Abstract
Arsenic trioxide (ATO) has been shown to induce differentiation and apoptosis in acute promyelocytic leukemia (APL) cells concomitant with down-regulation of the PML-RARα fusion protein, a product of the t(15:17) translocation characteristic of APL leukemic cells. However, ATO is also a potent inducer of apoptosis in a number of other cancer cells lacking the t(15:17) translocation. The exact mechanism of ATO-induced apoptosis in these cells is not yet clear. We tested the effect of ATO on 7 myeloma cell lines with varying p53 status and report that in cells with mutated p53, ATO induced rapid and extensive (more than 90%) apoptosis in a time- and dose-dependent manner concomitant with arrest of cells in G2/M phase of the cell cycle. Myeloma cells with wild-type (wt) p53 were relatively resistant to ATO with maximal apoptosis of about 40% concomitant with partial arrest of cells in G1 and up-regulation of p21. The use of caspase blocking peptides, fluorescence-tagged caspase-specific substrate peptides, and Western immunoblotting confirmed the involvement of primarily caspase-8 and -3 in ATO-induced apoptosis in myeloma cells with mutated p53 and primarily caspase-9 and -3 in cells expressing wt p53. We also observed up-regulation by ATO of R1 and R2 APO2/TRAIL (tumor necrosis factor–related apoptosis-inducing ligand) receptors. Most important, however, we observed a synergy between ATO and APO2/TRAIL in the induction of apoptosis in the partially resistant myeloma cell lines and in myeloma cells freshly isolated from myeloma patients. Our results justify the use of the combination of these 2 drugs in clinical setting in myeloma patients.
Introduction
Multiple myeloma (MM) is a clonal B-cell malignancy affecting both the immune system and bone destruction. It is the second most frequent hematologic malignancy, inflicting 40 000 people in the United States, with a 5-year survival of less than 20%.1-3 Therefore, new therapeutic modalities are needed for this disease.
Arsenic trioxide (ATO), like all-trans retinoic acid (ATRA), is a potent drug in the treatment of acute promyelocytic leukemia (APL)4 and, like ATRA, has been shown to induce differentiation and apoptosis of APL cells in vitro and in vivo in animal models.5 Most important, ATO is very effective in the treatment of APL patients, with very little toxicity.6ATO is a potent inducer of apoptosis in a number of other cell types such as acute myeloid leukemia (AML),7 gastric cancer,8 neuroblastoma,9,10 and in MC-CAR cells.11 The exact mechanism of ATO-induced apoptosis is not yet clear. Several mechanisms were proposed to explain ATO-induced differentiation and apoptosis of APL cells. One mechanism, similar to ATRA, involves down-regulation of the promyelocytic leukemia (PML) protein through binding to the RARα portion of the PML-retinoic acid receptor α (RARα) fusion protein, the product of the t(15:17) translocation. However, because ATO is effective in APL patients resistant to ATRA12,13 and is cytotoxic to cells lacking the PML-RARα fusion protein, other mechanisms of action were also attributed to ATO in cells lacking the t(15:17) translocation. These mechanisms include mitochondrial damage,5,14,15 activation of histone deacetylase,16 blocking of cells in G1 by induction of p21 in MC-CAR cells,11 and blocking of cell cycle in G2/M in U937 cells.17 Activation of caspases such as caspase-3,10,18 caspase-9,19 and caspase-820 was also reported for ATO-induced apoptosis.
TNF-related apoptosis-inducing ligand (APO2/TRAIL) belongs to the large family of tumor necrosis factor (TNF)–like signal-inducing proteins, and their corresponding receptors belong to the large family of TNF-like signal transduction receptor proteins. APO2/TRAIL induces a death signal following binding to the R1 or R2 APO2/TRAIL receptors.21-25 Normal cells escape APO2/TRAIL-induced apoptosis by virtue of coexpressing decoy receptor molecules such as R3 or R4, which are capable of binding of APO2/TRAIL but lack the intracellular death domains that transmit downstream cell death signals through activation of caspase-8.24 Tumor cells generally do not express these decoy receptor molecules.26-28 Other mechanisms for APO2/TRAIL resistance include mutations in caspase-8, the primary caspase involved in APO2/TRAIL death signaling upstream from caspase-3.29-31
We have previously shown that APO2/TRAIL and adenovirus-mediated delivery of p53 (Ad-p53) are potent inducers of apoptosis in myeloma cells.32-34 More recently, we have shown that Ad-p53 synergizes with APO2/TRAIL in the induction of apoptosis in myeloma cells through up-regulation of R1 and R2 APO2/TRAIL receptors.35,36 Others have recently shown that treatment with various chemotherapeutic drugs37-39 or ionizing radiation also results in the induction of APO2/TRAIL receptors and APO2/TRAIL decoy receptors.40 In addition, APO2/TRAIL has been shown to exert an antitumor effect in vivo in different xenograft models of cancer and exhibited very limited toxicity in monkeys.41
In this study we conducted a detailed analysis of the pathways of ATO-induced apoptosis in myeloma cells with different p53 status. We describe 2 distinct pathways for ATO-induced apoptosis in terms of the effect on cell cycle and involvement of initiator caspases, depending of p53 status. In addition, we report here, for the first time, that ATO synergizes with APO2/TRAIL in the induction of apoptosis in myeloma cells through up-regulation of R1 and R2 APO2/TRAIL receptors.
Materials and methods
Cell lines, cell culture, and induction of apoptosis by arsenic trioxide
The list of all myeloma cell lines used in this study, their source, and p53 status is depicted in Table1. Cells (4 × 105/mL) were cultured in 1 mL RPMI 1640 medium plus 15% fetal calf serum (FCS medium) (GIBCO, Grand Island, NY) in a 24-well plate in a CO2 incubator. Arsenic trioxide (ATO, Trisenox; Cell Therapeutics, Seattle, WA) was added at concentrations of 0 to 10 μM, and cultures were harvested after 24 and 28 hours and were tested for apoptosis by the annexin V method (see “Staining with annexin V and detection of apoptotic cells”).
All myeloma cell lines used in this study
| Cell line . | p53 status . | Reference no. . | Source . |
|---|---|---|---|
| ARP-1 | Null | 44, 46 | University of Arkansas |
| 8226 | Mutated | 45 | ATCC |
| U266 | Mutated | 45 | ATCC |
| ARH-77 | Mutated | 36 | ATCC |
| MC-CAR | W.t. | 36 | ATCC |
| HS-Sultan | W.t. | 36 | ATCC |
| IM9 | W.t. | 36 | ATCC |
| Cell line . | p53 status . | Reference no. . | Source . |
|---|---|---|---|
| ARP-1 | Null | 44, 46 | University of Arkansas |
| 8226 | Mutated | 45 | ATCC |
| U266 | Mutated | 45 | ATCC |
| ARH-77 | Mutated | 36 | ATCC |
| MC-CAR | W.t. | 36 | ATCC |
| HS-Sultan | W.t. | 36 | ATCC |
| IM9 | W.t. | 36 | ATCC |
Cell lines were maintained in culture as described in “Materials and methods.” All experiments were started with log phase cells.
ATCC indicates American Type Culture Collection (Manassas, VA).
Cell culture and induction of apoptosis by APO2/TRAIL
Cells (4 × 105/mL) were cultured in 1 mL FCS medium in a 24-well plate. Purified, recombinant, native-sequence, trimeric, human APO2/TRAIL (Genentech, South San Francisco, CA) was added at 0 to 100 ng/mL, and wells were harvested after 24 and 48 hours of treatment and tested for apoptosis as described below. For combination studies of ATO and APO2/TRAIL, 0 to 10 μM ATO was added to wells containing 0 to 100 ng/mL APO2/TRAIL.
Staining with annexin V and detection of apoptotic cells
Apoptosis was determined by staining of exposed phosphatidylserine with annexin V–fluorescein isothiocyanate (annexin V–FITC) (BioVision, Palo Alto, CA) as recommended by the manufacturer. Stained cells were analyzed by flow cytometry (FACSCalibur; Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA). Quantitation of apoptosis was done by the CellQuest program; 10 000 thousand cells were analyzed.34
Analysis of cell cycle
Cells (4 × 105/mL) were cultured in 1 mL FCS medium in a 24-well plate. The effect of ATO on the cell cycle was determined by staining of myeloma cells with propidium iodide as described before.43 Stained cells were analyzed by flow cytometry (FACSCalibur; BDIS) using the ModFit software (BDIS); 10 000 cells were collected in each sample.
Determination of surface APO2/TRAIL receptors by immunofluorescence staining
The effect of ATO on surface expression of APO2/TRAIL receptors was examined in cells treated with 4 μM ATO for 24 hours where apoptosis reached around 40%. Cells were harvested and stained by indirect immunofluorescence staining for APO2/TRAIL receptors as described before36 with antibodies specific for R1, R2, R3, and R4 APO2/TRAIL receptors. Primary antibodies to APO2/TRAIL R1 (clone M271), antibodies to APO2/TRAIL R2 (clone M413), antibodies to decoy receptor 1 (APO2/TRAIL R3) (clone M413), and antibodies to decoy receptor 2 (APO2/TRAIL R4) (clone M413) were obtained from Immunex (Seattle, WA). Antibodies (1 μg/106 cells) were incubated for 30 minutes at 4°C, after which unbound antibody was washed out. Secondary antibody, goat-antimouse FITC (Jackson Immunochemicals, Raritan, NJ) was added for 30 minutes, and unbound secondary antibody was washed out. Isotype-matched immunoglobulin (Ig) was used for background staining. Analysis of results was performed by the FACSCalibur (BDIS) as described in “Staining with annexin V and detection of apoptotic cells.” Dead cells were identified by light scatter and were gated out.
Blocking of ATO-induced apoptosis by caspase-specific blocking peptides
To identify the downstream caspases involved in ATO-induced apoptosis, we used caspase-specific blocking peptides (FMK derivatives; BioVision, Palo Alto, CA). We used 2 μM of each of the following peptides: YVAD (caspase-1 inhibitor), VDVAD (caspase-2 inhibitor), DEVD (caspase-3 inhibitor), LEVD (caspase-4 inhibitor), WEHD (caspase-5 inhibitor), VEID (caspase-6 inhibitor), IETD (caspase-8 inhibitor), LEHD (caspase-9 inhibitor), AEVD (caspase-10 inhibitor), and VAD (pancaspase inhibitor). FA-FMK at similar concentration was used as a control. Cells (4 × 105/mL) were cultured in 1 mL FCS medium in a 24-well plate. ATO was added at concentration of 7.5 μM with or without the blocking peptide or control (FA-FMK). Cultures were harvested after 48 hours and were tested for apoptosis by the annexin V binding method.
Determination of caspase activity by fluorescent-tagged caspase-specific substrate peptides
To positively identify the downstream caspase activation pathway utilized by ATO, we used caspase-specific fluorescent-tagged substrate peptides to monitor caspase activation. Typically, cells (4 × 105/mL) were cultured in 1 mL FCS medium in a 24-well plate. ATO was added at concentration of 7.5 μM, and cultures continued for 0, 16, 24, and 48 hours. For measurement of caspase activity, cells were cultured further for 1 hour at 37°C with fluorescence-tagged caspase-specific substrate peptide specific for caspase-3, -8, and -9, and fluorescence generated due to the hydrolysis of the caspase substrate peptide was analyzed by flow cytometry as determined above for annexin V. FITC–caspase-8 and FITC–caspase-9 substrate peptides (CaspaTag) were from Serologicals, Norcross, GA. FITC–caspase-3 substrate peptide was from BioVision (Palo Alto, CA).
Determination of p21 expression and activation of PARP, caspase-3, -8, and -9 by Western immunoblotting
Cells were treated with 7.5 μM ATO for 0, 16, 24, and 48 hours, and aliquots of 3 × 106 to 4 × 106cells were washed (× 2) with phosphate-buffered saline (PBS), and total cellular protein was extracted as described before.43 Equal amounts of protein (50 μg) were loaded onto each lane, and protein bands were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). For Western immunoblotting, loading controls were performed using the housekeeping protein, β-actin (Sigma, St Louis, MO). Alternatively, loading controls were performed by first electrophoresing 5 μg protein per lane and staining with Coomassie blue. Loading for the Western immunoblotting gel was corrected according to the Coomassie blue–stained gel following quantitation of protein bands by densitometry. Gel electrophoresis, immunoblotting, detection of specific bands, and quantitation of protein bands by densitometry were performed as previously described.43 Mouse monoclonal antibodies to p21/WAF-1 (clone 187), caspase-8 (clone H-134), caspase-3 (clone E8), and rabbit polyclonal anticaspase-9 (clone H-83) were from Santa Cruz Biotechnology (Santa Cruz, CA). A low molecular weight ladder of biotinylated protein markers (Biorad, Hercules, CA) was run for each gel. The p21/WAF-1, procaspase-3, -8, and -9 were tentatively identified according to their migration on the blot. In all experiments 4 myeloma cell lines were processed in the same SDS-PAGE and immunoblotted simultaneously on the same membrane. This approach minimized experimental variation due to variations in protein transfer, immunoblotting, detection, and exposure to x-ray film and allows comparison of protein bands between different cell lines and within the same cell line.
Staining for CD38bright CD45− myeloma cells and flow sorting of myeloma cells
Bone marrow samples were collected from MM patients under proper consent, and the mononuclear fraction was obtained by Ficoll-Hypaque separation. Staining for myeloma (CD38+/CD45−) cells and flow sorting were performed as described before using the FACSstar Plus (Turbo-Fast Sorter, BDIS).33Myeloma cells with more than 97% purity were obtained.
Results
ATO-induced apoptosis and cell cycle arrest is dependent of p53 status
To determine the role of p53 in ATO-induced apoptosis, we used myeloma cell lines with different p53 status. Table 1 outlines the myeloma cell lines used in this study. Thus, U266, RPMI 8226, and ARH-77 cells express mutated p53,44 whereas IM9, MC-CAR, and HS-Sultan express wild-type (wt) p53.36 ARP-1 cells are p53 null cells.45 46
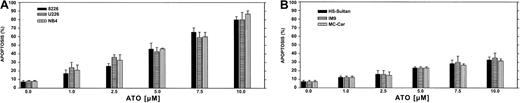
We first performed a time-dose titration of ATO to determine optimal conditions for maximal apoptosis in each cell line. The results obtained from U266 and RPMI 8226 myeloma cell lines expressing mutated p53 and the NB4 cells (acute promyelocytic leukemia cells) are depicted in Figure 1A. We observed a time- and dose-dependent apoptosis between 1 to 10 μM ATO, with apoptosis of more than 85% observed after 48 hours of treatment with 10 μM ATO in U266 and 8226 cells. The kinetics and extent of apoptosis for these myeloma cells were similar to the extent of apoptosis observed for the NB4, used here as a reference for ATO-sensitive target cells (Figure1A). In contrast, HS-Sultan, IM9, and MC-CAR cells (all express wt p53) were less sensitive to ATO at all time points tested, and maximal apoptosis was only 35% following 48 hours exposure to 10 μM ATO (Figure 1B).
ATO-induced apoptosis in myeloma cells expressing mutated (A) or wt (B) p53.
Cells were cultured for 2 days with 0 to 10 μM ATO. Apoptosis was determined following 48 hours of treatment, by the annexin V method. NB4 cells were used as a reference for an ATO-sensitive cell line. For more experimental details, see “Materials and methods.” Bars are ± SD of at least 3 experiments. Note the clear difference in sensitivity to apoptosis between cells expressing wt or mutated p53.
ATO-induced apoptosis in myeloma cells expressing mutated (A) or wt (B) p53.
Cells were cultured for 2 days with 0 to 10 μM ATO. Apoptosis was determined following 48 hours of treatment, by the annexin V method. NB4 cells were used as a reference for an ATO-sensitive cell line. For more experimental details, see “Materials and methods.” Bars are ± SD of at least 3 experiments. Note the clear difference in sensitivity to apoptosis between cells expressing wt or mutated p53.
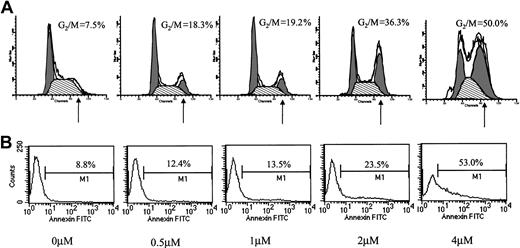
Conflicting results regarding the effect of ATO on cell cycle distribution have been reported. Whereas Park et al reported blocking of MC-CAR cells at G1,11 they also reported a G2/M arrest by ATO in U937 cells.17 We hypothesized that ATO might have a differential effect on cell cycle in cells expressing wt versus mutated p53. We therefore examined the effect of varying doses of ATO on the cell cycle and apoptosis in myeloma cells with mutated or wt p53. We first tested the effect of 24-hour treatment with 0 to 4 μM ATO on the cell cycle (Figure2A) and apoptosis (Figure 2B) of RPMI 8226 cells expressing mutated p53. ATO induced a dose-dependent apoptosis (8.8% to 53%) concomitant with arrest of cells in G2/M of the cell cycle (7.5% to 50% of cells in G2/M).
Effect of ATO on the cell cycle and apoptosis of RPMI 8226 cells.
(A) Cell cycle. (B) Apoptosis. RPMI 8226 cells expressing mutated p53 were cultured for 48 hours with 0 to 4 μM ATO. Cell cycle distribution was determined by the propidium iodide staining method, and stained cells were analyzed by flow cytometry using the ModFit software; 10 000 cells were analyzed. Apoptosis was determined by the annexin V method. For additional experimental details, see “Materials and methods.” Note the time- and dose-dependent arrest of cells in G2/M and the parallel kinetics of cell cycle arrest and extent of apoptosis. At least 3 experiments were performed, and 1 representative experiment is shown.
Effect of ATO on the cell cycle and apoptosis of RPMI 8226 cells.
(A) Cell cycle. (B) Apoptosis. RPMI 8226 cells expressing mutated p53 were cultured for 48 hours with 0 to 4 μM ATO. Cell cycle distribution was determined by the propidium iodide staining method, and stained cells were analyzed by flow cytometry using the ModFit software; 10 000 cells were analyzed. Apoptosis was determined by the annexin V method. For additional experimental details, see “Materials and methods.” Note the time- and dose-dependent arrest of cells in G2/M and the parallel kinetics of cell cycle arrest and extent of apoptosis. At least 3 experiments were performed, and 1 representative experiment is shown.
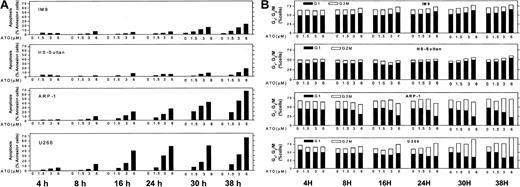
Similar results were obtained for other myeloma cells with mutated p53. The combined results obtained from U266 and ARP-1 cells (mutated and null p53, respectively) and IM9 and HS-Sultan cells (wt p53) are depicted in Figure 3A (apoptosis) and Figure 3B (cell cycle). Cells were cultured for 0, 4, 8, 16, 24, 30, and 38 hours without or with 1.5, 3, and 6 μM ATO and assayed at this time points for apoptosis and cell cycle distribution. ATO induced apoptosis in a time- and dose-dependent manner in all cell lines tested (Figure 1). As was the case for RPMI 8226 cells (Figure 2), U266 and ARP-1 cells were sensitive to low doses of ATO, reaching more than 50% apoptosis within the first 16 hours of treatment with ATO (Figure 3A). Furthermore, ATO induced G2/M cell cycle arrest in ARP-1 and U266 cells in a time/dose-dependent manner, in a similar kinetics observed for apoptosis, reaching a maximum of 70% to 80% apoptosis and more than 65% of cells arrested at G2/M phase of the cell cycle following 38 hours of treatment with 6 μM ATO (Figure 3). These results suggest that treatment with ATO of cells expressing mutated p53 results in apoptosis from G2/M phase of the cell cycle. In contrast to cells with mutated p53, HS-Sultan and IM9 cells expressing wt p53 were not blocked at G2/M; instead, a small increase (15% to 20%) in the proportion of cells arrested at G1 was observed following 30 to 38 hours of treatment with 3 to 6 μM ATO. At these time/dose points apoptosis was around 20% to 30% (Figure 3). Similar results were observed for MC-CAR cells expressing wt p53 (results not shown). These results further support a different apoptosis pathway for cells expressing wt p53.
ATO-induced apoptosis and cell cycle arrest in IM9, HS-Sultan, U266, and ARP-1 cells.
(A) Apoptosis. (B) Cell cycle arrest. Cells were cultured for 0, 4, 8, 16, 24, 30, and 38 hours with 0 to 6 μM ATO. Apoptosis was determined by the annexin V method and cell cycle distribution by the propidium iodide staining method. For additional experimental details, see legends to Figures 1 and 2 and “Materials and methods.” Representative results from at least 3 different experiments are shown.
ATO-induced apoptosis and cell cycle arrest in IM9, HS-Sultan, U266, and ARP-1 cells.
(A) Apoptosis. (B) Cell cycle arrest. Cells were cultured for 0, 4, 8, 16, 24, 30, and 38 hours with 0 to 6 μM ATO. Apoptosis was determined by the annexin V method and cell cycle distribution by the propidium iodide staining method. For additional experimental details, see legends to Figures 1 and 2 and “Materials and methods.” Representative results from at least 3 different experiments are shown.
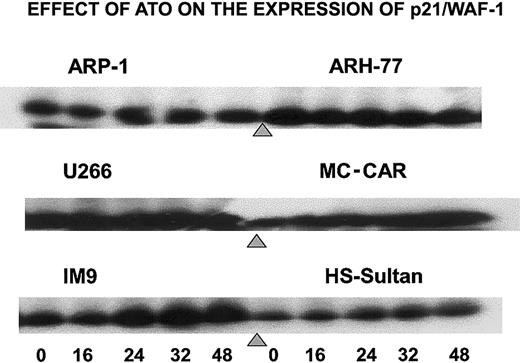
To further establish the differential effect of ATO on the cell cycle, we tested the effect of ATO on p21 levels in cells exhibiting G1 arrest versus cells exhibiting G2/M arrest. Indeed, treatment of IM9, HS-Sultan, and MC-CAR cells (wt p53) with 6 μM ATO resulted in a time-dependent increase in p21 protein expression reaching 5- to 10-fold increase following 32 to 48 hours of treatment. In contrast, U266, ARP-1, and ARH-77 cells (mutated or null p53), which were blocked in G2/M instead of G1 phase of the cell cycle, did not exhibit up-regulation of p21 (Figure 4). These results further confirm a differential effect of ATO on myeloma cells depending on p53 status.
ATO induces up-regulation of p21 expression in cells expressing wt p53.
Cells were cultured as above for 0, 16, 24, 32, and 48 hours with 6 μM ATO, and aliquots were taken for Western immunoblotting; 50 μg protein extract was loaded onto each lane. Loading control was performed according to the results obtained from a prerun of gels stained for protein by Coomassie blue and quantitation of protein bands by densitometry. For further experimental details, see the legend to Figure 8 and “Materials and methods.”
ATO induces up-regulation of p21 expression in cells expressing wt p53.
Cells were cultured as above for 0, 16, 24, 32, and 48 hours with 6 μM ATO, and aliquots were taken for Western immunoblotting; 50 μg protein extract was loaded onto each lane. Loading control was performed according to the results obtained from a prerun of gels stained for protein by Coomassie blue and quantitation of protein bands by densitometry. For further experimental details, see the legend to Figure 8 and “Materials and methods.”
Downstream caspases involved in ATO-induced apoptosis in cells expressing wt or mutated p53
We then compared the effect of ATO on caspase activation in myeloma cells expressing wt p53 or mutated p53. This was accomplished by 3 different methods: by using caspase-specific inhibitory peptides, by using fluorescence-tagged caspase-specific substrate peptides, and by Western immunoblotting. We first assayed caspase activation by caspase-specific blocking peptides. U266 cells (mutated p53) and HS-Sultan cells (wt p53) were cultured for 48 hours with 7.5 μM ATO, with or without 2 μM of the caspase-specific blocking peptide, or control peptide (FA-FMK), and apoptosis was estimated by the annexin V method. The results obtained with caspase blocking peptides are depicted in Figure 5. Ten different caspase blocking peptides were employed. Caspase-3 and the pancaspase blocking peptide, Z-VAD, substantially blocked ATO-induced apoptosis in both cell lines. In U266 cells caspase-8 and caspase-10 blocking peptide completely blocked apoptosis, whereas caspase-9 blocking peptide only partially blocked apoptosis (25%) in these cells. In contrast to U266 cells, caspase-9 inhibitor completely blocked apoptosis in HS-Sultan cells, whereas caspase-8 and -10 blocking peptide had minimal effect (Figure 5). Interestingly, WEHD (caspase-5 inhibitor) and VEID (caspase-6 inhibitor) partially blocked apoptosis in U266 cells but were ineffective in blocking of apoptosis in HS-Sultan cells. Inhibitors of caspase-1 and -2 were practically ineffective in blocking apoptosis in both cell lines (Figure 5). Nonspecific toxicity of the control peptide (CP) was low, and toxicity of the blocking peptide was less than 5% (results not shown). These results taken together suggest that the extrinsic apoptotic pathway, involving caspase-8 and -10, is the major pathway engaged in ATO-induced apoptosis in cells with mutated p53. In contrast, the dominant caspase activated by ATO in cells expressing wt p53 is caspase-9, a prominent caspase in the intrinsic mitochondrial apoptotic pathway.
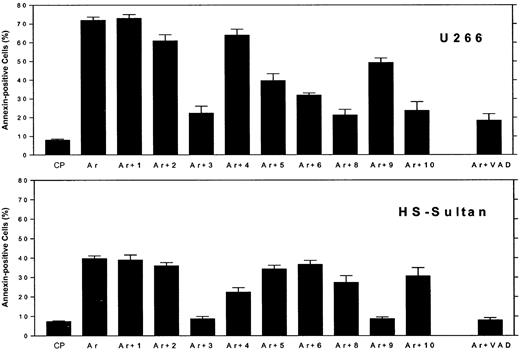
Differential blocking of apoptosis by caspase-specific blocking peptides in myeloma cells expressing wt or mutated p53.
U266 cells (mutated p53, upper panel) and HS-Sultan (wt p53, lower panel) cells were cultured for 48 hours with 7.5 μM ATO, with or without 2 μM of each caspase inhibitory peptide, or control peptide (FA-FMK). Nonspecific toxicity of the control peptide (CP) was around 5%. The toxicity of the blocking peptides was 2% to 5% above untreated control cells (results not shown). Apoptosis was determined by the annexin V method. Ar is ATO alone; Ar+1 is ATO plus caspase-1 inhibitor; Ar+2 is ATO plus caspase-2 inhibitor; and so on. For more experimental details, see “Materials and methods.” Bars are ± SD of at least 3 experiments. Note the differential blocking of apoptosis by caspase-9 and caspase-8 and -10, depending on the status of p53.
Differential blocking of apoptosis by caspase-specific blocking peptides in myeloma cells expressing wt or mutated p53.
U266 cells (mutated p53, upper panel) and HS-Sultan (wt p53, lower panel) cells were cultured for 48 hours with 7.5 μM ATO, with or without 2 μM of each caspase inhibitory peptide, or control peptide (FA-FMK). Nonspecific toxicity of the control peptide (CP) was around 5%. The toxicity of the blocking peptides was 2% to 5% above untreated control cells (results not shown). Apoptosis was determined by the annexin V method. Ar is ATO alone; Ar+1 is ATO plus caspase-1 inhibitor; Ar+2 is ATO plus caspase-2 inhibitor; and so on. For more experimental details, see “Materials and methods.” Bars are ± SD of at least 3 experiments. Note the differential blocking of apoptosis by caspase-9 and caspase-8 and -10, depending on the status of p53.
To confirm the results obtained from using caspase blocking peptides, fluorescence-tagged caspase-specific substrate peptides were employed. In this assay activated caspases are capable of degrading the fluorescence-tagged substrate peptide, thereby generating a fluorescent hydrolysis product. RPMI 8226 cells (mutated p53) were treated with 7.5 μM ATO for 0, 16, 24, and 48 hours, after which the cells were harvested and assayed for caspase activity and for apoptosis by the annexin V method. An example of the flow cytometry histograms obtained for apoptosis and for caspase-3, -8, and -9 activity is depicted in Figure 6. A time-dependent activation of caspase-3 and -8 was observed concomitant with apoptosis. Caspase-8 activation in RPMI 8226 cells was evident early, following 16 hours of treatment with ATO. In contrast, caspase-9 was less active in each time point tested, with percent cells expressing active caspase-9 lagging behind the percent of cells undergoing apoptosis by annexin V. Furthermore, caspase-9 activity was much lower in these cells, with mean fluorescence intensity (MFI) of 63 units compared with an MFI of 210 units for caspase-8 (Figure 6).
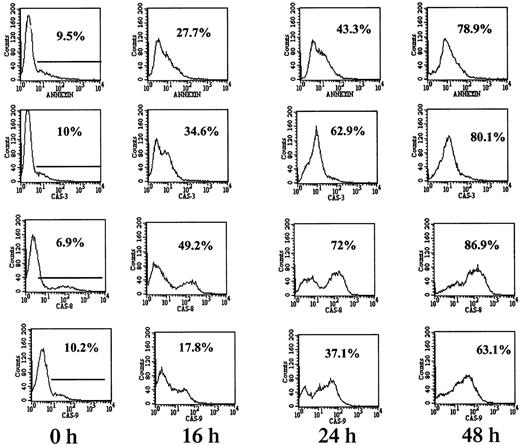
Caspase-8 is the dominant activated caspase in cells with mutated p53.
RPMI 8226 cells were cultured for 0, 16, 24, and 48 hours with or without 7.5 μM ATO followed by 1 hour of incubation with FITC-tagged caspase peptides specific for caspase-3, -8, and -9 as recommended by the manufacturer. FITC–caspase substrate peptides had 5% to 10% background staining (0 h). For comparison, apoptosis was determined in separate tubes by the FITC–annexin V. Fluorescence was quantitated by flow cytometry as described in “Materials and methods”; 10 000 cells were analyzed for each sample. Representative results from at least 3 different experiments are shown. For additional experimental details, see “Materials and methods.” Note the similar kinetics between apoptosis and caspase activity. Note also the preferential activation of caspase-8 over caspase-9 in RPMI 8226 cells.
Caspase-8 is the dominant activated caspase in cells with mutated p53.
RPMI 8226 cells were cultured for 0, 16, 24, and 48 hours with or without 7.5 μM ATO followed by 1 hour of incubation with FITC-tagged caspase peptides specific for caspase-3, -8, and -9 as recommended by the manufacturer. FITC–caspase substrate peptides had 5% to 10% background staining (0 h). For comparison, apoptosis was determined in separate tubes by the FITC–annexin V. Fluorescence was quantitated by flow cytometry as described in “Materials and methods”; 10 000 cells were analyzed for each sample. Representative results from at least 3 different experiments are shown. For additional experimental details, see “Materials and methods.” Note the similar kinetics between apoptosis and caspase activity. Note also the preferential activation of caspase-8 over caspase-9 in RPMI 8226 cells.
The combined results obtained from ARP-1 cells (p53 null), U266 cells (mutated p53), and the wt p53-expressing IM9 and HS-Sultan cells are presented in Figure 7. Whereas caspase-3 was activated in all cell lines, differential activation of caspase-8 was observed for ARP-1 and U266 cells expressing nonfunctional p53. In contrast, differential activation of caspase-9 was observed in IM9 and HS-Sultan cells expressing wt p53. Caspase-9 activation in these cells was higher and occurred faster than caspase-8 and preceded apoptosis despite a low level of apoptosis. A similar trend was observed when the MFI of these 2 caspases was compared (results not shown).
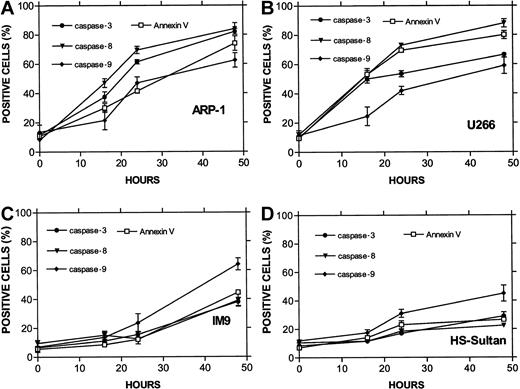
Differential activation of caspase-8 and -9 in myeloma cells expressing wt or mutated p53.
(A) ARP-1 (p53 null), (B) U266 (mutated p53) cells, and (C) IM9 and (D) HS-Sultan (wt p53) cells were cultured with ATO and assayed for apoptosis and for caspase activity using FITC-tagged caspase-specific substrate peptides as described above in the legend to Figure6. For more details, see “Materials and methods.” Bars are ± SD of at least 3 experiments. Note the parallel kinetics between apoptosis and caspase activity in the 4 cell lines and the preferential activation of caspase-9 over caspase-8 in cells with wt p53, although the overall extent of apoptosis in these cells was low.
Differential activation of caspase-8 and -9 in myeloma cells expressing wt or mutated p53.
(A) ARP-1 (p53 null), (B) U266 (mutated p53) cells, and (C) IM9 and (D) HS-Sultan (wt p53) cells were cultured with ATO and assayed for apoptosis and for caspase activity using FITC-tagged caspase-specific substrate peptides as described above in the legend to Figure6. For more details, see “Materials and methods.” Bars are ± SD of at least 3 experiments. Note the parallel kinetics between apoptosis and caspase activity in the 4 cell lines and the preferential activation of caspase-9 over caspase-8 in cells with wt p53, although the overall extent of apoptosis in these cells was low.
To further confirm the pattern of caspase activation in the various cell lines, we performed Western immunoblotting of these cells treated with 7.5 μM ATO for 0, 16, 24, and 48 hours, as described above. The immunoblots for polyadenosine-5′-diphosphate-ribose polymerase (PARP) cleavage (116 kDa and 85 kDa), procaspase-8 cleavage (55 kDa zymogene), and procaspase-9 cleavage (46 kDa zymogene) are shown in Figure 8. Cleavage of PARP occurred in all cell lines early on, 16 to 24 hours after treatment with ATO. On the other hand, caspase cleavage correlated with the extent of apoptosis for each cell line tested. Hence, ARP-1 and U266 cells, which are more sensitive to ATO than IM9 and HS-Sultan cells, exhibited extensive cleavage of caspase-8 24 hours after onset of treatment, whereas procaspase-9 was very low in these cells. In contrast to ARP-1 and U266 cells, IM9 and HS-Sultan cells expressed relatively high levels of procaspase-9 and activation occurred late, 48 hours after onset of ATO treatment, correlating well with the extent of apoptosis (Figures 7-8). Cleavage of caspase-8, on the other hand, was very minimal in these cells. Thus, the results obtained from studies with caspase blocking peptides, caspase substrate peptides, and from Western immunoblotting are in good agreement and collectively suggest a differential activation of downstream caspases depending on p53 status in the cells.
Western immunoblot analysis of ATO-induced PARP, caspase-3, -8, and -9 activation in wt and mutated p53-expressing myeloma cells.
ARP-1, IM9, U266, and HS-Sultan cells were cultured for 0, 16, 24, and 48 hours with or without 7.5 μM ATO. Extraction of cellular protein, SDS-PAGE, immunoblotting, and detection of specific protein bands were performed as described in “Materials and methods.” Representative results from at least 3 different experiments are shown. For loading controls, membranes were striped and reprobed for β-actin. For additional experimental details, see “Materials and methods.”
Western immunoblot analysis of ATO-induced PARP, caspase-3, -8, and -9 activation in wt and mutated p53-expressing myeloma cells.
ARP-1, IM9, U266, and HS-Sultan cells were cultured for 0, 16, 24, and 48 hours with or without 7.5 μM ATO. Extraction of cellular protein, SDS-PAGE, immunoblotting, and detection of specific protein bands were performed as described in “Materials and methods.” Representative results from at least 3 different experiments are shown. For loading controls, membranes were striped and reprobed for β-actin. For additional experimental details, see “Materials and methods.”
Synergy between ATO and APO2/TRAIL in the induction of apoptosis
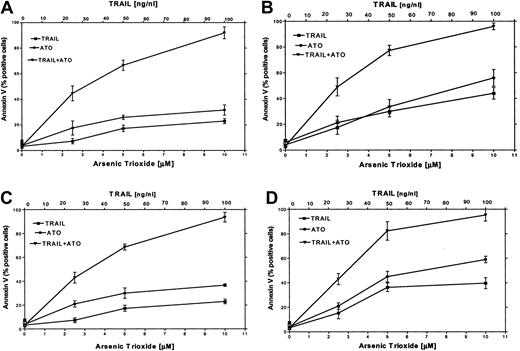
It has been reported that certain chemotherapeutic drugs induce APO2/TRAIL receptors and thereby can potentially engage both the intrinsic and the extrinsic apoptotic pathways. We hypothesized that treatment with ATO may also result in induction of APO2/TRAIL receptors. We therefore tested the combined effect of ATO (0 to 10 μM) and APO2/TRAIL (0 to 100 ng/mL) in myeloma cell lines that were sensitive (ARP-1, U266; null and mutated p53) or partially resistant to ATO (IM9, HS-Sultan; wt p53). The results are depicted in Figure 9. Treatment of HS-Sultan cells with ATO alone or with APO2/TRAIL alone for 48 hours resulted in 22% and 32% apoptosis, respectively, at the highest concentrations of both drugs (10 μM ATO; 100 ng/mL APO2/TRAIL). In contrast, the combination of the 2 drugs was clearly synergistic in all combinations tested, with maximal apoptosis of 89%. Similar results were obtained for IM9 cells, with 93% apoptosis with both drugs following 48 hours of treatment, compared with a maximum of 26% and 38% apoptosis following 48 hours of treatment with TRAIL alone or ATO alone, respectively (Figure 9). Similar experiments were performed with ARP-1 and U266 cells, which are more sensitive to apoptosis induced by individual treatment with ATO or APO2/TRAIL. Thus, treatment of ARP-1 cells with APO2/TRAIL alone or ATO alone for 48 hours resulted in 43% and 57% apoptosis, respectively, whereas the combination of the 2 drugs resulted in 97% apoptosis. Similar results were obtained for U266 where apoptosis with APO2/TRAIL and ATO resulted in a maximum of 97% apoptosis with both drugs, compared with a maximum of 40% and 59% with APO2/TRAIL or ATO alone, respectively, at 10 μM ATO and 100 ng/mL APO2/TRAIL (Figure 9). These results clearly indicated that in cell lines expressing wt p53, which are partially resistant to ATO, the combination of ATO and APO2/TRAIL results in a synergy between the 2 drugs.
ATO synergizes with APO2/TRAIL in the induction of apoptosis in partially resistant myeloma cells.
(A) HS-Sultan, (B) ARP-1, (C) IM9, and (D) U266 cells were cultured for 2 days with 0, 2.5, 5, and 10 μM ATO with or without 25, 50, and 100 ng/mL APO2/TRAIL. Apoptosis was determined by the annexin V method. For additional experimental details, see the legend to Figure 1 and “Materials and methods.” Bars represent SD from at least 3 independent experiments. Note the clear synergy between ATO and APO2/TRAIL in IM9 and HS-Sultan cells expressing wt p53.
ATO synergizes with APO2/TRAIL in the induction of apoptosis in partially resistant myeloma cells.
(A) HS-Sultan, (B) ARP-1, (C) IM9, and (D) U266 cells were cultured for 2 days with 0, 2.5, 5, and 10 μM ATO with or without 25, 50, and 100 ng/mL APO2/TRAIL. Apoptosis was determined by the annexin V method. For additional experimental details, see the legend to Figure 1 and “Materials and methods.” Bars represent SD from at least 3 independent experiments. Note the clear synergy between ATO and APO2/TRAIL in IM9 and HS-Sultan cells expressing wt p53.
An isobologram analysis of the synergy between ATO and APO2/TRAIL for 4 cell lines, IM9 and HS-Sultan (wt p53) and U266 and ARP-1 (mutated p53), was performed according to the model of Laska et al.47 We tested for synergy using 2, one-sided, 2 sample t tests to determine whether apoptosis, as measured by the percent of cells positive for annexin V, was lower in cells treated with a full dose of a single agent (APO2/TRAIL 100 ng/mL, ATO 10 μM) than in cells treated with a 50:50 combination (APO2/TRAIL 50 ng/mL plus ATO 5 μM).47 Tests were conducted at an α of 0.025 level of significance, and the maximum of the 2 one-sided tests is reported as the P value for the overall test. The results indicate clear synergy of the combination of 5 μM ATO and 50 ng/mL APO2/TRAIL compared with 10 μM ATO and 100 ng/mL APO2/TRAIL, as single drugs, in all 4 cell lines tested. The P values were more than .0001, .0005, .0008, and .001 for HS-Sultan, IM9, ARP-1, and U266, respectively.
In our studies, APO2/TRAIL was present throughout the experiment. However, we have evidence that the increase in the expression of R1 and R2 TRAIL receptors is not permanent and a substantial internalization of the receptors occurs 36 to 48 hours after removal of TRAIL (Y.G., unpublished results, 2002).
Induction of APO2/TRAIL receptors by ATO
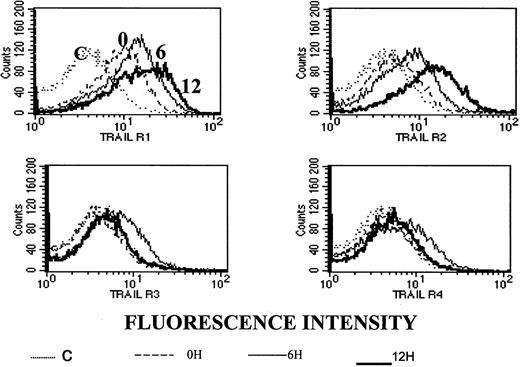
To test whether the observed synergy between ATO and APO2/TRAIL is due to up-regulation of APO2/TRAIL receptors, we analyzed surface expression of APO2/TRAIL receptors after treatment with ATO using mouse monoclonal antibodies specific for the external epitopes of APO2/TRAIL receptors. We first tested the kinetics of induction by ATO of APO2/TRAIL receptors. The results obtained from HS-Sultan cells (wt p53) are depicted in Figure 10. A more than 40% increase in the expression of R1 and R2 APO2/TRAIL agonist receptors was observed concomitant with a small decrease in the expression of R3/R4 APO2/TRAIL decoy receptors as early as 6 to 12 hours after addition of 4 μM ATO.
Kinetics of ATO-induced up-regulation of R1 and R2 APO2/TRAIL receptors in HS-Sultan cells.
HS-Sultan cells were cultured (0.4 × 106/mL) in RPMI medium plus 15% FCS for 0, 6, and 12 hours with 4 μM ATO. Surface expression of APO2/TRAIL receptors was determined by indirect staining with monoclonal antibodies specific for R1, R2, R3, and R4 APO2/TRAIL receptors. The thin dotted line is the immunoglobulin isotype-matched control (C); the dashed line represents time 0; the thin solid line represents 6 hours with ATO; and the thick solid line represents 12 hours with ATO. R1 to R4 are antibodies to the various APO2/TRAIL receptors; 10 000 cells were analyzed in the live cells gate determined by light scatter. For additional experimental details, see “Materials and methods.” It is clear that ATO up-regulates the expression of R1 and R2 APO2/TRAIL receptors as early as 6 to 12 hours after induction.
Kinetics of ATO-induced up-regulation of R1 and R2 APO2/TRAIL receptors in HS-Sultan cells.
HS-Sultan cells were cultured (0.4 × 106/mL) in RPMI medium plus 15% FCS for 0, 6, and 12 hours with 4 μM ATO. Surface expression of APO2/TRAIL receptors was determined by indirect staining with monoclonal antibodies specific for R1, R2, R3, and R4 APO2/TRAIL receptors. The thin dotted line is the immunoglobulin isotype-matched control (C); the dashed line represents time 0; the thin solid line represents 6 hours with ATO; and the thick solid line represents 12 hours with ATO. R1 to R4 are antibodies to the various APO2/TRAIL receptors; 10 000 cells were analyzed in the live cells gate determined by light scatter. For additional experimental details, see “Materials and methods.” It is clear that ATO up-regulates the expression of R1 and R2 APO2/TRAIL receptors as early as 6 to 12 hours after induction.
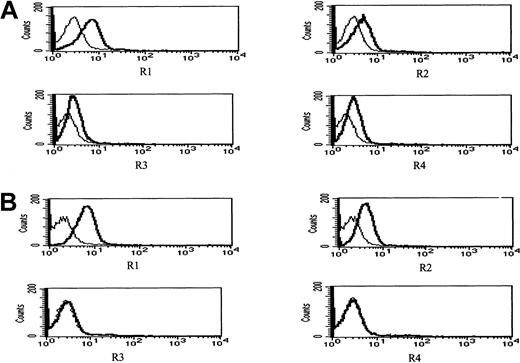
Figure 11 depicts surface expression in IM9 cells of R1 and R2 agonist APO2/TRAIL receptors and R3 and R4 APO2/TRAIL decoy receptors before (Figure 11A) and following 24 hours of treatment with 4 μM ATO (Figure 11B). Before treatment, both R1 and R2 APO2/TRAIL receptors were expressed on the surface of IM9 cells with relatively low expression of R3 and R4 APO2/TRAIL decoy receptors. However, treatment with ATO resulted in enhanced expression of R1/R2 APO2/TRAIL receptors from 46% to 82% and from 21% to 58% for R1 and R2 TRAIL receptors, respectively. Concomitant with the observed increase in the expression of R1 and R2 TRAIL receptors, we observed a decrease in the expression of R3/R4 decoy receptors from 22% and 28% to 3% and 2% for R3 and R4 TRAIL decoy receptors, respectively. We conclude that the synergy between ATO and APO2/TRAIL is indeed the result of early modulation of APO2/TRAIL receptors.
ATO enhances the expression of R1 and R2 APO2/TRAIL receptors in IM9 cells.
IM9 cells were cultured for 24 hours with 4 μM ATO. Surface expression of APO2/TRAIL receptors was determined by indirect staining with monoclonal antibodies specific for R1, R2, R3, and R4 APO2/TRAIL receptors. (A) Untreated cells; (B) ATO-treated cells. The thick line is the corresponding antibody, and the thin line is the corresponding immunoglobulin isotype control. For additional experimental details, see “Materials and methods” and the legend to Figure 10. ATO up-regulates the expression of R1 and R2 APO2/TRAIL receptors and down-regulates the expression of R3 and R4 decoy receptors, resulting in a net increase in the number functional TRAIL receptors.
ATO enhances the expression of R1 and R2 APO2/TRAIL receptors in IM9 cells.
IM9 cells were cultured for 24 hours with 4 μM ATO. Surface expression of APO2/TRAIL receptors was determined by indirect staining with monoclonal antibodies specific for R1, R2, R3, and R4 APO2/TRAIL receptors. (A) Untreated cells; (B) ATO-treated cells. The thick line is the corresponding antibody, and the thin line is the corresponding immunoglobulin isotype control. For additional experimental details, see “Materials and methods” and the legend to Figure 10. ATO up-regulates the expression of R1 and R2 APO2/TRAIL receptors and down-regulates the expression of R3 and R4 decoy receptors, resulting in a net increase in the number functional TRAIL receptors.
To test whether freshly isolated myeloma cells behave as myeloma cell lines, we isolated (CD38+ CD45−) myeloma cells from the bone marrow of 6 MM patients by flow sorting, and myeloma cells with more than 95% purity were obtained.33 Freshly isolated myeloma cells were cultured for 2 days with 4 μM ATO, 100 ng/mL APO2/TRAIL, or both, and apoptosis was scored after 24 and 48 hours. The results are depicted in Table2. As was the case for myeloma cell lines, the combination of ATO and APO2/TRAIL was better than additive in the induction of apoptosis in freshly isolated myeloma cells obtained from all 6 patients tested. Thus, following 24 hours of treatment, a mean percent apoptosis of 68.6% ± 8.4% (± SD) was obtained for the combination of APO2/TRAIL plus ATO, whereas a mean of 31.8% ± 9% and 23% ± 6.2% was obtained for ATO alone and APO2/TRAIL alone, respectively. Following 48 hours of treatment, the combination of ATO and APO2/TRAIL resulted in 95.6% ± 4.4% compared with 48.5% ± 11.5% for ATO alone and 37.8% ± 6.8% for APO2/TRAIL alone (Table 2).
Synergy between APO2/TRAIL and ATO in the induction of apoptosis in freshly isolated myeloma cells
| MM patients (% myeloma cells in BM before sorting) . | % apoptotic cells . | |||||
|---|---|---|---|---|---|---|
| TRAIL, day 1 . | ATO, day 1 . | TRAIL plus ATO, day 1 . | TRAIL, day 2 . | ATO, day 2 . | TRAIL plus ATO, day 2 . | |
| M1 (69) | 30 | 34 | 71 | 37 | 39 | 94 |
| M2 (41) | 17 | 33 | 69 | 38 | 47 | 95 |
| M3 (55) | 18 | 20 | 55 | 29 | 42 | 87 |
| M4 (36) | 30 | 26 | 70 | 42 | 45 | 92 |
| M5 (63) | 29 | 43 | 68 | 38 | 66 | 97 |
| M6 (77) | 20 | 46 | 77 | 44 | 64 | 99 |
| MM patients (% myeloma cells in BM before sorting) . | % apoptotic cells . | |||||
|---|---|---|---|---|---|---|
| TRAIL, day 1 . | ATO, day 1 . | TRAIL plus ATO, day 1 . | TRAIL, day 2 . | ATO, day 2 . | TRAIL plus ATO, day 2 . | |
| M1 (69) | 30 | 34 | 71 | 37 | 39 | 94 |
| M2 (41) | 17 | 33 | 69 | 38 | 47 | 95 |
| M3 (55) | 18 | 20 | 55 | 29 | 42 | 87 |
| M4 (36) | 30 | 26 | 70 | 42 | 45 | 92 |
| M5 (63) | 29 | 43 | 68 | 38 | 66 | 97 |
| M6 (77) | 20 | 46 | 77 | 44 | 64 | 99 |
Fresh myeloma cells were purified by flow sorting of CD38bright/CD45 bone marrow (BM) cells. Myeloma cells with a purity of more than 95% were obtained.33Sorted cells were cultured for 2 days with 4 μM ATO and/or 100 ng/mL APO2/TRAIL.
Apoptosis was determined by the annexin V method in cells treated with APO2/TRAIL alone, ATO alone, or with both drugs.
MI through M6 represent myeloma cells sorted from the BM of 6 MM patients. Background apoptosis in nontreated cells was less than 10%.
Discussion
ATO-induced apoptosis and cell cycle arrest in myeloma cells expressing wt or mutated p53
The mechanism of ATO-induced apoptosis is not yet clear. Most important, different mechanisms were proposed for different type of cells, and inconsistent results were reported by different groups for different types of cells. We performed detailed studies in myeloma cells with varying p53 status in order to delineate the effect of p53 function on the sensitivity to apoptosis, G1 or G2/M cell cycle arrest, and on downstream caspases involved in ATO-induced apoptosis.
Using 7 myeloma cell lines with wt or mutated p53, we found that ATO induces apoptosis in 2 distinct pathways, depending on p53 status. In myeloma cells with mutated p53 or p53 null cells (U266, ARH-77, 8226, ARP-1), low concentrations of ATO (2 to 4 μM) induce rapid apoptosis, reaching more than 50% in less than 16 hours. Percent apoptosis closely correlated with the percent of cells arrested in the G2/M phase of the cell cycle in a time- and dose-dependent fashion. This pattern of apoptosis was very similar to the one observed in the APL cell line, NB4, used is this study as a reference for a “classical” ATO-sensitive cells. In contrast to myeloma cells expressing mutated p53, myeloma cells expressing wt p53 such as MC-CAR, IM9, and HS-Sultan demonstrated partial or full resistance to ATO following long exposure (48 hours) to a high dose of ATO (6 to 10 μM), with apoptosis ranging between 5% and 10% at 16 hours and about 35% at 48 hours. Most important, however, treatment of these cells with ATO did not result in cell cycle arrest at G2/M but, instead, a slight arrest of cells in G1 was observed. Concomitant with G1 arrest, we observed substantial up-regulation of p21 in these cells, which explains the small increase in G1 arrest. These results explain the apparent contradictory results reported by different groups for different cell types. For example, Park et al reported ATO-induced G1arrest in MC-CAR cells11 but G2/M arrest in U937 cells.17 Because MC-CAR cells express wt p53 whereas U937 expresses mutated p53, the results of Park et al are in agreement with our results. Further support was found in studies of ATO-induced apoptosis in T-cell blasts. Normal T cells transformed with phytohemagglutinin (PHA) were treated with 0 to 20 μM ATO for 24 hours, resulting in a maximum of 30% apoptosis by annexin V at 20 μM ATO, concomitant with slight arrest of the cells in G1 and activation of caspase-9 (Y.G., manuscript in preparation). Hence, normal T cells behave like myeloma cells with wt p53.
In the presence of functional p53, ATO acts like a DNA-damaging agent (eg, UV and ionizing radiation), most likely inducing DNA breaks that trigger p53-dependent DNA repair apparatus involving up-regulation of gadd45, p21, and blocking of G1 cyclins followed by G1 arrest and eventually leading to differentiation and/or apoptosis.5,48,49 Apoptosis in cells with functional p53 could also be induced by ATO as a result of p53-dependent transactivation of the apoptosis-inducing protein, p53AIP1, leading to mitochondrial damage and apoptosis via the intrinsic mitochondrial pathway.50 This apoptotic pathway could be activated if one assumes that ATO directly or indirectly can induce phosphorylation of Ser46 on the p53 molecule.50 This possibility is now under investigation in our laboratory. On the other hand, in the absence of functional p53 and G1 arrest, DNA damage can result in a G2 arrest independent of p53 but involving other DNA damage-sensing proteins such as Atm and Atr through a downstream activation of Chk1 and Chk2 kinases, which phosphorylate the Cdc25 phosphatase, and thus blocking Cdc25-regulated dephosphorylation of Cdc2. This can lead to blocking of the formation of the mitotic cyclin B/Cdc2 complex, effectively blocking cells in G2.51 Preliminary studies in our laboratory suggest that a p53-independent differential blocking of cyclins indeed occurs in cells undergoing apoptosis by ATO and in cells expressing nonfunctional p53 (Y.G., unpublished results, 2002). Our interpretation of the effect of ATO in vitro could explain the low toxicity observed in patients undergoing treatment with ATO, because G1block, unlike G2 block, is less toxic to the cells and can be reversible.
p53-dependent differential caspase activation in ATO-induced apoptosis
Our findings suggesting a p53-dependent apoptosis in cells treated with ATO are further supported by the results obtained from studies of the caspase cascade activation by ATO in cells expressing wt versus mutated p53. Thus, using caspase blocking peptides, caspase-specific substrate peptides, and Western immunoblotting, we clearly show that in cells expressing functional p53 the initiator caspase-9 is the principal caspase activated by ATO leading to caspase-3 activation and apoptosis. In contrast, in the absence of functional p53, caspase-8 and -10 are the principal caspases activated by ATO leading to caspase-3 activation and apoptosis. Interestingly, Seol et al reported activation of caspase-9 in PCI-1 cells,19 whereas Kitamura et al reported activation of caspase-8 in ATO-induced apoptosis in gastric cancer cells.20 If the difference between these 2 cell lines is the status of p53, our results explain the seemingly conflicting reports by these 2 groups for caspase activation by ATO. Caspase-8 and -10 are the primary caspases involved in the well characterized extrinsic apoptosis pathways attributed to the fasL and TRAIL,24,52,53 whereas caspase-9 is the primary caspase involved in the intrinsic mitochondrial pathway.54 In rare cases the 2 apoptotic pathways could be linked through activation by caspase-8 of the proapoptotic protein, Bid, resulting in its translocation to the mitochondria and apoptosis via mitochondrial damage.55 It is possible that ATO might be involved in triggering both apoptotic pathways, because results from our laboratory clearly suggest that depolarization of mitochondrial membrane potential occurs early on in cells expressing mutated p53, preceding apoptosis as measured by annexin V (Y.G., manuscript in preparation).
ATO synergizes with TRAIL in the induction of apoptosis
Of a particular interest is our finding that treatment with ATO results in enhancement of the expression of APO2/TRAIL receptors and in a decrease in the expression of APO2/TRAIL decoy receptors. These findings are supported by the synergy observed between TRAIL and ATO in cell lines that are less sensitive to APO2/TRAIL or ATO. These changes in the expression of APO2/TRAIL receptor were evident as early as 6 to 12 hours after treatment with ATO, much earlier than any measurable apoptosis by ATO. In cells expressing mutated (or null) p53, apoptosis is very rapid, including caspase-8 activation; therefore, when such cells were treated with APO2/TRAIL plus ATO, substantial apoptosis occurred very early, and therefore it is hard to dissect the contribution of each component. In contrast, in cells expressing wt p53, apoptosis is very slow, so APO2/TRAIL's effect is evident both in the kinetics and in the extent of apoptosis.
Up-regulation of R1/R2 APO2/TRAIL receptors was reported by Sun et al for ATRA and, similar to our results, ATRA synergized with APO2/TRAIL in the induction of apoptosis in lung cancer cells.56 This effect of ATO on surface APO2/TRAIL receptors is similar to the effect reported for various other chemptherapeutic drugs38-40 and for adenovirus delivery of p53, reported by us.35 36
These results taken together suggest that ATO is a potent inducer of apoptosis in myeloma, particularly in cells expressing mutated p53, and synergizes with APO2/TRAIL in the induction of apoptosis in all myeloma cell lines tested. The fact that freshly isolated myeloma cells have increased susceptibility to the combination of ATO and APO2/TRAIL suggests that our findings are likely to be clinically relevant and that these 2 drugs might work in a similar way in vivo. In this regard, it is important to note that in APL patients treated with 0.15 mg/kg/d ATO, a maximum concentration (Cmax) of 1 to 2 μM ATO in the plasma was documented.4 6 However, multiple myeloma patients receive between 0.25 to 0.4 mg/kg/d ATO. Therefore, the Cmax of ATO in these patients is expected to be much higher than in APL patients and well within the effective doses of 3 to 6 μM used in our study. Finally, it is important to mention that ATO has been used in myeloma patients in phase 1-2 clinical trials as a single agent or in combination with ascorbic acid or in combination with thalidomide. Given the low toxicity of TRAIL and ATO and given the synergy we observed between the 2 drugs, our results justify the use of the combination of these 2 drugs for the treatment of MM patients.
Fluorescence-activated cell-sorting (FACS) analyses were performed in the Institutional Flow Cytometry Core Facility. The authors thank Mr Charles Thomas for performing these analyses and Dr Cagla Akay for her technical help in the caspase assays. We also thank Genentech for supplying APO2/TRAIL and Immunex for supplying the anti-APO2/TRAIL receptor antibodies and Cell Therapeutics for supplying ATO (Trisenox). We also thank Dr Dan Douer for supplying the NB4, promyelocytic leukemia cell line.
Prepublished online as Blood First Edition Paper, January 16, 2003; DOI 10.1182/blood-2002-10-3231.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yair Gazitt, Department of Medicine/Hematology, University of Texas Health Science Center, 7703 Floyd Curl Dr, San Antonio, TX 78284; e-mail:gazitt@uthscsa.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal