Abstract
Platelet factor 4 (PF4) is expressed during megakaryocytic differentiation. We previously reported that GATA-1 and ETS-1 regulate the rat PF4 promoter and transactivate the PF4 gene. For the present study, we investigated the regulatory elements and their transcription factors responsible for the lineage-specific expression of the PF4 gene. The promoter activities of deletion constructs were evaluated, and a novel regulatory element termed TME (tandem repeat of MEIS1 binding element) (–219 to –182) was defined. Binding proteins to TME were strongly detected in HEL nuclear extracts by electrophoresis mobility shift assay (EMSA), and they were purified by DNA affinity chromatography. By performing Western blottings and supershift assays, the binding proteins were identified as homeodomain proteins, MEIS1, PBX1B, and PBX2. These factors are expressed in megakaryocytes differentiated from CD34+ cells in human cord blood. MEIS1 and PBXs bind to the TME as MEIS1/PBX complexes and activate the PF4 promoter. In nonmegakaryocytic HepG2 cells, GATA-1 and ETS-1 activate the PF4 promoter approximately 10-fold. Surprisingly, we found that additional expression of both MEIS1 and PBX2 multiplied this major activation another 2-fold. This activation was not observed when MEIS1 binding sites in the TME were disrupted. Furthermore, inhibition of the binding of endogenous MEIS1/PBX complexes to the TME decreased the promoter activity by almost one half, in megakaryocytic HEL cells. Thus, these studies demonstrate that the homeodomain proteins, MEIS1, PBX1B, and PBX2, play an important role in megakaryocytic gene expression.
Introduction
Megakaryocytes are the hematopoietic precursors of platelets, which play an essential role in thrombosis and hemostasis. Platelet factor 4 (PF4) is expressed exclusively in megakaryocytes and platelets.1 Because the PF4 gene is activated during the late stages of megakaryocytopoiesis, it is useful as a specific marker of megakaryocytic differentiation.2 We have previously sequenced and characterized the rat PF4 promoter.3,4 We identified a number of upstream enhancer and repressor elements. In addition, we demonstrated that GATA-1 and ETS-1 regulate the PF4 promoter and activate PF4 gene expression.5,6 Promoters of several megakaryocyte-specific genes, including glycoprotein (GP) IIb, GPIbα, thrombopoietin receptor (Mpl), GPIX, and GPV,7-11 also contain cis-acting regulatory elements such as GATA and ETS binding sites, and these genes are regulated by GATA-1 and Ets family proteins such as ETS-1, PU.1, and Fli-1.12-15 Furthermore, other transcription factors participating in megakaryocytic differentiation have been reported. For example TAL-1 (SCL) and NF-E2p45 have been implicated in early- and late-stage differentiation of megakaryocytes, respectively.15,16 Although these factors may be important for megakaryocytopoiesis, they are not involved in mediating constitutive expression of the megakaryocyte-specific genes described earlier.
Homeobox genes are broadly classified into 2 subclasses: Hox and non-Hox homeobox genes. Many genes in both classes are expressed in a variety of hematopoietic cells. Meis1 was first cloned from a site of viral integration in tumors arising in BXH-2 leukemic mice.17,18 Three genes, Meis1, Hoxa7, and Hoxa9 are activated in BXH-2 mice. Among these 3 genes, the overexpression of HOXA9 and MEIS1 in bone marrow cells induces leukemia in mice.19,20 In humans, Meis1 was found to be active in a subset of myeloid leukemia cell lines with the highest expression levels seen in the cell lines with a megakaryocytic-erythroid phenotype.21 Among the non-Hox homeobox genes, Pbx1 is normally expressed in hematopoiesis in a cell type–specific manner.22 In contrast Pbx2 and Pbx3, closely related genes to Pbx1, are expressed ubiquitously.22 Both in vitro and in vivo data suggest that MEIS and PBX mainly function in combination with HOX. MEIS1 and PBX1 are known to form a ternary complex with HOXA9 in myeloid leukemic cells.23 The homeodomain protein complexes, such as MEIS1/PBX, HOX/PBX, HOX/MEIS, and HOX/PBX/MEIS, recognize a more extended sequence than each individual binding motif and bind DNA more tightly than MEIS, PBX, and HOX by themselves.24-28 Although various homeodomain protein complexes have been identified and the role of homeodomain proteins in hematopoiesis has been demonstrated by overexpression, antisense, or gene-targeting experiments,29-32 their physiologic target genes in hematopoietic cells have not been identified.
The present study identifies a novel regulatory element in the rat PF4 promoter by comparing the transcriptional activity of PF4 promoter deletion constructs in megakaryocytic and nonmegakaryocytic cell lines. Furthermore, we demonstrate that homeodomain proteins, including MEIS1 and PBXs (PBX1B and PBX2) bind to this element in HEL cells. In addition, we describe expression of homeodomain proteins in human megakaryocytes. Surprisingly, MEIS1/PBX complexes strongly activate PF4 gene expression in a manner that is cooperative with GATA-1 and ETS-1. From these results, we propose that PF4 is a target gene of MEIS1/PBX complexes and that these transcription factors promote megakaryocytic gene expression.
Materials and methods
Plasmid construction
To make the P2-PF4 construct, a 1.1-kb fragment of the rat PF4 promoter (-1104 to +20) was isolated. The preparation of the full-length PF4 promoter was previously described.6 To make a series of reporter plasmids from P2-A to P2-G, the 1.1-kb promoter fragment was digested with EcoRI-EcoT22I (-1104 to -818) for P2-A, SmaI-HinfI (-847 to -638) for P2-B, PstI-BamHI (-563 to -222) for P2-D, NspI-ScaI (-345 to -114) for P2-E, PvuII-SmaI (-190 to -61) for P2-F, and ScaI-BanII (-114 to +20) for P2-G. For the preparation of P2-C (-736 to -389) polymerase chain reaction (PCR) was used because there were no convenient restriction sites. These fragments were blunted by T4 DNA polymerase and inserted into the SmaI site of the PGV-P2 (Toyo Ink, Tokyo, Japan) vector containing the SV40 promoter and luciferase reporter gene. For making the PF4mut plasmid, a BamHI-blunt double-stranded oligonucleotide containing mutations (the same sequence as Mut-2 in Figure 2A) was prepared and inserted between the BamHI-PvuII sites of PF4luc. The mutations in this plasmid were verified by DNA sequencing.
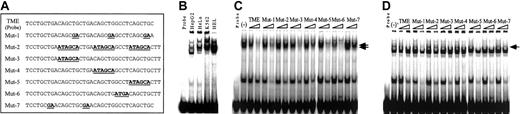
Detection of binding proteins to the TME by EMSA. (A) Sense sequences of the double-stranded oligonucleotides used in EMSA. TME indicates the wild-type TME sequence. Mutant oligonucleotides, Mut-1 to Mut-7, have various mutations in the TME and were used as the competitors. Bold underlined text indicates mutated sequences. (B) Wild-type TME EMSA performed with 6 μg nuclear extracts from 4 cell lines. (C-D) Competition experiments performed with nuclear extracts from HEL (C) and HepG2 (D) cells. (–) indicates no competitor. Arrows indicate the specific competed bands.
Detection of binding proteins to the TME by EMSA. (A) Sense sequences of the double-stranded oligonucleotides used in EMSA. TME indicates the wild-type TME sequence. Mutant oligonucleotides, Mut-1 to Mut-7, have various mutations in the TME and were used as the competitors. Bold underlined text indicates mutated sequences. (B) Wild-type TME EMSA performed with 6 μg nuclear extracts from 4 cell lines. (C-D) Competition experiments performed with nuclear extracts from HEL (C) and HepG2 (D) cells. (–) indicates no competitor. Arrows indicate the specific competed bands.
cDNAs of MEIS1, PBX1B, and PBX2 were obtained from a cDNA pool derived from HEL cells by PCR. These PCR products were cloned into the pGEM-T Easy vector (Promega, Madison, WI). Correct clones were identified by DNA sequencing. The coding sequences of MEIS1, PBX1B, and PBX2 were then removed by EcoRI digestion of cloned plasmids and were inserted into the EcoRI site of pcDNA3 (Invitrogen, San Diego, CA). The preparations of the 2 expression vectors, pcDNA3-hEts-1 and pXM-mGATA-1, were described previously.6
Cell culture and isolation of nuclear extract
HEL and K562 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 IU/mL penicillin and 100 μg/mL streptomycin (P/S). HeLa and HepG2 cells were maintained under the same conditions except that Dulbecco modified Eagle medium (DMEM) was used. The isolation method for nuclear extracts was described previously.6
Transient transfection assay
For identifying regulatory elements, reporter plasmids were transfected into K562 and HEL cells by electroporation and into HepG2 and HeLa cells by lipofection. The method of electroporation was described by Ravid et al.3 Lipofectin and lipofectamine reagents (GIBCO BRL, Gaithersburg, MD) were used for lipofection according to the manufacturer's instructions. For the electroporation, 8 μg reporter plasmid was used with 5 μg pβactin-lacZ as an internal control plasmid. For the experiment described in Figure 6B, 7 μg of each of the reporter and internal control plasmids was used. In the lipofection method, 0.5 μg reporter plasmid was used with 0.5 μg control plasmid. Each assay was carried out more than 3 times.
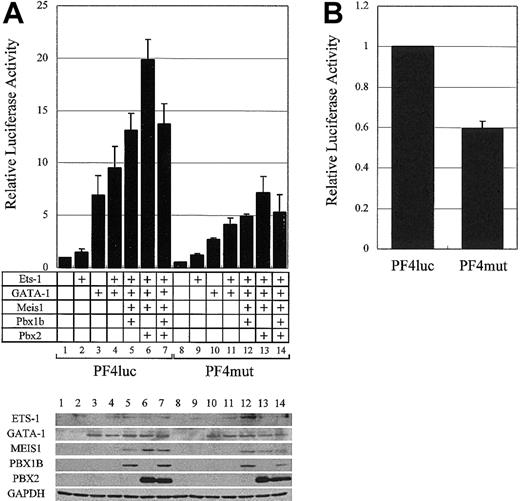
The synergistic activation of the PF4 promoter by GATA-1, ETS-1, and the PBX/MEIS complex. (A) The plasmids for the expression of ETS-1 and GATA-1 were transfected into HepG2 cells with the plasmids for MEIS1, PBX1B, and PBX2 expression. Transcriptional activities were evaluated with PF4luc or PF4mut plasmids. The expressions of all transcription factors and GAPDH (as a control) were confirmed by Western blotting (bottom panel). (B) PF4luc or PF4mut reporter-plasmids were transfected into HEL cells, and transcriptional activities were evaluated. Error bars indicate SDs.
The synergistic activation of the PF4 promoter by GATA-1, ETS-1, and the PBX/MEIS complex. (A) The plasmids for the expression of ETS-1 and GATA-1 were transfected into HepG2 cells with the plasmids for MEIS1, PBX1B, and PBX2 expression. Transcriptional activities were evaluated with PF4luc or PF4mut plasmids. The expressions of all transcription factors and GAPDH (as a control) were confirmed by Western blotting (bottom panel). (B) PF4luc or PF4mut reporter-plasmids were transfected into HEL cells, and transcriptional activities were evaluated. Error bars indicate SDs.
In the case of the reporter assays using expression plasmids, PF4luc plasmid was used as the reporter plasmid.6 PF4luc (0.5 μg) was transfected into HepG2 cells with 0.5 μg pβactin-lacZ by using Lipofectamine2000 reagent (GIBCO BRL). For the overexpression of the transcription factors, 1 μg of each expression plasmid was used. In all assays, cells were harvested approximately 48 hours after transfection. Both assays for luciferase and β-galactosidase were carried out more than 3 times.
EMSA and supershift assay
The double-stranded DNA fragment (TME frg) constructed by 2 oligonucleotides, 5′-TCCTGCTGACAGCTGCTGACAGCTGGCCTCAGCTGC-3′ (sense strand) and 5′-CGCAGCTGAGGCCAGCTGTCAGCAGCTGTCAGCAGGA-3′ (antisense strand), was labeled with Klenow polymerase and used as the probe. Double-stranded fragments, sense strands of which are described as Mut-1 through Mut-7 in Figure 2A, were used as competitors in 5- or 50-fold molar excess to the labeled probe in this study. The TME mutation probe in Figure 5B (MutTGACAG) was constructed by 2 oligonucleotides, 5′-TCCTGCAGCGATCTGCAGCGATCTGGCCTCAGCTGC-3′ (sense strand) and 5′-CGCAGCTGAGGCCAGATCGCTGCAGATCGCTGCAGGA-3′ (antisense strand). For the most part, electrophoresis mobility shift assay (EMSA) was almost the same as described previously.6 The binding reaction was carried out at 4°C for 45 minutes. In the case of the supershift assay, 6 μg nuclear extract from each cell line was incubated with antibodies at room temperature for 20 minutes before the probe was added. All antibodies (Pbx1/2/3 [C-20] Pbx1 [P-20], Pbx2 [G-20], Pbx3 [D-17], Meis1 [N-17], and HoxD9 [H-342]) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Each antibody against PBX1, PBX2, or PBX3 recognizes the specific N-terminal sequence of each protein and does not cross-react with the other proteins. The antibody against PBX1/2/3 recognizes a common C-terminal sequence of these 3 PBXs. However, in the case of PBX1B the common C-terminal sequence does not exist in it. Therefore, αPBX1/2/3 does not recognize PBX1B. For HOXA9, the antibody against HOXD9, which also recognized HOXA9, was used.
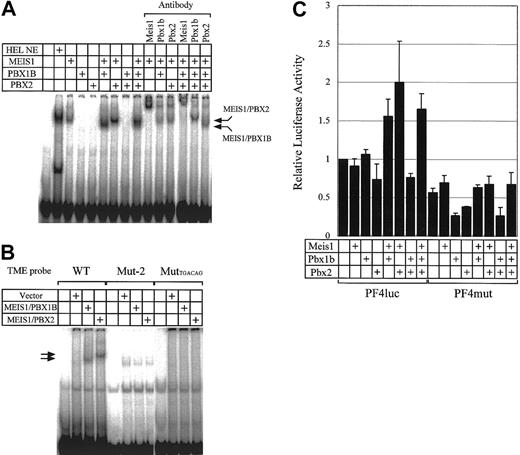
Binding activities of various combinations of MEIS1, PBX1B, and PBX2 to the TME and their effect on transcriptional activities of the PF4 promoter. (A) HEL NE (nuclear extract) and MEIS1, PBX1B, and PBX2 prepared by in vitro translation were used in EMSA. Supershift assays were also performed with antibodies to MEIS1, PBX1B, and PBX2. (B) EMSA was performed with in vitro translated proteins. The TME and 2 mutant TME probes were used. Mut-2 probe is the same sequence as Mut-2 described in Figure 2A. The MutTGACAG probe is the mutant TME probe in which 2 Meis1 binding sites are disrupted (TGACAG to AGCGAT). (C) The plasmids for the expression of MEIS1, PBX1B, and PBX2 were transfected into HepG2 cells with PF4luc or PF4mut reporter-plasmids. PF4luc contains 1.1 kb PF4 promoter in front of the luciferase reporter gene. PF4mut contains mutations in the Meis1 binding sites in the TME. Error bars indicate SDs.
Binding activities of various combinations of MEIS1, PBX1B, and PBX2 to the TME and their effect on transcriptional activities of the PF4 promoter. (A) HEL NE (nuclear extract) and MEIS1, PBX1B, and PBX2 prepared by in vitro translation were used in EMSA. Supershift assays were also performed with antibodies to MEIS1, PBX1B, and PBX2. (B) EMSA was performed with in vitro translated proteins. The TME and 2 mutant TME probes were used. Mut-2 probe is the same sequence as Mut-2 described in Figure 2A. The MutTGACAG probe is the mutant TME probe in which 2 Meis1 binding sites are disrupted (TGACAG to AGCGAT). (C) The plasmids for the expression of MEIS1, PBX1B, and PBX2 were transfected into HepG2 cells with PF4luc or PF4mut reporter-plasmids. PF4luc contains 1.1 kb PF4 promoter in front of the luciferase reporter gene. PF4mut contains mutations in the Meis1 binding sites in the TME. Error bars indicate SDs.
In vitro transcription/translation
The proteins for the EMSA experiments were prepared in vitro. The expression of plasmids was carried out by transcription and translation in vitro using the “TNT coupled transcription-translation reticulocyte lysate (T7 polymerase version)” (Promega) according to the manufacturer's instructions. The obtained translated products (MEIS1, PBX1B, and PBX2) were analyzed by Western blotting, and 3/50 of the total product of each protein was used for the EMSA.
Purification of DNA binding proteins
A biotinylated TME frg was prepared by annealing the 5′ biotinylated sense strand (5′-bio-TCCTGCTGACAGCTGCTGACAGCTGGCCTCAGCTGCTT-3′) with the antisense strand (5′-AACGCAGCTGAGGCCAGCTGTCAGCAGCTGTCAGCAGGA-3′). The annealed double-stranded oligonucleotide (6 nmol) was mixed with 20 mg nuclear extract prepared from 1 × 109 HEL cells in binding buffer (35 mM Tris (tris(hydroxymethyl)aminomethane)–HCl [pH 7.9] 1.5 mM DTT (dithiothreitol), 6.25 mM MgCl2, 15% glycerol, 1.5 mM EDTA (ethylenediaminetetraacetic acid), 50 mM KCl, 1 μg/μL poly (dI-dC)) at 4°C for 1 hour. This sample was then applied to a 500-μL NeutraAvidin Plus (Pierce, Rockford, IL) column, and the flow-through fraction was reloaded. The column was washed with about 30 mL binding buffer, and the binding proteins were eluted in 500-μL fractions with 10 mL elution buffer in a linear gradient of KCl from 5 mM to 500 mM.
Western blotting
Affinity purified fraction (20 μL; Figure 3B) and 20 μg of each nuclear extract (Figure 4E) were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were blotted onto nitrocellulose membranes. The membranes were blocked in Tris-buffered saline (TBS) containing 3% nonfat milk and 0.1% Tween 20 for 1 hour at room temperature and then incubated in the same solution containing the primary antibody (NF-E2p45 [C-19], TAL1 [E-14], E2A [Yae], GATA-1 [N6], GATA-2 [CG2-96], ETS-1 [N-276] [Santa Cruz Biotechnology] or glyceraldehyde-3-phosphate dehydrogenase [GAPDH; CHEMICON International, Temecula, CA]) at room temperature for 1 hour. The membranes were washed and then incubated in blocking solution containing the secondary antibody at room temperature for 1 hour. The immunoblots were visualized with the ECL Western blotting detection system (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's instructions.
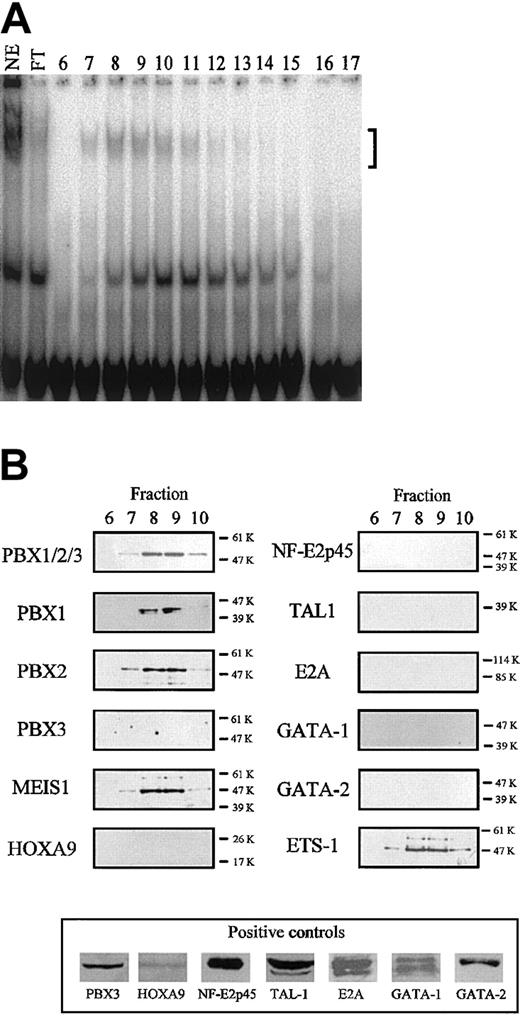
Identification of the TME binding proteins. (A) Binding of TME-DNA chromatographed fractions of nuclear extracts from HEL cells. In the lane labeled NE, 6 μg nuclear extract before the purification was used. In the lanes labeled FT and nos. 6 to 17, the flow through fraction and the fractions nos. 6 to 17 were used, respectively. The bracket indicates the specific shifted bands. (B) Identification of the binding proteins by Western blotting. The purified proteins were analyzed by Western blotting with antibodies to homeodomain proteins (left panels) and to transcription factors expressed in megakaryocytes (right panels). In the case of the antibodies for which no signal was detected, the activities of these antibodies were confirmed by Western blotting using recombinant proteins (HOXA9 and NF-E2p45) or cell extracts (bottom panel). Molecular weight is indicated to the right of each panel.
Identification of the TME binding proteins. (A) Binding of TME-DNA chromatographed fractions of nuclear extracts from HEL cells. In the lane labeled NE, 6 μg nuclear extract before the purification was used. In the lanes labeled FT and nos. 6 to 17, the flow through fraction and the fractions nos. 6 to 17 were used, respectively. The bracket indicates the specific shifted bands. (B) Identification of the binding proteins by Western blotting. The purified proteins were analyzed by Western blotting with antibodies to homeodomain proteins (left panels) and to transcription factors expressed in megakaryocytes (right panels). In the case of the antibodies for which no signal was detected, the activities of these antibodies were confirmed by Western blotting using recombinant proteins (HOXA9 and NF-E2p45) or cell extracts (bottom panel). Molecular weight is indicated to the right of each panel.
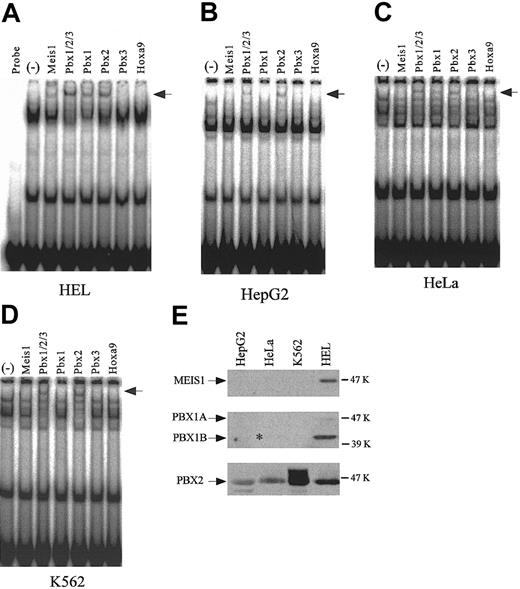
Supershift assays using antibodies to homeodomain proteins, and the detection of endogenous expression of homeodomain proteins. Supershift assays were performed with the nuclear extracts from HEL (A), HepG2 (B), HeLa (C), and K562 (D) cells. (–) indicates no antibody. Arrows indicate the supershifted bands. (E) Endogenous expression of MEIS1, PBX1, and PBX2 was detected by Western blotting using 20-μg nuclear extracts from each cell line. The asterisk denotes the existence of a faint band.
Supershift assays using antibodies to homeodomain proteins, and the detection of endogenous expression of homeodomain proteins. Supershift assays were performed with the nuclear extracts from HEL (A), HepG2 (B), HeLa (C), and K562 (D) cells. (–) indicates no antibody. Arrows indicate the supershifted bands. (E) Endogenous expression of MEIS1, PBX1, and PBX2 was detected by Western blotting using 20-μg nuclear extracts from each cell line. The asterisk denotes the existence of a faint band.
Preparation of rat megakaryocytes
Rat bone marrow cells were flushed from the femur and tibia using CATCH buffer (Ca2+/Mg2+-free Hanks balanced salt solution, 13.6 mM sodium citrate, 1 mM adenosine, 2 mM theophylline). The marrow cells were dispersed and centrifuged. The cell pellet was washed with CATCH medium and centrifuged again. The cell pellet was resuspended in lysis buffer (140 mM NH4Cl, 17 mM Tris-HCl [pH 7.2]) and incubated at 37°C for 10 minutes. Cells were centrifuged and washed twice with CATCH buffer. Cells were resuspended at a concentration of 1.5 × 106 cells/mL in Iscoves modified Dulbecco medium (IMDM) that contained 0.1 ng/mL thrombopoietin (kind gift from Kirin Brewery), 2% FBS, and 2% P/S and incubated for 3 days under the condition of humidified 5% CO2 at 37°C. Megakaryocytes were purified by a MACS Separation column (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer's instruction. Mouse antirat CD61 antibody (BD Biosciences, Erembodogem, France) and goat antimouse immunoglobulin G (IgG) microbeads (Miltenyi Biotech) were used. The purity of megakaryocytes was confirmed by staining for acetylcholinesterase (AchE).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using a Chromatin Immunoprecipitation Assay Kit (Upstate, Lake Placid, NY) according to the manufacturer's instruction. Antibodies (10 μg) for PBXs or subtype-matched normal rabbit IgG (Santa Cruz Biotechnology) were used. PCR was performed for 35 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds using the primers (forward 5′-CATACAGCATACCTTCTGCG-3′ and reverse 5′-AAGCAGCTGAGGCCAGCTGTCAGCA-3′). PCR products were separated on a 2% agarose gel and stained by ethidium bromide. The intensity of each band was quantified by Scion Image.
Isolation of CD34+ progenitor cells from cord blood and liquid suspension cultures
Isolation of CD34+ progenitor cells from cord blood and subsequent cell culture for megakaryocytopoiesis was performed according to the method of Sato et al.33 Umbilical cord blood samples from healthy full-term newborn infants were obtained from Tokyo Metropolitan Bokutou Hospital, after informed consents of the mothers. Cord blood samples were diluted with phosphate-buffered saline (PBS) and then centrifuged on Ficoll-Paque (d = 1.077 g/mL) (Pharmacia Biotech). CD34+ progenitor cells were purified from mononuclear cell preparations using the Dynal CD34 Progenitor Cell Selection System (Dynal AS, Oslo, Norway). Flow cytometric analysis of purified cells using a phycoerythrin (PE)–conjugated anti-CD34 monoclonal antibody (clone BIRMA-K3; DAKO, Glostrup, Denmark) showed that more than 95% of the selected cells were positive for CD34. These cells were cultured in Xvivo-20 (BioWhittaker, Walkersville, MD) containing 50 ng/mL thrombopoietin (PeproTech, London, United Kingdom) and 40 ng/mL c-kit ligand (Biosource), at a density of less than 1 × 105 cells/mL. Cultures were maintained at 37°C in humidified 5% CO2.
Flow cytometry
A total of 105 to 106 cells were suspended in 2% formaldehyde in PBS for 1 hour and were washed with PBS 3 times. Cells were incubated in 0.1% BSA containing PBS for 30 minutes with fluorescein isothiocyanate (FITC)–conjugated monoclonal antibody (mAb) CD41-FITC (5B12; DAKO), CD42b-FITC (SZ2; Beckman-Coulter, Tokyo, Japan), and isotype-matched antibodies (IgG1 FITC-conjugated [YLEM], IgG1 PE–conjugated [Leinco Technologies, Ballwin, MO]) that served as controls, respectively. After incubation, cells were washed with PBS containing 0.1% BSA 3 times, then suspended in 1 mL PBS containing 0.1% BSA, 200 μg/mL RNase A (Sigma, St Louis, MO), and 15 μg/mL propidium iodide (CALBIOCHEM, San Diego, CA). Cell-associated immunofluorescence was analyzed by FACScan (Becton Dickinson, San Jose, CA).
RNA preparation and RT-PCR analysis
Total RNA samples were isolated using Trizol reagent (GIBCO BRL). Total RNA (5 μg) was reverse-transcribed by the SUPER SCRIPT First-Strand Synthesis System for reverse transcription (RT)–PCR (GIBCO BRL), according to the manufacturer's instructions. After the RT reaction, 1/20 volume of the cDNA template and a set of primers (PF4, forward 5′-AGCATGAGCTCCGCAGCCGGGTTCT-3′, reverse 5′-GTAGGCAGCTAGTAGCTAACTCTCC-3′; HPRT (hypoxanthine phosphoribosyltransferase), forward 5′-GGCGTCGTGATTAGTGATGATGAACC-3′, reverse 5′-CTTGCGACCTTGACCATCTTTGGA-3′; NF-E2p45, forward 5′-CGACTCAGGATTATCCCTCAAC-3′, reverse 5′-CTGGTCTAGAGAACTCAGCTCCTT-3′; 5′-Meis1, forward 5′-ATGGCGCAAAGGTACGACGATCTAC-3′, reverse 5′-TTACATGTAGTGCCACTGCCCCTCC-3′; Pbx1a, Pbx1b, forward 5′-CCACGTGATGAATCTCCTGCGAGAG-3′, reverse 5′-TCACTGTATCCTCCTGTCTGGCTGA-3′; Pbx2, forward 5′-CTGGTTTGGCAACAAGAGGATTCGC-3′, reverse 5′-TGGAGGTATCAGAGTGAACACTCCC-3′) were mixed and used for PCR amplification. The following describes the temperature and time of each step of the PCR (denaturing, annealing, extension), the number of PCR cycles and PCR product length obtained: PF4, 95°C 1 minute, 52°C 1 minute, 72°C 2 minutes, 30 cycles, 323 bp; HPRT, 95°C 1 minute, 52°C 1 minute, 72°C 2 minutes, 30 cycles, 467 bp; NF-E2p45, 95°C 1 minute, 52°C 1 minute, 72°C 2 minutes, 36 cycles, 841 bp; Meis1, 95°C 1 minute, 65°C 30 seconds, 72°C 1 minute, 35 cycles, 1172 bp; Pbx1a and Pbx1b, 94°C 30 seconds, 55°C 30 seconds, 72°C 1 minute, 35 cycles, 627 and 514 bp; and Pbx2, 94°C 30 seconds, 55°C 30 seconds, 72°C 1 minute, 35 cycles, 410 bp.
Results
–182 to –219 region of the PF4 promoter is a candidate region regulating megakaryocyte-specific gene expression
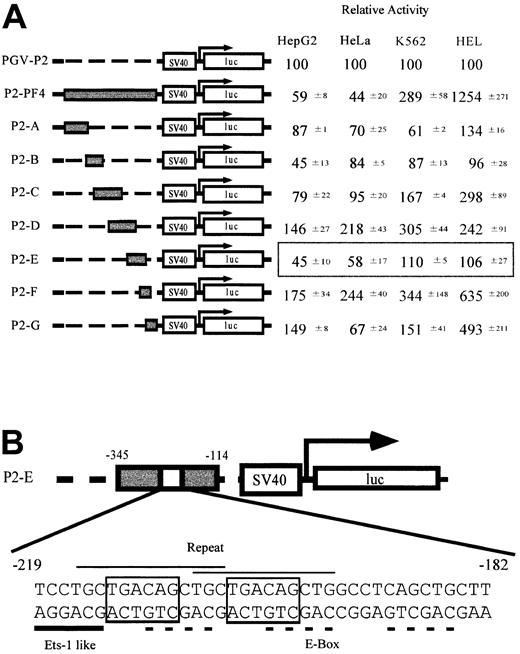
To identify the sequences regulating megakaryocyte-specific gene expression, we made a series of constructs in which full (P2-PF4) or partial PF4 promoter fragments (P2-A to P2-G) were linked to a minimal SV40 promoter and luciferase cDNA (Figure 1A). These constructs and PGV-P2, a control vector not including the PF4 promoter, were transfected into 4 cell lines, constitutive (HEL cells) and inducible (K562 cells) megakaryocytic cell lines,34 as well as nonmegakaryocytic cell lines (HepG2 and HeLa cells), and their luciferase activities were assayed. Each activity of the test promoter was compared with PGV-P2 activity, and the percentages were described as relative activities. The luciferase activity of P2-PF4 was used as the standard for megakaryocyte specificity. In other words, the PF4 promoter was maximally active in the megakaryocytic HEL cells and was maximally suppressed in nonmegakaryocytic HepG2 and HeLa cells. Although the D and F regions activated the promoter in all cells tested, the E region of the PF4 promoter suppressed the activity specifically in the nonmegakaryocytic cell lines, HepG2 and HeLa cells. These results suggest that the E region contains the sequences regulating cell type–specific gene expression. We focused on the specific element in the E region (-182 to -219) that did not overlap with D and F. We termed this E-region element the TME (tandem repeat of MEIS1 binding element). The sequence of the TME is shown in Figure 1B. This sequence includes 2 overlapping copies of a repeat (CTGCTGACAGCTG) and an Ets-1–like binding site (Ets-1 binding sequence, (C/G)(A/C)GGA(A/T)G(C/T)). Furthermore, each repeated unit contains an E-box motif (CAGCTG) and a Meis1 binding site (TGACAG). A third E-box motif is located downstream of the repeated sequences.
Screening of the regulatory elements related to the tissue-specific expression of the PF4 gene. (A) Relative activities of the constructs containing PF4 promoter fragments in the 4 cell lines. HepG2 and HeLa cells are nonmegakaryocytic cell lines. K562 cells are inducible, and HEL cells are constitutive megakaryocytic cell lines. Reporter plasmids containing either a full-length PF4 promoter (P2-PF4) or each of 7 fragments (P2A-P2G) linked to the SV40 minimal promoter were transfected into the 4 cell lines, and the luciferase assay was performed. Luciferase activities are expressed as relative activities ± SD, where the activity of the PGV-P2 was regarded as 100. (B) Structure of the novel regulatory element found only in the P2-E. Characteristic sequences are indicated by the lines above (repeated sequence, thin line) and below (ETS-1–like motif, thick line) the DNA sequence. Dotted lines and boxed sequences indicate E-box motifs and MEIS1 (TGACAG) binding motifs, respectively.
Screening of the regulatory elements related to the tissue-specific expression of the PF4 gene. (A) Relative activities of the constructs containing PF4 promoter fragments in the 4 cell lines. HepG2 and HeLa cells are nonmegakaryocytic cell lines. K562 cells are inducible, and HEL cells are constitutive megakaryocytic cell lines. Reporter plasmids containing either a full-length PF4 promoter (P2-PF4) or each of 7 fragments (P2A-P2G) linked to the SV40 minimal promoter were transfected into the 4 cell lines, and the luciferase assay was performed. Luciferase activities are expressed as relative activities ± SD, where the activity of the PGV-P2 was regarded as 100. (B) Structure of the novel regulatory element found only in the P2-E. Characteristic sequences are indicated by the lines above (repeated sequence, thin line) and below (ETS-1–like motif, thick line) the DNA sequence. Dotted lines and boxed sequences indicate E-box motifs and MEIS1 (TGACAG) binding motifs, respectively.
Binding proteins to the TME exist in both megakaryocytic and nonmegakaryocytic cell lines
EMSA was performed to study the transcription factors that bind to the TME. Specific shifted bands were detected with nuclear extracts from all cell lines (Figure 2B). The shifted bands were strongest with nuclear extracts from HEL cells and weakest with nuclear extracts from HeLa and HepG2 cells. To identify the binding motifs in the TME, competition assays were performed. Wild-type and 7 different mutated double-stranded oligonucleotides were used as competitors (Figure 2A), and EMSA was performed with nuclear extracts from megakaryocytic HEL (Figure 2C) and nonmegakaryocytic HepG2 (Figure 2D) cells. In both cases, Mut-2 did not compete, Mut-7 competed slightly, and Mut-5 and Mut-6 competed strongly against the TME probe. These results indicate that the TGACAGCTGCTGACAGCTG sequence includes a binding element that is recognized by nuclear proteins from both megakaryocytic and nonmegakaryocytic cells.
Homeodomain proteins, MEIS1 and PBXs, are present in the TME binding complexes
For further analysis of the role of the TME in megakaryocytic cells, we investigated the binding proteins of the TME. To identify the TME binding proteins, DNA affinity chromatography was carried out. The nuclear extracts from HEL cells and the TME double-stranded oligonucleotides were used for this experiment. Affinity binding proteins were eluted and their binding location was assayed by EMSA (Figure 3A). Binding activity was detected in several fractions, however, mainly in fraction no. 7-10. In the TME, there existed E-box motifs (CAGCTG), Meis1 binding motifs (TGACAG), and an NF-E2–like binding sequence (TGCTGA-CagcT). We have already pointed out that the TGACAGCTGCTGACAGCTG sequence is important for protein binding (Figure 2C-D). From this information, transcription factors, including E-box binding proteins, MEIS1, NF-E2p45, and other transcription factors related to megakaryocytic differentiation, were selected as candidates for the TME binding proteins. We used Western blotting to investigate whether or not these factors are included in the active binding fractions. Figure 3B shows that MEIS1, PBX1B, PBX2, and ETS-1 were detected in the active fractions. Meis1 is a homeobox gene and its gene product recognizes the TGACAG motif. PBXs are also homeobox gene products that recognize the TGAT sequence and these factors interact with MEIS1.
Both Meis1 and Pbxs are strongly expressed in HEL cells and bind to the TME
To confirm the identities of the putative TME binding proteins and to investigate any differences between them, with respect to their presence in the cell lines studied, supershift assays were performed with nuclear extracts from 4 cell lines and antibodies to the candidate homeodomain proteins (Figure 4A-D). PBX2 was detected as the supershifted band in all 4 cell lines. PBX1, however, was detected only in HEL and HeLa cells. MEIS1 was detected only in HEL cells. The endogenous expression levels of MEIS1, PBX1, and PBX2 in 4 cell lines were investigated by Western blotting (Figure 4E). An asterisk indicates a very weak band of PBX1B in HeLa cells. In all cell lines PBX2 was expressed, but PBX1B and MEIS1 were strongly expressed only in HEL cells. These endogenous expression levels correlate with the results of supershift assays.
MEIS1 binds directly to the TME and activates the PF4 promoter in cooperation with PBX1B or PBX2
To investigate the relationship between MEIS1, PBX1B, and PBX2, we prepared recombinant proteins using an in vitro translation system and performed EMSA (Figure 5A). MEIS1 bound to the TME by itself, but PBX1B and PBX2 did not. However, PBXs bound to the TME as part of a MEIS1/PBX complex. In Figure 5A, MEIS1/PBX1B and MEIS1/PBX2 complexes are indicated with arrows. The complexes were confirmed by supershift assays. When Mut-2 (in Figure 2) and MutTGACAG (containing 2 disrupted MEIS1 binding sites [TGACAG to AGCGAT]) were used as the probes, MEIS1/PBX complexes did not bind to the probes (Figure 5B). From these results and a report about the interaction between MEIS1 and PBXs, we considered it likely that MEIS1 binds to the TME and PBXs bind to MEIS1. However, the direct interactions of PBXs with the TME in the complex with MEIS1 are not clear. The bands of MEIS1/PBX complexes were detected below the band of MEIS1 in Figure 5A. A similar result using EMSA with MEIS1 and HOXA9 proteins was previously reported.26 This finding remains puzzling.
To investigate the functions of the TME-binding proteins in mediating PF4 gene expression, we overexpressed these transcription factors together with the PF4 promoter-reporter plasmid (PF4luc) in HepG2 cells and assayed reporter gene activity (Figure 5C). Interestingly, the overexpression of the single proteins, MEIS1, PBX1B, or PBX2, did not increase PF4 promoter-regulated gene expression. However, the coexpression of MEIS1 and PBX increased the activity. This increase was not observed using the PF4mut construct that contains the Mut-2 sequence instead of the TME. Therefore, the results from EMSA and the reporter assays indicate that the binding of the MEIS1/PBX complex to the TME is required for the activation of the PF4 promoter.
MEIS1/PBX complexes synergistically activate PF4 gene expression when added to GATA-1 and ETS-1
To investigate the role of the MEIS1/PBX complexes in megakaryocytic gene expression, GATA-1 and ETS-1 expression vectors were transfected into nonmegakaryocytic HepG2 cells with combinations of MEIS1, PBX1B, and PBX2 expression vectors (Figure 6A). Western blotting in Figure 6A shows that, although the expression levels of the various factors were relatively unaffected by the coexpression of other factors, additional expression of the MEIS1/PBX2 complex increased PF4 promoter-driven gene expression 2-fold higher than that produced by expressing GATA-1 and ETS-1. We point out that this activation of the PF4 promoter with these 4 factors in nonmegakaryocytic cells is 20-fold higher than the control, and this enhancement is not observed using PF4mut. Furthermore, the inhibition of the binding of endogenous MEIS1/PBX complexes to the TME by using PF4mut decreased the PF4 promoter activity by approximately 40% in megakaryocytic HEL cells (Figure 6B). These results indicate that MEIS1/PBX complexes coactivate the PF4 promoter with GATA-1 and ETS-1 and that these proteins may play an important role in megakaryocytic gene expression.
To investigate whether MEIS1/PBX complexes bind to the TME in vivo, ChIP assays were performed using rat megakaryocytes. The endogenous TME and MEIS1/PBX complexes were coimmunoprecipitated using antibodies for PBX1 and PBX2. The precipitated TME was detected by PCR amplification of the PF4 promoter fragment (-300 to -182) that includes the TME (-219 to -182). Specific amplification was detected when antibodies for PBXs were used but was not detected in the case of the control IgG (Figure 7). This result strongly suggests the binding of MEIS1/PBX complexes to the TME in vivo.
Analysis of TME binding proteins in rat megakaryocytes. ChIP assays were performed using the purified rat megakaryocytes. Antibodies for PBXs or isotype-matched control were used. Precipitated DNA fragments were amplified by PCR with primers specific for the rat PF4 promoter (-300 to -182), including the TME (-219 to -182). PCR products were separated on a 2% agarose gel and stained by ethidium bromide. The intensity of each band was evaluated by Scion Image. (–) indicates no antibody.
Analysis of TME binding proteins in rat megakaryocytes. ChIP assays were performed using the purified rat megakaryocytes. Antibodies for PBXs or isotype-matched control were used. Precipitated DNA fragments were amplified by PCR with primers specific for the rat PF4 promoter (-300 to -182), including the TME (-219 to -182). PCR products were separated on a 2% agarose gel and stained by ethidium bromide. The intensity of each band was evaluated by Scion Image. (–) indicates no antibody.
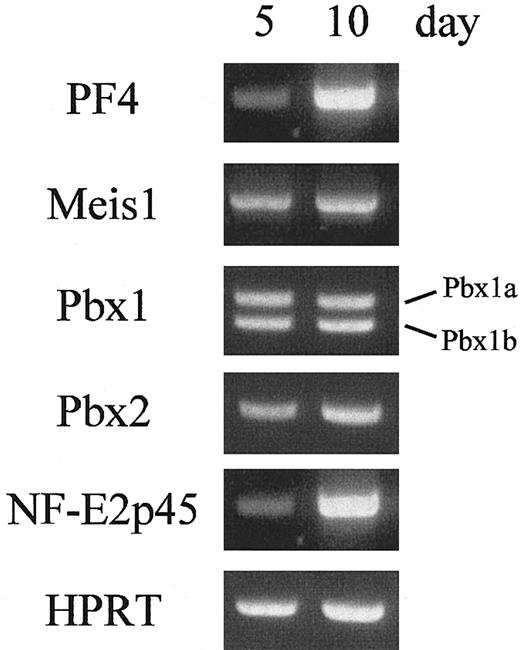
Meis1, Pbx1b, and Pbx2 are expressed during megakaryocytopoiesis
Our studies have documented the binding activities of MEIS1, PBX1B, and PBX2 to the TME, and the transcriptional activities of the activation complexes have been demonstrated. To confirm their functions in megakaryocytes, the expression levels of these homeobox genes were evaluated in cultured megakaryocytes. Megakaryocytes were prepared from CD34+ progenitor cells in human cord blood. After 18 days of incubation, about 90% of the cultured cells were differentiated into megakaryocytes and platelets (Table 1). Furthermore, in the cultured cells, erythrocytes were hardly detected, that is glycophorin A (CD 235a), an erythrocyte-specific marker, was not observed (data not shown). These results indicate that this differentiation system is suitable for analyzing megakaryocyte-specific gene expression. The cultured cells were harvested, and their total RNAs were isolated. RT-PCR was performed with the specific primers for PF4, Meis1, Pbx1a, Pbx1b, Pbx2, or NF-E2p45 (Figure 8). The results showed that the 3 homeobox genes as well as the PF4 gene were expressed 5 and 10 days after incubation. These data indicate that Meis1, Pbx1b, and Pbx2 are expressed in human megakaryocytes and may contribute to megakaryocytic gene expression.
Differentiation of cultured cells into megakaryocytes and platelets
. | 3 day . | 8 day . | 13 day . | 18 day . |
|---|---|---|---|---|
| GPllb, CD41+ cells, % | 3.7 | 74.1 | 82.6 | 88.9 |
| GPlbα, CD42b+ cells, % | 2.0 | 60.2 | 82.6 | 90.1 |
. | 3 day . | 8 day . | 13 day . | 18 day . |
|---|---|---|---|---|
| GPllb, CD41+ cells, % | 3.7 | 74.1 | 82.6 | 88.9 |
| GPlbα, CD42b+ cells, % | 2.0 | 60.2 | 82.6 | 90.1 |
CD34+ progenitor cells from cord blood were isolated and cultured under conditions promoting megakaryocytopoiesis. About 90% of the cultured cells differentiated into megakaryocytes that were positive for CD41 or CD42b.
The expression of homeobox genes during megakaryocytopoiesis. The expression levels of homeobox genes in human megakaryocytes were evaluated. The cultured cells were harvested, and the total RNAs were isolated. RT-PCR was performed with specific primers for PF4, Meis1, Pbx1a, Pbx1b, Pbx2, NF-E2p45, or HPRT (as a control).
The expression of homeobox genes during megakaryocytopoiesis. The expression levels of homeobox genes in human megakaryocytes were evaluated. The cultured cells were harvested, and the total RNAs were isolated. RT-PCR was performed with specific primers for PF4, Meis1, Pbx1a, Pbx1b, Pbx2, NF-E2p45, or HPRT (as a control).
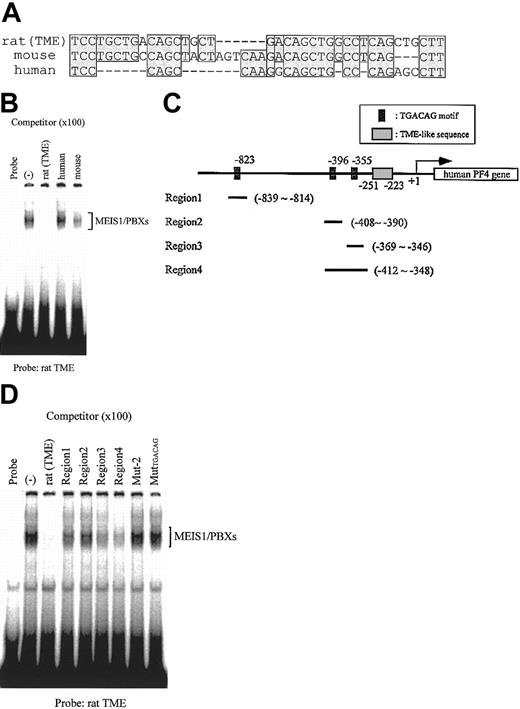
Homologous sequences to the rat TME exist in both human and mouse PF4 promoters
The TME is located immediately upstream of the T-cluster region in the rat PF4 promoter.4 We compared the sequences of this region in the rat PF4 promoter to those in the human and mouse PF4 promoters and found that homologous sequences to the rat TME exist in both human and mouse (Figure 9A). EMSA using these TME-like sequences as competitors showed that MEIS1/PBX complexes did not bind to the human TME-like sequence (Figure 9B). In the human TME-like sequence, the TGACAG motif was not conserved. We searched for this motif in the 1 kb of the 5′-flanking region of the human PF4 gene and found 3 such regions (Figure 9C). We checked the binding properties of these regions and found that MEIS1/PBX complexes did in fact bind to the region 4, which includes the 2 TGACAG motifs (Figure 9D). These results suggest that the upstream TGACAG motifs in the human PF4 gene might be necessary for the regulation of PF4 gene expression.
Comparison of the rat TME with TME-like sequences in the 5′-flanking region of mouse and human PF4 genes. (A) Mouse and human TME-like sequences that exist immediately upstream of the T-cluster region were compared with the rat TME sequence. Homologous sequences are indicated by shaded boxes. Dashes indicate deletion sequences. (B) EMSA was performed with the rat TME probe; MEIS1 and PBXs proteins were prepared by in vitro translation. Mouse and human TME-like sequences described in panel A were used as the competitors. (C) Locations of the 3 TGACAG motifs found within the 1-kb 5′-flanking region of the human PF4 gene are delineated. Fragments, including TGACAG motifs, region 1 to region 4, were used as the competitors in EMSA shown in panel D. (D) EMSA was performed with the rat TME probe; MEIS1 and PBXs proteins were prepared by in vitro translation. Fragments from the human PF4 gene described in panel C and the rat TME mutated fragment (MutTGACAG) were used as the competitors. (–) indicates no competitor.
Comparison of the rat TME with TME-like sequences in the 5′-flanking region of mouse and human PF4 genes. (A) Mouse and human TME-like sequences that exist immediately upstream of the T-cluster region were compared with the rat TME sequence. Homologous sequences are indicated by shaded boxes. Dashes indicate deletion sequences. (B) EMSA was performed with the rat TME probe; MEIS1 and PBXs proteins were prepared by in vitro translation. Mouse and human TME-like sequences described in panel A were used as the competitors. (C) Locations of the 3 TGACAG motifs found within the 1-kb 5′-flanking region of the human PF4 gene are delineated. Fragments, including TGACAG motifs, region 1 to region 4, were used as the competitors in EMSA shown in panel D. (D) EMSA was performed with the rat TME probe; MEIS1 and PBXs proteins were prepared by in vitro translation. Fragments from the human PF4 gene described in panel C and the rat TME mutated fragment (MutTGACAG) were used as the competitors. (–) indicates no competitor.
Discussion
PF4 gene is a target of MEIS1/PBX complexes
We have shown that MEIS1/PBX complexes are involved in the regulation of PF4 gene expression. Meis1 and Pbx were originally cloned as leukemia-related genes.17,18,35 In fact, overexpression of these genes induces leukemia in mice.19,20 Recently, it has been reported that PBX1 plays an important role in hematopoiesis.32 Yet, the physiologic functions and target genes of MEIS1 and PBXs in hematopoietic cells have still not been identified. This study identified the TME in the rat PF4 promoter and demonstrated that MEIS1/PBX complexes bind to the TME. This then is the first demonstration of a direct relationship between homeodomain proteins, MEIS1 and PBXs, and megakaryocytic gene expression.
MEIS1/PBX complexes are important for megakaryocytic gene expression
PF4 is a lineage-specific marker that appears during late stages of megakaryocytic differentiation.2 We previously cloned and characterized the rat PF4 promoter.3 More recently we reported that GATA-1 and ETS-1 bind to the PF4 promoter and transactivate the PF4 gene.6 Other studies have implicated a role for GATA-1 and ETS-1 in the regulation of megakaryocyte-specific genes, such as GPIIb, Mpl, GPIX, GPIbα, and GPV.7-11 In this study, we found that MEIS1/PBX complexes synergistically transactivated the PF4 gene in combination with GATA-1 and ETS-1 (Figure 6A). Furthermore, the inhibition of the binding of endogenous MEIS1/PBX complexes to the TME by disrupting Meis1 binding sites decreased PF4 promoter activity by approximately 40% in megakaryocytic HEL cells (Figure 6B). These results indicate that MEIS1/PBX complexes are as essential as GATA-1 and ETS-1 for complete megakaryocytic gene expression.
In the human PF4 promoter, MEIS1/PBX complexes did not bind to the human TME-like sequence but rather to the TGACAG motifs in the 5′-flanking region (Figure 9C-D). These results suggest that the TGACAG motifs and MEIS1/PBX complexes are also important for the regulation of human PF4 gene expression.
Are there different functions for PBX1B and PBX2?
In this study, we found that both PBX1B and PBX2 bind to the TME and transactivate the PF4 gene coordinately with MEIS1. The MEIS1/PBX2 complex activates the PF4 promoter more strongly than the MEIS1/PBX1B complex (Figures 5C and 6A). These results indicate that PBX1B and PBX2 may have different functions as transcription factors. In a previous report, Pbx1 has been shown to be expressed in specific tissues, whereas Pbx2 and Pbx3 are ubiquitously expressed. For example, Pbx1 is not expressed in lymphoid cell lines, but Pbx2 is expressed.22 The differences in the expression patterns of PBX1 and PBX2 in tissues may influence critical aspects of tissue function. In Figure 4E, we detected both MEIS1 and PBX1 only in the nuclear extracts from HEL cells. However, this result did not imply that the expressions of MEIS1 and PBX1B are restricted only to megakaryocytes, because in previous reports the expression of either Meis1 and Pbx1 in K562 cells or Pbx1 in HeLa cells was detected by Northern blotting and RT-PCR, respectively.21,22 It has also been speculated that these factors do not localize to the nucleus of expressing cell lines. It was reported that the coexpression of PBX1 with PREP1, a MEIS1 homolog protein, induced the nuclear localization of PBX1, although PBX1 by itself was observed in the cytoplasm.36 Therefore, interactions between MEIS1 and each one of the PBXs may decide both their localization and functions in megakaryocytes.
Differences in the binding complexes from megakaryocytic and nonmegakaryocytic cells
MEIS1, PBX1B, and PBX2 are expressed in HEL cells (Figure 4E). Although PBXs were also detected in HeLa (PBX1B and PBX2), HepG2 (PBX2), and K562 (PBX2) cells, MEIS1 was not detected in these cells (Figure 4B-E). Because PBXs do not bind to the TME by themselves (Figure 5A), another factor must be taking the place of MEIS1 for their binding to the TME in HeLa, HepG2, and K562 cells. It was previously reported that PREP1 bound to the TGACAG motif as a complex with PBX1B or PBX2 in HeLa cells.37 We propose that another factor like PREP1 binds to the TME in nonmegakaryocytic cells and are now investigating this possibility.
Is the MEIS/HOX/PBX ternary complex necessary for megakaryocytic gene expression?
In BXH-2 murine myeloid leukemia, Hoxa9 is activated along with Meis1.29,30 Retrovirally driven coexpression of Hoxa9 and Meis1 is sufficient to induce myeloid leukemia in mice.19,20 It has been suggested that HOXA9 forms ternary complexes with PBX2 and MEIS1 in myeloid leukemic cells.23 These results indicate that the ternary complex, including HOXA9, plays an important role in inducing leukemia. We investigated whether HOXA9 exists in the binding complex to the TME by Western blotting and supershift assays (Figures 3B and 4A-D). Unexpectedly, we did not detect HOXA9 in the TME complex. What then is the requirement of Hoxa9 for megakaryocytic gene expression physiologically? HOXA9 is expressed in CD34+ cells and is inactivated as cells leave the CD34+ compartment.38 In HEL cells, we did not detect the mRNA of Hoxa9 by RT-PCR (data not shown). From these data it seems likely that the expression of the Hoxa9 gene gradually decreases throughout megakaryocytic differentiation and finally only MEIS1/PBX complexes play this important role for PF4 gene expression. However, the mechanism of switching these gene expressions is not clear because the physiologic function of HOXA9 as well as those of MEIS1 and PBX in hematopoiesis has not been sufficiently clarified. On this point, further investigation is needed.
Prepublished online as Blood First Edition Paper, February 27, 2003; DOI 10.1182/blood-2002-02-0380.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Stuart H. Orkin for providing the GATA-1 expression vector and Kirin Brewery for the gift of thrombopoietin. We also thank Drs William C. Aird and Robert W. Jackman for helpful suggestions and review of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal