Abstract
We report in this paper that glycophorin C (GPC) is the receptor for PfEBP-2 (baebl, EBA-140), the newly identified erythrocyte binding ligand of Plasmodium falciparum. PfEBP-2 is a member of the Duffy binding–like erythrocyte binding protein (DBL-EBP) family. Although several reports have been published characterizing PfEBP-2, the identity of its erythrocytic receptor was still unknown. Using a combination of enzymatically treated red blood cells (RBCs) and rare, variant RBCs lacking different surface proteins, we have shown that PfEBP-2 does not bind to cells lacking GPC. Additionally, we found that PfEBP-2 binds differentially to variants of GPC lacking exon 2 or exon 3, and determined that the binding domain on GPC is potentially restricted to amino acid residues 14 through 22 within exon 2. Thus PfEBP-2 is involved in a sialic acid–dependent pathway of invasion, which does not involve glycophorin A or glycophorin B and represents a novel route of entry into the RBCs.
Introduction
Invasion of erythrocytes by the malaria parasite is a multistep process involving several specific interactions between receptors on the red blood cells (RBCs) and parasite ligands.1 Inhibition of RBC invasion would prevent infection and consequently disease. Therefore, considerable effort has gone into the identification and characterization of the molecules involved in this process. Plasmodium falciparum is not completely dependent on a single receptor for the invasion of human erythrocytes.2 This heterogeneity was highlighted by the different invasion rates obtained when distinct strains of the parasite were used to invade enzymatically modified RBCs and RBC mutants.3, 4, 5, 6 Based on this receptor heterogeneity, the invasion process was divided into 3 pathways: glycophorin A (GPA)– and sialic acid–dependent; GPA-independent but sialic acid dependent; and GPA- and sialic acid–independent but trypsin-sensitive. The GPA-independent pathways are referred to as the alternative pathways of invasion.4 In P falciparum, only that involving GPA and the 175-kDa erythrocyte-binding antigen (EBA-175) has been well characterized.7 Recently, PfEBP-2 as well as 2 additional potential parasite ligands8 containing the cysteine-rich Duffy binding–like (DBL) motifs found in EBA-1757 of P falciparum and Duffy binding protein-1 in Plasmodium vivax9 were identified from the genome sequencing project, all of which are postulated to have a role in invasion.10, 11, 12 In this study, we report the identification of glycophorin C (GPC) as a receptor for PfEBP-2 and show that the binding on GPC is limited to amino acids (aa's) 14-22 in exon 2.
Study design
Antibodies against PfEBP-2
Gene fragments encoding 3 different regions of PfEBP-2 (chr13_4000047.phat_1)13 were amplified from genomic DNA and expressed as glutathione S-transferase (GST) fusion polypeptides14 : EBP-2-1 (aa 333-452 and part of the DBL domain), EBP-2-2 (aa 798-962), and EBP-2-3 (aa 968-1137). Antibodies were raised against all 3 recombinant proteins in mice (2 μg per immunization per mouse) using the repetitive immunization schedule15,16 and in rabbits (100 μg per immunization using complete Freunds adjuvant/incomplete Freunds adjuvant [CFA/IFA]). All antibodies recognized the native protein similarly; however, all binding studies presented are those done with mouse anti–EBP-2-3.
Parasites
P falciparum 3D7 strain was grown in culture in human erythrocytes.17 To obtain biosynthetically labeled secreted parasite proteins containing the pool of putative binding ligands,6 parasites were synchronized18 and the schizonts purified19 and cultured (107 parasites per milliliter) in the presence of 0.1 mCi (3.7 MBq) TRAN35 S in methionine- and cysteine-deficient medium2 for 12 to 16 hours. The labeled supernatant was spun for 1 minute at 14 000g and stored in aliquots at –70°C until use.
Erythrocytes
Erythrocytes collected in 10% citrate-phosphate-dextrose were washed 3 times in phosphate-buffered saline (PBS) and treated with the various enzymes as described.10 Efficacy of each enzyme treatment was assessed in the Laboratory of Immunohematology, New York Blood Center, by assaying for loss of RBC agglutinability using a panel of monoclonal antibodies (mAbs) against suitable antigenic determinants on different blood group proteins.
RBCs deficient in blood group antigen Kell and Dombrock GPA (Ena-), glycophorin B (GPB; S-s-), GPC (Leach phenotype), or GPC variants (Gerbich and Yus phenotypes),20 along with age- and storage-matched control RBCs stored in liquid nitrogen as frozen pellets, were thawed before use in binding assays directly into PBS at 37°C and washed 3 times with RPMI 1640.
Erythrocyte binding assay
An aliquot of 500 μL of labeled parasite supernatant, confirmed as having PfEBP-2 by immunoprecipitation, was mixed with 100 μL target erythrocytes for 30 minutes at room temperature. The mixture was then spun at 6000g for 1 minute through sodium phthalate to remove unbound material. RBCs pelleted at the bottom of the tube, along with parasite proteins bound to them, were recovered by puncturing the tube. Cells were lysed in NETT (0.5% Triton X100, 150 mM NaCl, 10 mM EDTA [ethylenediaminetetraacetic acid], 50 mM Tris [tris(hydroxymethyl)aminomethane], pH 7.4) for 30 minutes at 4°C and spun at 14 000 rpm for 5 minutes. The soluble extract containing the PfEBP-2 ligand-receptor complex was immunoprecipitated with mouse antibodies against EBP-2-3. All experiments were repeated at least 3 times with identical pattern of binding.
Antibody-mediated inhibition of binding
The binding specificity of PfEBP-2 to GPC on RBCs was tested by examining the effect of 4 mAbs against different regions of GPC21,22 : 2B-39 (aa 1-6), 2B-41 (aa 16-22), 3-23 (aa 13-20), and 2B-42 (aa 45-50; Figure 2A), along with a control mAb against the Kell antigen (MIMA 40) on the binding. Each mAb was preincubated at 100 μg/mL with normal RBCs for 30 minutes before 500 μL parasite supernatant was added and processed as described above. For comparison, mouse anti–EBP-2-3 serum and the preimmune serum were used to confirm the ability of specific antibodies to block binding of the ligand to the receptor.
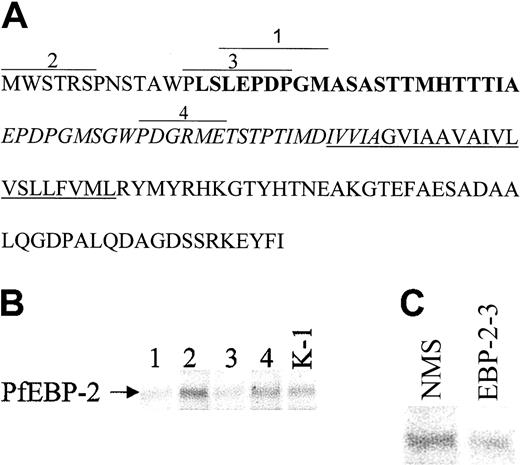
PfEBP-2 binds to aa 14-22 in exon 2 of GPC. (A) The amino acid sequence of GPC with the epitopes (overlined) corresponding to the monoclonal antibodies: Segment 1, 2B-41: antibody against GPC aa 16-22; Segment 2, 2B-39: antibody against GPC aa 1-6; Segment 3, 3-23: antibody against GPC aa 13-20; and Segment 4, 2B42: antibody against GPC aa 45-50. Exon 1 is represented by plain text; exon 2, bold text; exon 3, italicized text; and exon 4, plain text. The underlined amino acids represent the transmembrane domain. (B-C) mAbs against amino acid residues 13-20 and 16-22 of GPC inhibit binding of EBP-2 to red cells. Labeled supernatant proteins of P falciparum were mixed with normal RBCs preincubated with the different antibodies, followed by immunoprecipitations of the soluble extract. The bound PfEBP-2 is shown. Lane 1, antibody against GPC aa 16-22; lane 2, antibody against GPC aa 1-6; lane 3, antibody against GPC aa 13-20; lane 4, antibody against GPC aa 45-50; lane K-1, control mAb against the Kell protein; lane NMS, mouse preimmune serum; lane EBP-2, mouse anti–EBP-2-3.
PfEBP-2 binds to aa 14-22 in exon 2 of GPC. (A) The amino acid sequence of GPC with the epitopes (overlined) corresponding to the monoclonal antibodies: Segment 1, 2B-41: antibody against GPC aa 16-22; Segment 2, 2B-39: antibody against GPC aa 1-6; Segment 3, 3-23: antibody against GPC aa 13-20; and Segment 4, 2B42: antibody against GPC aa 45-50. Exon 1 is represented by plain text; exon 2, bold text; exon 3, italicized text; and exon 4, plain text. The underlined amino acids represent the transmembrane domain. (B-C) mAbs against amino acid residues 13-20 and 16-22 of GPC inhibit binding of EBP-2 to red cells. Labeled supernatant proteins of P falciparum were mixed with normal RBCs preincubated with the different antibodies, followed by immunoprecipitations of the soluble extract. The bound PfEBP-2 is shown. Lane 1, antibody against GPC aa 16-22; lane 2, antibody against GPC aa 1-6; lane 3, antibody against GPC aa 13-20; lane 4, antibody against GPC aa 45-50; lane K-1, control mAb against the Kell protein; lane NMS, mouse preimmune serum; lane EBP-2, mouse anti–EBP-2-3.
Results and discussion
Receptor identity of PfEBP-2
Binding of PfEBP-2 was totally abolished by treatment of normal RBCs with neuraminidase or trypsin (Figure 1A, lanes N and T). However, treatment with α-chymotrypsin appeared to enhance the binding of PfEBP-2 (Figure 1A, lane CT) relative to normal, untreated erythrocytes (Figure 1A, lane WT). This suggested that the RBC receptor for PfEBP-2 contains sialic acid residues in addition to a trypsin-sensitive moiety. Resistance to α-chymotrypsin ruled out GPB as a candidate receptor.20 The enhanced binding to cells treated with chymotrypsin might be due to the receptor's being more accessible for PfEBP-2 binding after treatment. For example, GPB, Dombrock, band 3, and some other blood group antigens are sensitive to treatment with chymotrypsin and are present on RBCs in high.20 Any change in their presence or their conformational structure after treatment could rearrange the surface topology of the RBC and thus potentially increase the access of the ligand to its specific receptor.
The erythrocyte receptor for PfEBP-2 is GPC. (A) The receptor is neuraminidase- and trypsin-sensitive but chymotrypsin-resistant. 35S-labeled proteins derived from parasite culture supernatants were mixed with red blood cells treated with neuraminidase (N), trypsin (T), or chymotrypsin (CT), or with untreated cells (WT). Separated cells were then lysed and the soluble fraction immunoprecipitated with anti–EBP2-3 antibodies. The immune complex was dissociated in SDS-PAGE buffer and run on 8% gels. The specific bound protein band in the autoradiograph corresponding to PfEBP-2 is shown. (B) PfEBP-2 does not bind to GPC null (Leach phenotype) cells. 35S-labeled proteins were mixed with rare, variant red blood cells lacking GPB (S-s-), GPA [En(a-)], Kell (K0), GPC (Leach), Dombrock [Gy(a-)], or control wild-type (WT) RBCs. The bound parasite ligand was immunoprecipitated with anti–EBP-2-3 antibodies. (C) PfEBP-2 binds differentially to GPC variant cells. Labeled proteins were mixed with identical amounts of Leach, Gerbich (Ge), Yus, or control (WT) RBCs. The bound parasite ligand was immunoprecipitated with anti–EBP-2-3 antibodies.
The erythrocyte receptor for PfEBP-2 is GPC. (A) The receptor is neuraminidase- and trypsin-sensitive but chymotrypsin-resistant. 35S-labeled proteins derived from parasite culture supernatants were mixed with red blood cells treated with neuraminidase (N), trypsin (T), or chymotrypsin (CT), or with untreated cells (WT). Separated cells were then lysed and the soluble fraction immunoprecipitated with anti–EBP2-3 antibodies. The immune complex was dissociated in SDS-PAGE buffer and run on 8% gels. The specific bound protein band in the autoradiograph corresponding to PfEBP-2 is shown. (B) PfEBP-2 does not bind to GPC null (Leach phenotype) cells. 35S-labeled proteins were mixed with rare, variant red blood cells lacking GPB (S-s-), GPA [En(a-)], Kell (K0), GPC (Leach), Dombrock [Gy(a-)], or control wild-type (WT) RBCs. The bound parasite ligand was immunoprecipitated with anti–EBP-2-3 antibodies. (C) PfEBP-2 binds differentially to GPC variant cells. Labeled proteins were mixed with identical amounts of Leach, Gerbich (Ge), Yus, or control (WT) RBCs. The bound parasite ligand was immunoprecipitated with anti–EBP-2-3 antibodies.
We thus focused our binding studies on RBC surface proteins that are sialylated and sensitive to cleavage by trypsin by using mutant RBCs, deficient in one of a number of such blood group antigens in binding assays with the native PfEBP-2. RBCs lacking GPA [En(a-)], GPB (S-s-U-), Kell (K0), and Dombrock [Gy(a-b-)] all bound PfEBP-2 efficiently (Figure 1B). However, erythrocytes lacking GPC (GPC null, Leach phenotype) did not bind PfEBP-2 at all. These results supported the enzymatic profile of the receptor obtained in Figure 1A, as GPC is a membrane sialoglycoprotein that is cleaved by trypsin between residues 48 and 49.20 The moderate increase of binding to the mutant S-s-U- and K0 cells (Figure 1B) could also be due to changes in the ability of the ligand to bind to the GPC receptor, as was discussed above. GPB is sensitive to chymotrypsin and therefore the pattern of binding of the S-s-U-cells (GPB null) is in accordance with those of chymotrypsin RBCs.
Glycophorins have long been postulated to serve as receptors for P falciparum merozoites.23 However, most of the initial studies focused on GPA as the primary receptor and GPB as the secondary one. Although PfEBP-2 binding to human RBCs is dependent on sialic acid, we have clearly shown that it does not bind to either GPA or GPB. Hence, this receptor-ligand interaction involving GPC represents a third sialic acid–dependent pathway of invasion.
All P falciparum members of the Duffy binding–like erythrocyte binding protein (DBL-EBP) superfamily of proteins are characterized by the presence of 2 cysteine-rich domains that are homologous to the binding domain first reported in the Duffy-binding protein (DBP) of P vivax.24 EBA-175 has 2 such domains, F1 and F2, of which the F2 domain has been shown to bind to GPA.7 PfEBP-2 is very similar in structure to EBA-175, with the highest identity (∼60%) within the F2 binding domain.8 This homology in the binding domain could thus be linked to the absolute requirement of both ligands for sialic acid in their corresponding receptor molecules.
Although several reports characterizing PfEBP-2 have been published in the last year10, 11, 12 and all agree that the binding of PfEBP-2 to human RBCs is neuraminidase sensitive, conflicting results have been reported on the effects of other protease treatments of the RBCs on the binding of PfEBP-2. Thompson et al11 suggested that the receptor is not a protein, as PfEBP-2, in their hands, bound to trypsin-, pronase-, and proteinase K–treated cells. Narum et al12 found a decreased binding of the ligand to trypsin-treated cells, as opposed to the lack of binding in our results and those of Mayer et al.10 However, Mayer et al, when describing the use of variant red cells for binding, reported contradictory results: binding to GPC null cells (Leach phenotype) but not to Gerbich phenotype cells, which lack just the peptide encoded by exon 3 of GPC. The method used in their study was an indirect way of assaying for PfEBP-2 binding and could be subject to excess residual protein present in the supernatant. Thus, our study is the first to offer direct evidence for a role for GPC in parasite invasion into the RBC.
Characterization of binding domains on GPC
To confirm and further characterize the regions of GPC that were responsible for binding PfEBP-2, we carried out a binding analysis of the 3 naturally occurring variants of GPC19 : Leach phenotype (GPC null), Gerbich phenotype (lacking exon 3, aa 36-63), and Yus phenotype (lacking exon 2, aa 14-35; Figure 2C). Leach cells did not bind PfEBP-2 and Gerbich cells bound the ligand with moderately higher efficiency than normal erythrocytes, while the Yus cells showed a severely diminished capacity for binding PfEBP-2. These results focused the binding region in GPC to amino acids encoded by exon 2 (aa 14-35), although they did not exclude the possibility that exon 1 of GPC could potentially also play a role, as a lack of exon 2 could affect the conformation and folding of the 5′ end of the GPC molecule, thus inhibiting binding. The moderate increase in binding to Ge cells could be due to changes in the secondary structure of GPC that influence the accessibility of the binding domain in exon 2 of GPC to PfEBP-2.
By using monospecific mAbs against different regions of GPC in an inhibition of binding assay, along with a mAb against Kell, a nonrelated RBC surface protein, we were able to delineate even further the binding domain on GPC. The amino acid sequence of GPC and the epitopes of the different mAbs21,22 are depicted in Figure 2A. A strong inhibition of binding was observed when mAbs against amino acid residues 13-20 and 16-22 of GPC (lanes 1 and 3) were added prior to the binding assay, as compared to the binding of PfEBP-2 in the presence of mAbs against amino acid residues 1-6 (lane 2) and 45-50 (lane 4) of GPC or anti-Kell (lane K-1). The GPC-specific antibodies do not interact with red cells treated with neuraminidase21,22 ; hence, their corresponding epitopes are sialylated. When antibodies against recombinant EBP-2-3 were used, the antibodies also inhibited its binding to normal erythrocytes (Figure 2C, lane EBP-2-3), as compared with the control preimmune serum (lane NMS). Thus, this confirms that the specific interaction can be blocked either by antibodies to the receptor or by antibodies to the ligand.
The above results strongly implicate the region between residues 13 and 22 of GPC as the binding domain for PfEBP-2, of which aa 14-22 lie within exon 2. The EBA-175 interaction with GPA was shown to involve both the sialic acid residues and the aa 1-64 peptide backbone of GPA.7
Our studies validate the existence of multiple invasion pathways and have identified a new ligand-receptor interaction, between PfEBP-2 and GPC. Access to numerous invasion pathways provides redundancy and adaptability to P falciparum merozoites for this critical step in their life cycle—the parasite has a survival advantage when faced with receptor heterogeneity in humans and/or specific immune responses against a particular parasite ligand. Moreover, RBC polymorphisms seem to have emerged by natural selection, protecting the host from severe malaria infection, as there is a significant geographic overlap between malaria endemicity and RBC disorders.25 In the western Pacific, Southeast Asian ovalocytosis (SAO) is found with high frequency and was thought to afford a survival advantage against malaria. SAO had been associated with a 27–base pair (bp) deletion in band 3 and also linked to a GPC polymorphism. The Gerbich phenotype in these regions was thought to be associated with the reduced frequency of malaria infections caused by both P falciparum and P vivax.26 However, most recent studies in a population in the same endemic area that was genotyped for a deletion in exon 3 of GPC have found that this polymorphism afforded no significant protection from infection.27 Our results suggest that polymorphism in exon 2, in addition to that in exon 3, of GPC should be more closely examined in order to find a molecular basis for the selection of GPC variants in these malaria-endemic areas. Of note is the fact that no GPA-deficiency polymorphisms have been reported in malaria-endemic regions, suggesting that the GPA–EBA-175 interaction may not be the primary route of parasite invasion, although EBA-175 appears to be synthesized at much higher levels than PfEBP-2 (data not shown). On the other hand, other nonmalaria selective forces might be responsible for the lack of GPA polymorphism in malaria-endemic areas. The importance of elucidating the GPA “alternative” pathways of invasion was recently highlighted by studies on field isolates in endemic regions of India28 where a majority of strains were reported to use non–GPA-mediated pathways of invasion.
Another point of view relating to the existence of multiple invasion pathways is that multiple ligand-receptor interactions have to take place so invasion can occur. Unless appropriate signals from all components of the molecular recognition cascade are in place, the parasite cannot enter the RBC. Some proteins may play a role early in the invasion process, whereas members of the EBP family, because of their homology to DBP-1 of P vivax, are postulated to be involved in junction formation.24 Now that the genome of P falciparum is completely sequenced,29 the entire family of erythrocyte binding ligands12 along with their respective erythrocyte receptors, can be characterized. The molecular arsenal that Plasmodium uses to invade the RBC should then be explored by the study of the use of these different ligands by field isolates in different malaria-endemic areas of the world. This would confirm the relevance of their inclusion in future vaccine design aimed at global distribution.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-10-3076.
Supported by National Institutes of Health grant P50 HL 54459-07.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the numerous colleagues and local patients who provided the blood samples and antibodies used in this testing and Jill Storry for running the enzyme efficacy assays on the RBCs.

![Figure 1. The erythrocyte receptor for PfEBP-2 is GPC. (A) The receptor is neuraminidase- and trypsin-sensitive but chymotrypsin-resistant. 35S-labeled proteins derived from parasite culture supernatants were mixed with red blood cells treated with neuraminidase (N), trypsin (T), or chymotrypsin (CT), or with untreated cells (WT). Separated cells were then lysed and the soluble fraction immunoprecipitated with anti–EBP2-3 antibodies. The immune complex was dissociated in SDS-PAGE buffer and run on 8% gels. The specific bound protein band in the autoradiograph corresponding to PfEBP-2 is shown. (B) PfEBP-2 does not bind to GPC null (Leach phenotype) cells. 35S-labeled proteins were mixed with rare, variant red blood cells lacking GPB (S-s-), GPA [En(a-)], Kell (K0), GPC (Leach), Dombrock [Gy(a-)], or control wild-type (WT) RBCs. The bound parasite ligand was immunoprecipitated with anti–EBP-2-3 antibodies. (C) PfEBP-2 binds differentially to GPC variant cells. Labeled proteins were mixed with identical amounts of Leach, Gerbich (Ge), Yus, or control (WT) RBCs. The bound parasite ligand was immunoprecipitated with anti–EBP-2-3 antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/11/10.1182_blood-2002-10-3076/5/m_h81134399001.jpeg?Expires=1765899795&Signature=PxzyUTG8v8qdv64fxroFAN-Wa0A-N6~ANLWNCulR1LDZQCgI8LcihtQDH8OP2B~n3IeIWBOG8BFydpHzJDA4iMcm1tNBH1AlmR96gWkGxIFRcL6DhK~UwVTarWexIHG5IXvzH9kQEvmffAG69ZfAKHZr7XySRliWE092vULv9sBmGsNPqGxzd8jzZOgjap4QeZA8~W1ODis133eQfIBRroV22fVu1sNQZJP3T3fuGJUXPxyQ7eyo9XZkeFwOHld~jvSEuJqwwcA6cs4ETJbF3VjDUijNPq8HGN8HDbGtw95M2D3LMciPfSNMjYtAb5ljHGN008CX~ZcZnHJwfNwe~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal