Abstract
Interferon (IFN) consensus sequence-binding protein (ICSBP) is an important transcription factor regulating proinflammatory cytokine production and the development of mononuclear phagocytes in vitro. Here we analyzed the role of ICSBP in the in vivo differentiation of 3 major subsets of murine dendritic cells (DCs). We found that ICSBP is predominantly expressed by the CD8α+ subset, and more important, that ICSBP−/− mice have a profound and selective deficiency in CD8α+ DEC205+ DCs in lymphoid tissues. Studies using wild-type/ICSBP−/−chimeras revealed that this defect in CD8α+ DC development is intrinsic to bone marrow–derived progenitors and not dependent on ICSBP expression in the nonhemopoietic compartment. Because DC precursor frequencies are unaltered in the bone marrow of ICSBP−/− mice, ICSBP appears to function by regulating CD8α+ DC differentiation downstream from the generation of common DC progenitors. Although CD8α− DCs are present in normal numbers in ICSBP−/− animals, up-regulation of CD40, CD80, and major histocompatibility complex (MHC) class II expression was found to be impaired in this subset after in vivo microbial stimulation. Together these results demonstrate that ICSBP is critically required for the in vivo differentiation of CD8α+ DCs and may also influence the functional maturation of the CD8α− subsets.
Introduction
Dendritic cells (DCs) are thought to exert a pivotal role in the induction of antigen-specific immune responses.1 Recent studies have demonstrated that the functional diversity of DCs can be attributed in part to the existence of distinct subsets of this important class of antigen-presenting cells.2 Murine DCs have been classified into 3 major subsets (CD8α+CD4−, CD8α−CD4+, and CD8α−CD4−) based on their expression of the surface markers CD4 and CD8α. It was originally thought that these cells represent distinct ontogenetic lineages with CD8α+DCs deriving from “lymphoid” progenitors and CD4+ and double-negative (DN) cells arising from a distinct set of “myeloid” precursors.3 This concept of dual lineages for DC development has recently been overturned by a series of reports demonstrating that all 3 subsets can be generated from either common myeloid or common lymphoid progenitors.4,5 It has also been suggested that different DC subsets represent no more than stages of DC differentiation rather than distinct lineages.6 At present, the nature of the factors that regulate murine DC subset differentiation are poorly understood and their delineation remains an important area of investigation in DC biology.
A major issue raised by the existence of distinct populations of DCs concerns whether these subsets possess distinct immunologic functions or whether their function is determined largely by environmental cues.7 CD8α+ DCs are thought to be closely associated with the induction of cell-mediated immunity. Thus, cells belonging to this subset preferentially produce the cytokine interleukin 12 (IL-12) on stimulation with a number of microbial agents such as bacterial lipopolysaccharide (LPS),8 bacterial CpG-containing oligonucleotides,9 andToxoplasma gondii soluble tachyzoite antigen (STAg)10 known to promote potent TH1 responses. Moreover, on in vivo transfer, antigen-pulsed CD8α+ DCs induced CD4+ T cells with a TH1 cytokine profile, while similarly treated CD8α− DCs promoted a TH2-dominated response.11 In addition, the CD8α+ subset has been shown to play a preferential role in the induction of cytotoxic T cells by cross-priming.12 However, this association of CD8α+ DCs with the induction of cell-mediated immunity is not absolute because additional evidence indicates that under some circumstances some CD8α− DCs can produce IL-12 and induce TH1 responses.13
In the present study, we have investigated host factors that control the differentiation of CD8α+ DCs in vivo. Our approach was to isolate CD8α+ and CD8α− DCs from mouse spleen and to screen by representational difference analysis (RDA) for genes expressed uniquely in the CD8α+ subset. One gene that showed a preferential association with CD8α+ DCs was that encoding interferon consensus sequence binding protein (ICSBP), a member of the interferon regulatory factor (IRF) gene family. Interestingly, this transcription factor had previously been shown to play a preferential role in the induction of IL-12p40 expression in macrophages and spleen cells stimulated with microbial products and to be essential for IL-12–dependent host resistance to both T gondii and Leishmania major.14-16 Thus, ICSBP controls a cytokine preferentially expressed by CD8α+ DCs. We now demonstrate that this same transcription factor is critical for the development of CD8α+ DCs in vivo.
Materials and methods
Animals
Male and female ICSBP−/− mice17backcrossed for 7 generations onto a C57Bl/6 background were generated by heterozygote mating. Wild-type littermates from the same breeding were used as controls. All mice were bred and maintained in a shared NIAID/NICHD (Association for the Assessment and Accreditation of Laboratory Animal Care [AALAC] accredited) animal facility under pathogen-free conditions and used in experiments at 4 to 7 weeks of age.
FACS staining and immunohistochemistry
Cell suspensions from lymphoid organs were obtained as previously described for splenic DC purification.18Briefly, lymphoid organs (spleen, lymph nodes, Peyer patches, and thymus) were harvested and incubated with a solution containing Liberase CI (Roche Biochemicals, Indianapolis, IN) for 30 minutes at 37°C. After incubation tissues were forced through a 100-μm cell strainer and washed with 5 mM phosphate-buffered saline–ethylenediaminetetraacetic acid (PBS-EDTA). Cells were resuspended in 30% PBS-bovine serum albumin (PBS-BSA; 1 mL/spleen) covered with 1 mL PBS and centrifuged (9500g) for 15 minutes at 4°C. The interface was collected and washed with PBS. The resulting cell suspensions were incubated with anti-DEC205–fluorescein isothiocyanate (FITC; Cedar Lane laboratories, Hornby, ON, Canada), anti-CD11c–phycoerythrin (PE; Becton Dickinson, San Diego, CA), anti-CD4–peridinin chlorophyll protein (PerCP; Becton Dickinson) and anti–CD8α-allophycocyanin (APC; Becton Dickinson) for 30 minutes at 4°C. Cells were then washed and acquired in a fluorescence-activated cell sorter (FACS Calibur; Becton Dickinson). Flow cytometry analysis was performed using FlowJo software (Tree Star, San Carlos, CA). The distribution of DCs in spleen before and after LPS injection was assessed by staining of frozen sections using biotin-conjugated monoclonal antibody (mAb) anti-CD11c (Becton Dickinson) as previously described.18
Semiquantitative reverse transcription–polymerase chain reaction
Total RNA was isolated from sorted DC subset samples using the RNeasy mini kit (Qiagen, Crawley, United Kingdom) combined with a DNA digestion step (DNAse set, Qiagen). Single-stranded cDNA was synthesized using the SuperScript preamplification system (Gibco BRL, Paisley, United Kingdom) and polymerase chain reaction (PCR) was carried out according to standard protocols on a PTC-100 thermal cycler (MJ Research, Watertown, MA). PCR products were electrophoresed on 1.5% agarose gels and visualized by ethidium bromide staining. The following primer pairs were used: β-actin: forward: GTTTGAGACCTTCAACACCCC, reverse: GTGGCCATCTCCTGCTCGAAGTC, product size 320 base pair (bp); ICSBP: forward: TCAGCTTTCTCCCAGATGGT, reverse: TAGAATTGCTGCAGCTCTCG, product size 403 bp.
Bone marrow chimeras
Bone marrow from tibia and femurs of wild-type (B6.SJL CD45.1) or ICSBP−/− (CD45.2) mice were recovered by flushing with PBS-EDTA and dissociated by repeated passage through a 20-gauge needle. The resulting cell suspensions were counted and 1:1 mixtures of wild-type and ICSBP−/− cells prepared. The donor mixtures were then injected intravenously into previously irradiated (900 rad) wild-type or ICSBP−/− recipients at a dose of 1 × 106 cells/mouse (0.2 mL). After 8 weeks the animals were killed and spleen cell suspensions prepared for DC subset analysis. To determine the donor origin of the DCs, cells were labeled with anti-CD45.2–FITC (Becton Dickinson), anti-CD11c–PE, anti-CD8α–PerCP, and CD45.1-biotin (Becton Dickinson) followed by streptavidin-APC (Becton Dickinson) and subjected to FACS analysis.
Bone marrow precursor analysis
Quantitation of hematopoietic stem cell (HSC), common myeloid precursor (CMP), and common lymphoid precursor (CLP) in bone marrow was performed using a published protocol.4 For depletion of lineage-positive (Lin+) cells, bone marrow cell suspensions (prepared as described in “Bone marrow chimeras”) were incubated with anti– Ter-192 biotin, anti-CD11b biotin, anti-B220 biotin, anti-CD3εbiotin, anti-Ly6G biotin (all from Becton Dickinson) for 30 minutes at 4°C. Cells were then washed and labeled with streptavidin-conjugated magnetic beads (Miltenyi Biotec, Auburn, CA). After further washing with 5 mM PBS-EDTA, Lin+cells were depleted using a negative-selection column (Miltenyi Biotec). The Lin− cells were then incubated with anti-CD90–FITC (Becton Dickinson), anti-CD127–PE (Becton Dickinson), anti–Sca-1–tetrahodamine isothiocyanate (TRITC; Caltag, Burlingame, CA) and anti-CD117–APC (Becton Dickinson) for HSC and CLP analysis. For CMP analysis a sample of bone marrow suspension was depleted of Lin+ cells (except for those carrying the CD11b marker) and incubated with anti-CD34–FITC (Becton Dickinson), anti-CD16/32–PE (Becton Dickinson), anti–Sca-1–TRITC (Caltag), and anti-CD117–APC (Becton Dickinson). FACS analysis was then performed as described above. The population with the phenotype CD117+Sca-1+CD90+CD127−Lin−was designated as HSC; CD117+Sca-1+CD127+Lin−as CLP, and CD117+CD34+CD16/32loSca-1−as CMP. Absolute cell numbers were determined by reference to the total cells in the starting bone marrow populations.
DC functional response assays
Wild-type and ICSBP−/−animals were injected intraperitoneally with PBS or 1 μg Oligo-CpG-DNA 166819or Escherichia. coli LPS and 6 hours later spleens harvested and low-density (LOD) cells purified as described above. The resulting populations were labeled with anti-CD40–FITC or anti-CD80–FITC or anti–I-A–FITC and with anti-CD11c–PE, anti-CD8α–PerCP as well as anti-CD4–APC. The cells were acquired using a FACS Calibur and analyzed using FlowJo software.
Results
ICSBP is preferentially expressed by the murine CD8α+CD4−CD11c+ DC subset
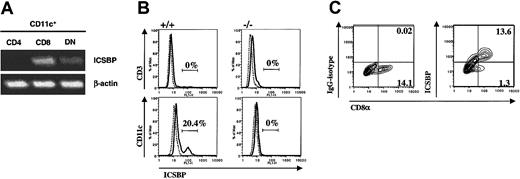
To study ICSBP expression by different DC subsets CD8α+CD11c+ and CD8α−CD11c+ cells were isolated from spleen by FACS; cDNA was prepared and subjected to RDA20 using CD8α+ DC cDNA as tester and CD8α− DC cDNA as driver (D.J.P. et al, unpublished data). Cloning and sequencing of one of the bands from the sample containing CD8α+ DC-specific cDNA revealed that it corresponded to nucleotides 98-461 of murine ICSBP (data not shown). Expression of ICSBP mRNA in CD8α+ DCs was confirmed by reverse transcription–PCR (RT-PCR) with specific primers (Figure1A). ICSBP message was also found to a lesser extent in DN DCs but was absent from CD4+ DCs, which represent most CD8α− cells (Figure 1A). Intracellular staining for ICSBP on CD11c+ DCs further revealed a unique subpopulation of cells expressing the transcription factor (Figure 1B). As expected, these cells were absent in DCs derived from ICSBP−/− animals (Figure 1B). Further analysis confirmed that the ICSBP+ subpopulation detected in wild-type mice corresponds to CD8α+ DCs (Figure 1C).
Selective constitutive expression of ICSBP in CD8α+CD11c+ DCs.
(A) Murine splenic CD11c+CD4+CD8α− (CD4), CD11c+CD4−CD8α+ (CD8), and CD11c+CD4−CD8α− (DN) cells were sorted, mRNA extracted, and RT-PCR performed using ICSBP- and β-actin–specific probes. PCR products were resolved by electrophoresis on agarose gels and products visualized by ethidium bromide staining. (B) Wild-type (+/+) or ICSBP−/− (−/−) splenic CD11c+ or CD3+ cells were purified by positive selection using CD11c- or CD3-conjugated magnetic beads. The resulting cells were then permeabilized, labeled with anti-ICSBP, and analyzed by FACS. (C) Wild-type CD11c+ spleen cells were obtained as in panel B and stained with CD8α+, permeabilized, and incubated with isotype-control or anti-ICSBP. Contour plots represent CD8α versus ICSBP or control-isotype expression; numbers represent the event frequency of the quadrant. Experiments were performed using at least 5 animals per group and are representative of at least 2 independent experiments.
Selective constitutive expression of ICSBP in CD8α+CD11c+ DCs.
(A) Murine splenic CD11c+CD4+CD8α− (CD4), CD11c+CD4−CD8α+ (CD8), and CD11c+CD4−CD8α− (DN) cells were sorted, mRNA extracted, and RT-PCR performed using ICSBP- and β-actin–specific probes. PCR products were resolved by electrophoresis on agarose gels and products visualized by ethidium bromide staining. (B) Wild-type (+/+) or ICSBP−/− (−/−) splenic CD11c+ or CD3+ cells were purified by positive selection using CD11c- or CD3-conjugated magnetic beads. The resulting cells were then permeabilized, labeled with anti-ICSBP, and analyzed by FACS. (C) Wild-type CD11c+ spleen cells were obtained as in panel B and stained with CD8α+, permeabilized, and incubated with isotype-control or anti-ICSBP. Contour plots represent CD8α versus ICSBP or control-isotype expression; numbers represent the event frequency of the quadrant. Experiments were performed using at least 5 animals per group and are representative of at least 2 independent experiments.
ICSBP-deficient mice display a systemic and selective loss of the CD8α+DEC205+CD11c+ subset of DCs
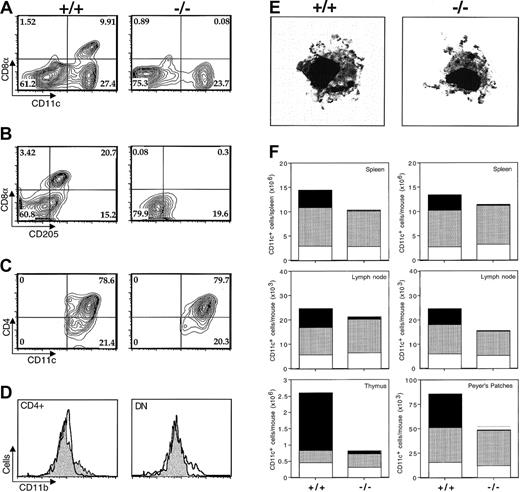
Because of its selective expression in CD8α+DCs, we asked whether ICSBP might have a role in the development of this DC subset. A phenotypic analysis of splenic low-density cells enriched for DCs revealed a dramatic (> 94%) deficiency in CD8α+CD11c+ double-positive cells (Figure 2A) in ICSBP−/−mice versus wild-type littermates. Although this finding suggested that ICSBP selectively directs CD8α+ DC development, it was possible that the gene instead preferentially regulates the expression of the CD8α molecule on the same cells. To rule out this hypothesis we examined the expression of a second marker, DEC205, selectively expressed by CD8α+ DCs under resting conditions.21 Consistent with our previous observation, the frequency of DEC205+CD11c+ cells was profoundly reduced in spleens of ICSBP−/− mice versus littermate controls (Figure 2B). In contrast, no significant changes in the frequencies of CD8α− populations (CD4+and DN) were observed in ICSBP−/− mice (Figure 2C). Moreover, the CD8α− DCs arising in the ICSBP−/− mice showed normal expression of CD11b and were indistinguishable from wild-type DCs in their morphology (Figure 2D,E). To determine whether these findings reflect a systemic defect in CD8α+ DC development in ICSBP−/− animals, we determined the frequency of this subset in other lymphoid tissues (thymus, inguinal, axillary and mesenteric lymph nodes, and Peyer patches) where CD8α+ DCs are generally found. The tissues from ICSBP−/− mice displayed marked reductions of 60% or greater in the levels of CD8α+DEC205+CD11c+ cells detected without major changes in the proportion of CD4+ and DN populations present (Figure 2F). Interestingly, despite the fact that Langerhans cells have been suggested to share a common lineage with CD8α+ DCs, their numbers were not reduced in the skin of ICSBP−/− mice (S. Stol, personal communication, 2001). Together the above findings suggest that ICSBP plays a critical and selective role in the development of CD8α+DCs in vivo.
ICSBP-deficient mice present a systemic and selective loss of CD11c+CD8α+DEC205+ DCs.
(A) CD8α and CD11c staining in low-density (LOD) mouse spleen cells from wild-type (left panels, +/+) or ICSBP−/− (right panels, −/−) animals. (B) DEC-205 and CD8α staining of the same populations after gating on CD11c+ cells. (C) CD4 expression in LOD cells gated on CD11c+CD8α−. Experiment shown is representative of 3 performed. (D) CD11b expression on CD4+(left panel) and DN (right panel) CD11c+ populations in wild-type (filled pattern) and ICSBP−/− (open pattern) spleens. (E) Representative Giemsa-stained purified CD11c+DCs from wild-type (left panel) versus ICSBP−/− spleen (right panel). Original magnification, × 400 for both panels in 2E. (F) Analysis of CD11c+ cell levels in spleen, lymph node, Peyer patches, and thymus of wild-type (left-hand bars in each panel) or ICSBP−/− (right-hand bars) animals. The different segments of each bar represent the proportion of CD8α+ (black pattern), CD4+ (dotted pattern), and double-negative (DN, open pattern) cells present in each population. The difference in the ratio of CD8α+ cells in this figure versus that in Figure 2A may be related to the higher total cell numbers in ICSBP−/−(55 ± 3.9 × 107) versus wild-type (15 ± 1.9 × 107) spleen resulting in a smaller apparent CD8α+ cell reduction. Bars represent means ± SDs of absolute numbers of DCs per mouse (n = 5).
ICSBP-deficient mice present a systemic and selective loss of CD11c+CD8α+DEC205+ DCs.
(A) CD8α and CD11c staining in low-density (LOD) mouse spleen cells from wild-type (left panels, +/+) or ICSBP−/− (right panels, −/−) animals. (B) DEC-205 and CD8α staining of the same populations after gating on CD11c+ cells. (C) CD4 expression in LOD cells gated on CD11c+CD8α−. Experiment shown is representative of 3 performed. (D) CD11b expression on CD4+(left panel) and DN (right panel) CD11c+ populations in wild-type (filled pattern) and ICSBP−/− (open pattern) spleens. (E) Representative Giemsa-stained purified CD11c+DCs from wild-type (left panel) versus ICSBP−/− spleen (right panel). Original magnification, × 400 for both panels in 2E. (F) Analysis of CD11c+ cell levels in spleen, lymph node, Peyer patches, and thymus of wild-type (left-hand bars in each panel) or ICSBP−/− (right-hand bars) animals. The different segments of each bar represent the proportion of CD8α+ (black pattern), CD4+ (dotted pattern), and double-negative (DN, open pattern) cells present in each population. The difference in the ratio of CD8α+ cells in this figure versus that in Figure 2A may be related to the higher total cell numbers in ICSBP−/−(55 ± 3.9 × 107) versus wild-type (15 ± 1.9 × 107) spleen resulting in a smaller apparent CD8α+ cell reduction. Bars represent means ± SDs of absolute numbers of DCs per mouse (n = 5).
The function of ICSBP in CD8α+ DC development is intrinsic to the hemopoietic compartment
To determine whether ICSBP function in the differentiation of CD8α+ DCs is intrinsic to the bone marrow–derived lineage from which these cells originate or instead represents an indirect influence of the gene in the nonhemopoietic compartment, we analyzed CD8α+ DC development in reciprocal bone marrow chimeras constructed with wild-type and ICSBP−/− mice. In the experimental design used, lethally irradiated wild-type or ICSBP−/− recipients were reconstituted with a 1:1 mixture of bone marrow cells from ICSBP−/− (CD45.2) and B6.SJL (CD45.1) mice. Because the 2 donor populations differ in their expression of CD45 alleles, it was possible to assess their individual development in the recipient animals. When examined 8 weeks after bone marrow cell transfer, complete leukocyte repopulation was observed in the lethally irradiated wild-type and ICSBP−/− recipients (data not shown). Frequency analysis of the splenic CD8α− DCs arising in these animals revealed that cells of both ICSBP+/+ and ICSBP−/− origin were able to develop in recipients of either strain. In striking contrast, when CD8α+ DCs were examined only cells of wild-type origin were detected in either bone marrow recipient (Figure3A,B). Thus, CD8α+ DC development depends on the expression of ICSBP in the hemopoietic but not the nonhemopoietic compartment and therefore the transcription factor would appear to function intrinsically in the differentiation of this subset.
CD8α+CD11c+ DC deficiency is due to the lack of ICSBP expression within the hemopoietic compartment.
Bone marrow chimeras were made by intravenous injection of a mixture of bone marrow cell suspensions (1:1 ratio, 1 × 106cells/animal in a volume of 0.2 mL) from wild-type mice congenic for the CD45.1 marker and ICSBP−/− animals, which are CD45.2+. Wild-type (A) or ICSBP−/− (B) hosts were irradiated (9 Gy [900 rads]) prior to bone marrow reconstitution. Eight weeks later, LOD cells were obtained and stained with anti-CD45.2–FITC, anti-CD11c–PE, anti-CD8α–PerCP, and anti-CD45.1–APC. Cells CD11c+CD8α+ (upper gate and panel) or CD11c+CD8α− (lower gate and panel) were analyzed for CD45.1 (wild-type) versus CD45.2 (ICSBP−/−) expression. The values expressed at the corners of each panel represent the relative frequency of cells within that specific quadrant (n = 3 or 4 animals/group).
CD8α+CD11c+ DC deficiency is due to the lack of ICSBP expression within the hemopoietic compartment.
Bone marrow chimeras were made by intravenous injection of a mixture of bone marrow cell suspensions (1:1 ratio, 1 × 106cells/animal in a volume of 0.2 mL) from wild-type mice congenic for the CD45.1 marker and ICSBP−/− animals, which are CD45.2+. Wild-type (A) or ICSBP−/− (B) hosts were irradiated (9 Gy [900 rads]) prior to bone marrow reconstitution. Eight weeks later, LOD cells were obtained and stained with anti-CD45.2–FITC, anti-CD11c–PE, anti-CD8α–PerCP, and anti-CD45.1–APC. Cells CD11c+CD8α+ (upper gate and panel) or CD11c+CD8α− (lower gate and panel) were analyzed for CD45.1 (wild-type) versus CD45.2 (ICSBP−/−) expression. The values expressed at the corners of each panel represent the relative frequency of cells within that specific quadrant (n = 3 or 4 animals/group).
ICSBP deficiency does not affect the frequency of common DC precursors in bone marrow
CD8α+ and CD8α− DCs had been shown to arise from 3 types of common progenitors present in bone marrow: hemopoietic stem cells (HSC), common myeloid precursors (CMP), and common lymphoid precursors (CLP).4 To examine whether the deficiency in CD8α+ DC in ICSBP−/− animals is due to the absence of one of these precursor lineages, a phenotypic analysis for markers associated with each progenitor type was performed on bone marrow from knockout versus wild-type animals. No differences were detected in absolute number or frequency (data not shown) of HSC, CMP, or CLP in the ICSBP−/− versus wild-type mice (Table1). Moreover, we found no evidence for enhanced apoptosis in any of the 3 progenitor populations (not shown). Thus, the deficiency in CD8α+ DCs in ICSBP−/− animals does not appear to be due to the absence of a distinct precursor population. Instead, these data argue that ICSBP acts further downstream in the differentiation of CD8α+ DCs from these common lineages.
ICSBP−/− mice are not deficient in common myeloid and lymphoid bone marrow progenitors
| . | CMP (P) . | CLP (P) . | HSC (P) . |
|---|---|---|---|
| WT | 15.1 ± 1.21 | 3.64 ± 0.36 | 12.7 ± 3.1 |
| ICSBP−/− | 13.3 ± 2.84 (.544) | 2.73 ± 0.69 (.121) | 19.1 ± 2.8 (.081) |
| . | CMP (P) . | CLP (P) . | HSC (P) . |
|---|---|---|---|
| WT | 15.1 ± 1.21 | 3.64 ± 0.36 | 12.7 ± 3.1 |
| ICSBP−/− | 13.3 ± 2.84 (.544) | 2.73 ± 0.69 (.121) | 19.1 ± 2.8 (.081) |
Numbers are cells/mouse (×104; n = 3/group).P values are significance of difference between ICSBP−/− and wild-type levels. CMP indicates common myeloid progenitor (CD117+CD34+CD16/3210Sca-1−); CLP, common lymphoid progenitor (CD117+Sca-1+CD127+Lin−); HSC, hemopoietic stem cell (CD117+Sca-1+CD90+CD127−Lin−); WT, wild-type.
Impaired responsiveness of ICSBP−/−CD8α− DCs to microbial stimulation in vivo
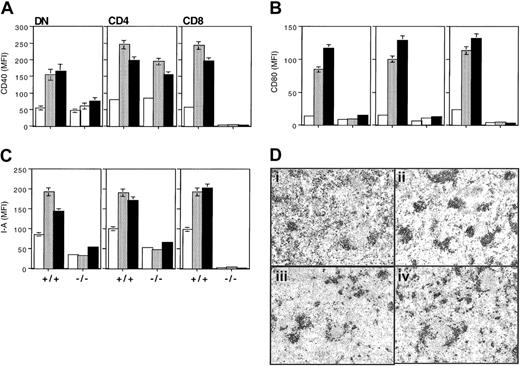
Although ICSBP was found to be constitutively expressed at highest levels in the CD8α+ DC subset and ICSBP−/−mice display a selective deficiency in this subset (Figure 1), it was still possible that the gene plays a more generalized role in the development of all DC subsets. Thus, the absence of CD8α+DCs in ICSBP−/− mice might reflect a block in a late step in DC differentiation, which in addition to preventing the expression of CD8α and DEC-205 also functionally impairs the remaining subsets. To examine this issue we asked whether ICSBP-deficient DCs belonging to the other 2 subsets respond normally to in vivo microbial stimulation with CpG-oligonucleotides or LPS. As shown in Figure4, panels A to C, DN, CD4+, and CD8α+ DC subpopulations purified from wild-type spleen all displayed significantly up-regulated CD40, CD80, and I-A expression in response to injected LPS or CpG-oligo. In contrast, all 3 subsets from ICSBP−/− mice failed to exhibit increased CD40, CD80, or I-A expression in response to the same stimuli with the exception of the CD4+ subset, which displayed a partial up-regulation in CD40 levels. In addition, immunohistochemical examination revealed a major defect in CD11c+ DC migration into T-cell areas of ICSBP−/− spleen in response to injected LPS (Figure 4D). These data suggest that while present in normal numbers in ICSBP-deficient mice, CD8α− DCs are functionally impaired in vivo.
Impaired functional maturation of ICSBP−/−CD8α− DCs in vivo.
Wild-type control and ICSBP−/− mice were injected intraperitoneally with PBS (0.2 mL/mouse) or 1 μg oligo-CpG DNA orE coli LPS. Six hours later low-density spleen cells were purified and the expression of CD40 (A), CD80 (B), and I-A (C) was analyzed by FACS within the cells of the DC subsets: CD11c+CD8α+CD4− (DN), CD11c+CD8α−CD4+ (CD4), CD11c+CD8α+CD4− (CD8) as indicated. Bars represent mean fluorescence intensity (MFI) ± SEM of each marker in each subset analyzed. (D) Representative micrographs of CD11c-stained spleen sections from wild-type (i and ii) or ICSBP−/− (iii and iv) mice injected with PBS (i and iii) or LPS (ii and iv) as described above. Orginal magnifications, × 200.
Impaired functional maturation of ICSBP−/−CD8α− DCs in vivo.
Wild-type control and ICSBP−/− mice were injected intraperitoneally with PBS (0.2 mL/mouse) or 1 μg oligo-CpG DNA orE coli LPS. Six hours later low-density spleen cells were purified and the expression of CD40 (A), CD80 (B), and I-A (C) was analyzed by FACS within the cells of the DC subsets: CD11c+CD8α+CD4− (DN), CD11c+CD8α−CD4+ (CD4), CD11c+CD8α+CD4− (CD8) as indicated. Bars represent mean fluorescence intensity (MFI) ± SEM of each marker in each subset analyzed. (D) Representative micrographs of CD11c-stained spleen sections from wild-type (i and ii) or ICSBP−/− (iii and iv) mice injected with PBS (i and iii) or LPS (ii and iv) as described above. Orginal magnifications, × 200.
Discussion
Although there is now considerable information concerning the functional diversity of DCs, little is known about the factors that determine the development of individual subsets in vivo. Flt-3 ligand22 as well as the transcription factors PU.123 and Ikaros24 have been shown to markedly affect DC generation in vivo. In addition, evidence has been presented for a selective influence of PU.1 and Ikaros on the ratio of CD8α− and CD8α+ DC subsets although it remains unclear to what extent these transcription factors act as determinants of differentiation.25,26 The one previously studied gene product that clearly appears to differentially direct DC subset development is the nuclear factor-κB (NF-κB) family member RelB.27 Mice deficient in this transcription factor generate CD8α+ DCs but neither of the other 2 murine subsets.27 We demonstrate here that ICSBP, another transcription factor associated with immune function, is critical in determining the differentiation of CD8α+ DCs, a functionally important subset of these antigen-presenting cells.
ICSBP (IRF-8) belongs to the IRF family of transcription factors but distinct from the other members of this family is expressed only in hemopoietic cells, including Lin− bone marrow cells28 as well as B and T lymphocytes and mononuclear phagocytes.29 Interferon (IFN)–γ stimulates ICSBP expression through signal transducer and activator of transcription 1 (STAT-1) that binds to the γ-activated site in the ICSBP gene promoter.29 Of direct relevance to the present study is our previous finding that spleen cells and elicited macrophages from ICSBP-deficient mice show impaired T gondii–induced production of IL-12p40 independent of the presence or absence of IFN-γ.15 This study15 along with others16,30 implicated ICSBP as an important transcription factor for IL-12p40 gene expression in macrophages. Later studies revealed that the major cell population producing IL-12p40 inT gondii–stimulated spleen is the CD8α+subset of DCs10 suggesting that the impaired IL-12 expression observed in splenocytes from ICSBP-deficient mice results from a defect in DCs rather than macrophages. The data presented here confirm the latter hypothesis but suggest that the defective IL-12p40 response seen in unfractionated spleen cells may have been due at least in part to the failure of the major DC subset expressing this cytokine to develop in the ICSBP-deficient animals. The concept that ICSBP plays a specific role in CD8α+ DC development is also supported by recent experiments investigating the generation of DC subsets in an in vitro system31 involving murine bone marrow cells stimulated with Flt-3 ligand plus LPS. In this study,32IL-12–producing CD8α+ DCs failed to develop in cultures from ICSBP−/− as opposed to wild-type mice. Retroviral transduction of knockout bone marrow with the ICSBP gene restored in vitro development of CD8α+ DCs and IL-12 production to normal levels. This finding extends previous observations indicating that ICSBP transduction corrects the defect in the in vitro generation of CD11bhigh F4/80+ macrophages displayed by ICSBP-deficient bone marrow progenitors.33Recent studies have elucidated the presence in mice of a plasmacytoid DC population that, in common with the “lymphoidlike” DC population studied here, can express the CD8α+marker.34-36 In experiments not presented here, one of us has shown that these cells are also absent in ICSBP−/−mice (H.T. et al, manuscript in preparation) suggesting an additional role for the transcription factor in the development of this DC lineage.
Although ICSBP is clearly required for the development of CD8α+ DCs both in vivo and in vitro, the precise point at which the gene functions to determine differentiation of this subset remains to be determined. Our finding that ICSBP−/− mice display no deficiencies in the bone marrow progenitor populations implicated in DC differentiation argues that the transcription factor is likely to affect CD8α+ DC generation at a later developmental stage. The latter hypothesis is consistent with previous studies demonstrating that the phenotypic determination of this subset occurs only after the relevant precursors arrive in lymphoid tissues.4 37
Because ICSBP appeared to be affecting a late step in the development of CD8α+ DCs, it was important to determine whether the gene might simultaneously influence some of the late maturational properties of the remaining 2 subsets. We found that although present in unaltered frequencies in ICSBP−/− spleen both CD8α− subsets failed to respond normally to in vivo microbial stimulation in terms of splenic migration and major histocompatibility complex (MHC) class II as well as costimulatory molecule expression. One interpretation of this observation is that the emergence of the CD8α+ DC subset is itself a late maturational step in DC development. Such an explanation is supported by a recent report in which CD8α− DCs were shown to give rise to CD8α+ DCs in lymphoid organs after in vivo injection.6 The latter findings, however, contradict previous studies demonstrating the simultaneous emergence of both subsets in lymphoid tissues37 as well as the observation that CD8α+ cells develop in the absence of CD8α− cells in RelB-deficient mice.27 The concept that CD8α+ DCs arise from CD8α−precursors is also inconsistent with our failure to observe a buildup of CD8α− DCs in the spleens of ICSBP-deficient mice where development of CD8α+ DCs is blocked. Instead we favor the hypothesis that CD8α+ and CD8α−DCs represent independent lineages and that ICSBP in addition to regulating CD8α+ DC development also affects the responsiveness of the other subsets that emerge in its absence, a conclusion supported by our recent experiments on ICSBP function in in vitro–derived DC populations.32 In that study, we noted that ICSBP, which is normally lacking in CD8α− DCs, is induced on microbial stimulation. Therefore, in its absence normal activation of the CD8α−DC population may not be possible. Further studies following both the expression and activity of ICSBP during in vivo DC development should help to resolve this issue.
We thank Ron Germain for his invaluable advice and criticism during the course of this project and Sabine Stoll for allowing us to quote her unpublished data on the frequency of Langerhans cells in ICSBP−/− mice.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2002-04-1088.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Julio Aliberti, Immunobiology Section/LPD/NIAID/NIH. 50 South Dr, Bethesda, MD 20892; e-mail:jaliberti@niaid.nih.gov.

![Fig. 3. CD8α+CD11c+ DC deficiency is due to the lack of ICSBP expression within the hemopoietic compartment. / Bone marrow chimeras were made by intravenous injection of a mixture of bone marrow cell suspensions (1:1 ratio, 1 × 106cells/animal in a volume of 0.2 mL) from wild-type mice congenic for the CD45.1 marker and ICSBP−/− animals, which are CD45.2+. Wild-type (A) or ICSBP−/− (B) hosts were irradiated (9 Gy [900 rads]) prior to bone marrow reconstitution. Eight weeks later, LOD cells were obtained and stained with anti-CD45.2–FITC, anti-CD11c–PE, anti-CD8α–PerCP, and anti-CD45.1–APC. Cells CD11c+CD8α+ (upper gate and panel) or CD11c+CD8α− (lower gate and panel) were analyzed for CD45.1 (wild-type) versus CD45.2 (ICSBP−/−) expression. The values expressed at the corners of each panel represent the relative frequency of cells within that specific quadrant (n = 3 or 4 animals/group).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/1/10.1182_blood-2002-04-1088/6/m_h80133587003.jpeg?Expires=1770045335&Signature=pvtiFb~sG7hVNXo8m9AgJnpvvHBIqlNAPoSrXMDLJ48I5ZpABT0zcxhVObobbE06C3KJ49KWRbBIbErSEBtVuYzKEl2U0UWj7Jn3wx2mxY4pAndQ4jMWtYYkSsVHuslreEh1NUU72uFDeeCjlWVNQI0dFLESxVNzRREJ7fXx33wNkTdv2v93u26oLyqM-fOl7EN-11qkqNJL6KOJGZBz5p81SCDuiu0O0Q8WcdOR9IJ2DbtXMi03hB3jkKMAyI-onMP5btEwGNOXQ62KlUOTJeZ8OzemHoR67X65Ic86ZQcxX8dDJTd2DY39wNNFO0BFyKwCmNEgmv3NbDPJrLppQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal