-regulation of TM mRNA levels. The down-regulation of TM mRNA levels, however, was not significantly influenced by pretreatment with these compounds (data not shown). Therefore, the extent of participation of receptors for ox-LDL in the modulation of TM expression is not apparent from the current study.
The present work was undertaken to evaluate the mechanism for down-regulation of TM mRNA levels in HUVECs in response to ox-LDL and ox-PAPC. The apparent half-life of TM mRNA in the cells treated with ox-LDL plus DRB (3.1 hours), a transcription inhibitor, and ox-LDL alone (3.0 hours) did not significantly differ from that observed in the control cells treated with DRB alone (2.7 hours) as reported previously.22,25 Furthermore, the half-life of TM mRNA in cells treated with ox-PAPC plus DRB or ox-PAPC alone was 2.8 hours and 3.1 hours, respectively. These also did not significantly differ from that observed in the cells treated with DRB or ox-LDL alone. The results suggest that ox-LDL and ox-PAPC, a representative oxidized phospholipid, could repress the transcriptional activity of the TM gene without affecting TM mRNA stability.
The present work further investigated the molecular basis for transcriptional regulation of the TM gene in HUVECs in response to ox-LDL or ox-PAPC. Treatment of HUVECs with retinol (vitamin A) or its derivatives (retinoids) such as retinal and retinoic acid increases transcriptional activity of the TM gene.13 Retinol and retinoids exert profound effects on the transcriptional regulation of various genes, mainly through 2 families of nuclear receptors, the RARs and the RXRs. These receptors are ligand-dependent transcription factors that bind to cis-acting DNA sequences, RAREs and RXR-responsive elements, located in the promoter region of their target genes.47 RARs usually must form a heterodimer with RXRs under physiologic conditions, and the heterodimer binds to a RARE sequence and promotes transcription.47 The 5′-flanking region of the human TM gene has the DR4 sequence at –1531 to –1516 bp and the 4 binding sites for the transcription factor Sp1 at –276 to –121 bp from the transcription site.10-12 Interactions between RAR-RXRα heterodimers and the DR4 sequence and between Sp1 and the second Sp1 binding sequence (–145 to –121 bp) are observed with nuclear proteins from untreated HUVECs, and up-regulation of TM mRNA levels in cells exposed to retinoids is accompanied by increased interaction between these nuclear receptors and their cognate binding sites in the TM promoter.12 It is further known that HUVECs contain predominantly RARβ48 and treatment of HUVECs with retinoic acid markedly increases nuclear expression of RARβ and Sp1.12 The present results suggest that the lower band observed in EMSA between the DR4 and nuclear proteins from untreated control HUVECs was predominantly composed of RARβ-RXRα heterodimer (Figure 5) and results from the experiments with LE540, a RARβ selective antagonist,39 indicate that TM mRNA levels in HUVECs are controlled by transcriptional activation via RARβ (Figure 6). The Sp family includes Sp1, Sp2, Sp3, and Sp4. In most promoters, Sp1 and Sp3 recognize the classical Sp1 consensus element with comparable affinity and specificity.49 The present results indicated that the complex 1 band observed in EMSA between the Sp1 binding element and nuclear proteins from untreated control HUVECs predominantly bind with Sp1 protein and the complex 2 and 3 bands can bind with Sp3 protein. In addition, Western blot analysis indicated that expression levels of RARβ, RXRα, Sp1, and Sp3 in the nuclei were significantly decreased by treatment of the cells with ox-LDL and ox-PAPC (Figure 7). The EMSA results indicated that treatment of HUVECs with ox-PAPC and oxidized phospholipid isolated from ox-LDL significantly decreased the binding of the nuclear proteins to both the DR4 sequence and the Sp1 binding sequence in the TM promoter (Figure 8). Furthermore, the transfection experiments with deletion mutants of the DR4 element and/or Sp1 binding element in the TM promoter demonstrated that both elements are essential to promote TM gene transcription and that ox-PAPC, but not n-PAPC, repress the promoter activity (Figure 9). Overall, the data indicate that the decreased TM transcription in HUVECs exposed to oxidized phospholipids is mediated, at least in part, by decreased binding of RXRα-RARβ heterodimers and Sp proteins including Sp1 and Sp3 to the their DNA recognition sequences in the TM promoter through suppression of nuclear expression levels of RARβ, RXRα, Sp1, and Sp3.
Transcriptional down-regulation of the TM gene is found also in HUVECs treated with lipopolysaccharide (LPS), TNF-α, or IL-1β50 as well as ox-LDL through unknown mechanisms. RXRα expression in rodent liver is repressed during an acute-phase response to inflammation after injection with LPS, TNF-α, or IL-1β. This occurs through decreased RXRα transcription and increased degradation of its mRNA, suggesting that the reduction in RXRα levels alone or in association with other nuclear receptors could be a mechanism for coordinately inhibiting the expression of multiple genes during the acute-phase response.51 The present work did not address whether ox-LDL–induced repression of RARβ and RXRα levels is derived from decreased transcriptional activities and/or increased degradation of their mRNA. The current study observed decreased binding of Sp1 and Sp3 to the Sp1 binding sequence and decreased nuclear contents of Sp1 and Sp3 in HUVECS treated with ox-LDL. Pan et al52 recently showed that transcription of the calmodulin gene in hepatoma cells is intimately related to synthesis of Sp1, and the cells treated with insulin showed a marked increase in Sp1 expression in the nuclei, although the insulin-dependent up-regulation of Sp1 synthesis was antagonized by TNF-α. The mechanism that results in a decrease in Sp1 expression by TNF-α as well as by ox-LDL is not known.
The present paper presents the novel observation that decreased expression of TM mRNA levels in endothelial cells exposed to ox-LDL is induced by transcriptional down-regulation of the TM gene mediated by ox-LDL phospholipids. The proposed mechanism involves a decrease in TM gene transcription mediated by reduced binding of RXRα-RAR heterodimers containing RXRα-RARβ to the DR4 sequence and Sp1 and Sp3 to the first and second Sp1 binding sites in promoter due to decreased content of the nuclear transcription factors RARβ, RXRα, Sp1, and Sp3. The down-regulation of TM, a physiologic antithrombotic factor, on the surface of endothelial cells exposed to ox-LDL may contribute significantly to thrombotic properties of the cells in an atherosclerotic lesion. The present study contributes to an understanding of the mechanisms of thrombus formation in atherosclerotic lesions and transcriptional down-regulation of proteins containing RAREs in the promoter site of genes.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-08-2428.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal