Abstract
The transcription factor PU.1 plays a pivotal role in normal myeloid differentiation. PU.1−/− mice exhibit a complete block in myeloid differentiation. Heterozygous PU.1 mutations were reported in some patients with acute myeloid leukemia (AML), but not in AML with translocation t(8;21), which gives rise to the fusion geneAML1-ETO. Here we report a negative functional impact of AML1-ETO on the transcriptional activity of PU.1. AML1-ETO physically binds to PU.1 in t(8;21)+ Kasumi-1 cells. AML1-ETO binds to the β3β4 region in the DNA-binding domain of PU.1 and displaces the coactivator c-Jun from PU.1, thus down-regulating the transcriptional activity of PU.1. This physical interaction of AML1-ETO and PU.1 did not abolish the DNA-binding capacity of PU.1. AML1-ETO down-regulates the transactivation capacity of PU.1 in myeloid U937 cells, and the expression levels of PU.1 target genes in AML French-American-British (FAB) subtype M2 patients with t(8;21) were lower than in patients without t(8;21). Conditional expression of AML1-ETO causes proliferation in mouse bone marrow cells and inhibits antiproliferative function of PU.1. Overexpression of PU.1, however, differentiates AML1-ETO–expressing Kasumi-1 cells to the monocytic lineage. Thus, the function of PU.1 is down-regulated by AML1-ETO in t(8;21) myeloid leukemia, whereas overexpression of PU.1 restores normal differentiation.
Introduction
The Ets family of transcription factors plays a key role in the growth, survival, differentiation, and activation of hematopoietic cells. This family of proteins is characterized by presence of an 85 amino acid,1 winged helix-turn-helix DNA-binding domain. PU.1 is one of the most important Ets transcription factors.2 Its expression is limited to hematopoietic cells, including primitive CD34+ cells, macrophages, B lymphocytes, neutrophils, mast cells, and early erythroblasts.2,3 In vitro studies suggest that PU.1 regulates the activity of a number of myeloid- and lymphoid-specific promoters and enhancers.4-10 PU.1 is a key transcription factor for normal myeloid development as demonstrated by a complete block of myeloid development in PU.1−/−mice.11,12 Fetal or newborn PU.1−/− mice have no detectable monocytes/macrophages or neutrophils.11,12 We have recently shown that PU.1 is mutated in patients with acute myeloid leukemia (AML).13These studies all point to the crucial role of PU.1 in both normal myeloid differentiation and leukemogenesis.
AML1 is a member of the Runt-like transcription factors (Runx-1, -2, and -3) named after the Runtprotein that regulates segmentation duringDrosophila embryogenesis.14-16 AML1 appears to act as an “organizing” factor for many promoters and enhancers by interacting with various coactivators and DNA-binding transcription factors.17-22 The AML1 gene is one of the most frequently translocated or mutated genes in human cancer.23-25 The t(8;21)(q22;q22) translocation fuses residues 1-177 of AML1 (including the DNA-binding domain) to nearly all of ETO (also known as CBF2T1).26 ETO is the human homolog ofDrosophila NERVY protein.27-29 The t(8;21) belongs to the most common chromosomal abnormalities in AML, accounting for 10% of all AML cases and 40% of the AML French-American-British (FAB) M2 phenotype.30-33 AML1 activates transcription from enhancer core motifs (TGT/cGGY), which are present in a number of genes relevant to myeloid development, including the macrophage colony-stimulating factor (M-CSF) receptor, granulocyte-macrophage colony-stimulating factor (GM-CSF), myeloperoxidase, and neutrophil elastase.34-39 Like AML1, AML1-ETO can act as a transcriptional activator,40-43 but it is also a transcriptional repressor in other contexts.44 Only one allele of AML1 is altered in leukemia cells expressing t(8;21), and AML1-ETO can efficiently repress AML1-dependent transcriptional activation. Therefore, AML1-ETO has been postulated to act as a dominant inhibitor of AML1 function.34,37 44
Recently, we have shown that AML1-ETO blocks CCAAT enhancer-binding protein (C/EBPα)–dependent activation of its own promoter thus blocking normal granulocytic differentiation of myeloid cells.36 Furthermore, AML1-ETO was shown to repress AML1 and MEF-2–dependent gene activation.45 In our earlier studies we demonstrated that c-Jun, a member of AP-1 transcription factor family, can interact with PU.1 at the β3β4 domain in PU.1 and coactivate the transcriptional activity of PU.1.46 Here we show that AML1-ETO blocks the transcriptional activity of PU.1 by displacing its coactivator c-Jun.
Materials and methods
Cell lines and cell culture
Human kidney 293T, mouse embryonal carcinoma F9, and ecotrophic Phoenix cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco, Aidenbach, Germany) supplemented with 10% fetal bovine serum (FBS), 1% glutamine, and 1% Penstrep ( all from Gibco). Human monoblastic U937 cells and t(8;21)+ Kasumi-1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS (both from Gibco).
Bone marrow cells were isolated from the femurs of Balb/C mice. The femurs were removed and stripped of the soft tissue and crushed to release cells within marrow cavity. The red blood cells were lysed with a 0.15-M solution of ammonium chloride. The pelleted cells were subjected to low-density mononuclear cell separation by incubating with density gradient (Histopaque 1083; Sigma, St Louis, MO) for 10 minutes and centrifuged at 600 rpm for 30 minutes, washed twice in phosphate-buffered saline (PBS), followed by culturing in Iscove modified Dulbecco medium (IMDM; Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 10% FBS (Stem Cell Technologies), 50 ng/mL stem cell factor (R & D Systems, Minneapolis, MN), 50 ng/mL interleukin 6 (IL-6; R & D Systems), and 50 ng/mL Flt-3 ligand (Flt-3L; R & D Systems).
Coimmunoprecipitation assay
Kasumi-1 cells (2 × 107) were lysed and 200 μg protein was used to perform immunoprecipitation as mentioned by Mao et al.45 The following antibodies were used: rabbit IgG (Santa Cruz Biotechnologies, Santa Cruz, CA; catalog no. sc2027), goat IgG (Santa Cruz Biotechnologies; catalog no. 2028), anti-AML1 antibody (Calbiochem, Schwalbach, Germany; catalog no. PC284), anti-PU.1 (Santa Cruz Biotechnologies; catalog no. sc352), and protein-A agarose beads (Santa Cruz Biotechnologies; catalog no. sc2001).
Western blot
After plating in 100-mm plates, 293T cells were transfected using the LipofectAMINE Plus kit (Gibco) as per the manufacturer's protocol. At 24 hours after transfection, cells were harvested and lysed in RIPA lysis buffer, and immunoblot for PU.1 was performed with 100 μg protein as described earlier.46-48 To generate protein lysates, 1 × 106 F9, Kasumi-1, or 293T cells were lysed and nuclear extracts were prepared and immunoblot was performed with 100 μg protein for c-Jun (Santa Cruz Biotechnologies; catalog no. sc54). Mouse bone marrow cells transduced with PU.1, AML1-ETO, or respective empty vectors were similarly lysed (RIPA lysis) and 100 μg protein was used for immunoblot analysis for PU.1 and AML1-ETO (anti-ETO antibody, Santa Cruz Biotechnologies; catalog no. sc9737). Mouse monoclonal anti–β-tubulin purchased from Roche (Mannheim, Germany; catalog no. 1111876) was used for immunoblot assay as internal control. Protein A–peroxidase–conjugated for antirabbit (Amersham Pharmacia, Freiburg, Germany; catalog no. NA 9120), or antigoat peroxidase–conjugated immunoglobulins (Dako, Hamburg, Germany; code no. p0449) were used as secondary antibodies.
Reporter constructs and expression plasmids
The human monocyte–specific M-CSF receptor promoter with or without AML1-binding site, p(PU.1)4TK, and p(mutPU.1)4TK (PU.1-binding sites and mutated PU.1-binding sites subcloned into pTK61luciferase) were described earlier.46 As an internal control plasmid for transient transfection assay, we used the pRL-null construct driving a Renilla luciferase gene (Promega, Madison, WI).49 Other vectors used were pECE-PU.1-murine, pcDNA.1-PU.1, pGEX-2TK-PU.1 or β3β4, pS3H-c-Jun, and pSP6-c-Jun, as described previously.46,50AML1B-pCMV5 and CBFβ-pCMV5 were described earlier.42AML1-ETO-pcDNA3 was constructed by enzymatic digestion of AML1-ETO-pCMV542 with XbaI and subcloning the resulting 2258-bp fragment into the XbaI site of pcDNA3 plasmid (Invitrogen, Karlsruhe, Germany).
Transfection assays
Transient transfections in 293T or F9 cells were carried out with LipofectAMINE transfection kit (Gibco) in 24-well plates as described earlier.46,47 49 U937 cells were transiently transfected by electroporation in RPMI medium at 980 μF and 280 V.Firefly luciferase activities from the constructs M-CSF receptor promoter luciferase, pXP2, p(PU.1)4TK, p(mutPU.1)4TK, andRenilla luciferase activity from internal control plasmid pRL-null were determined 24 hours after transfection using Dual Luciferase Reporter Assay System (Promega). Results are given as means + SEMs from at least 3 independent experiments.
Protein interaction assays
c-Jun and AML1-ETO were transcribed in vitro and translated in the presence of [35S]-methionine (Amersham Pharmacia) using the T7/SP6-coupled reticulocyte system (Promega) in accordance with the manufacturer's instruction. Glutathione-S-transferase (GST) precipitation assays were performed as described earlier.46 48
EMSA
γ32P-adenosine triphosphate (ATP; Amersham Pharmacia)–labeled double-stranded oligonucleotides of PU.1 DNA-binding site51 and AML1-binding site52for electrophoretic mobility shift assay (EMSA) were prepared. The assay was performed with in vitro–translated proteins as mentioned earlier.11 47 For supershift experiments 3 μL of either anti-PU.1 or anti-ETO antibodies were added to the reaction mixture.
Retroviral transduction assay
Ecotrophic Phoenix cells (5 × 106) were plated in 10-cm plates and transfected with 5 μg PINCO-GFP, PINCO-AML1-ETO-GFP, pGsam-PU.1-ires-NGFR, or pGsam-ires-NGFR vectors using LipofectAMINE transfection kit (Gibco). At 24 hours after transfection, the transfection medium was replaced with IMDM (supplemented with 10% FBS, 50 ng/mL stem cell factor, 50 ng/mL IL-6, and 50 ng/mL Flt-3L) for collection of the virus particles. After the viral particle production, freshly isolated mouse bone marrow cells were incubated with viral medium on fibronectin-coated plates and centrifuged for 30 minutes at 1000g (this step was repeated every 12 hours).53 At 60 hours after first transduction, nerve growth factor receptor–positive (NGFR+) or enhanced green fluorescence protein–positive (EGFP+) cells were isolated by fluorescence-activated cell sorting (FACS) analysis (Becton Dickinson, Heidelberg, Germany). To detect the expression of NGFR on the cell surface, cells were stained with mouse antihuman NGFR (Chemicon, Hofheim, Germany; catalog no. MAB5246) followed by phycoerythrin (PE)–conjugated rabbit antimouse immunoglobulins (mouse IgG R-phycoerythrin [RPE]; Dako; catalog no. R0439). Then, 1 × 104 transduced cells sorted for NGFR positivity were plated in 1.2 mL mouse colony-forming medium (Stem Cell Technologies). After 3, 6, and 12 days of plating live cells were counted by trypan blue staining.
Patient material and FACS analysis
Bone marrow cells from AML-M2 patients with or without t(8;21) were obtained after informed consent was given by the patients. Mononuclear cells were isolated from the bone marrow by density gradient centrifugation with Histopaque (Sigma). FACS analysis was performed with CD11b (Pharmingen, Hamburg, Germany; catalog no. 555388), CD14 (Pharmingen; catalog no. 555397), and CD64 (Pharmingen; catalog no. 555527).
Transfection of Kasumi-1 cells and FACS analysis
Kasumi-1 cells were electroporated as mentioned for U937 cells electroporation with pGsam-PU.1-ires-NGFR or pGsam-ires-NGFR vectors and sorted 24 hours after transfection for NGFR positivity by FACS (with anti-NGFR antibody from Chemicon, catalog no. MAB5246, and mouse IgG RPE from Dako, catalog no. R0439). Five days after sorting for NGFR expression, morphologic changes were observed by Wright-Giemsa staining of cells. Then, 1 × 106 NGFR+Kasumi-1 cells were incubated with 10 μL recombinant PE-conjugated mouse monoclonal CD11b (Pharmingen; catalog no. 555388) or fluorescein isothiocyanate (FITC)–conjugated mouse monoclonal CD14 (Pharmingen; catalog no. 555397) in 100 μL PBS for 60 minutes on ice, washed in PBS followed by analysis on a FACScan flow cytometer (Becton Dickinson) using Cellquest software. The cells were also analyzed for the isotype controls, PE-conjugated mouse IgG1κ (Pharmingen, catalog no. 554680) for CD11b-PE and FITC–conjugated mouse IgG1κ (Pharmingen; catalog no. 555748) for CD14-FITC. At 24 hours after transfection, 5 × 104 NGFR+ cells were plated in a 6-well plate and passaged with fresh medium every 24 hours. Cell count for live cells was performed by trypan blue staining every 24 hours.
Results
AML1-ETO interacts with PU.1 in vivo and inhibits its transcriptional activity
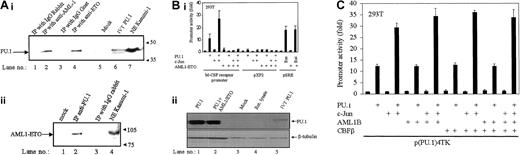
To determine whether PU.1 interacts with AML1-ETO, coimmunoprecipitation assays were performed in Kasumi-1 cells, a human cell line containing t(8;21). PU.1 coprecipitated with both AML1 and ETO antibodies but not with IgG control, suggesting that PU.1 interacts with AML1-ETO in vivo (Figure 1Ai). A similar experiment was performed using a PU.1-specific antibody: AML1-ETO coprecipitated with PU.1, but not with rabbit IgG control (Figure 1Aii).
To investigate the functional impact of this in vivo interaction, we performed transient transfection assays in 293T cells. An M-CSF receptor promoter luciferase reporter construct, which was transactivated 12-fold by PU.1 and 28-fold by PU.1/c-Jun, is completely down-regulated by AML1-ETO (Figure 1Bi). AML1-ETO had no effects on serum response element (pSRE)/Ras activity nor on the empty vector (pXP2) as negative controls. The expression levels of cotransfected PU.1 did not change in the presence of AML1-ETO indicating that the transactivating capacity, but not the expression of cotransfected PU.1, was down-regulated (Figure 1Bii).
AML1 does not affect transactivation capacity of PU.1 or PU.1/c-Jun
The M-CSF receptor promoter has adjacent AML1- and PU.1-binding sites.45 AML1-ETO retains the 177 N-terminus amino acids of AML1, suggesting that AML1 might also have an influence on transactivation of PU.1 or PU.1/c-Jun. Therefore, we addressed if AML1 had any functional impact on the transactivation capacity of PU.1 or PU.1/c-Jun using a promoter containing only PU.1-binding sites (p(PU.1)4TK). Transient transfection assays in 293T cells were performed with p(PU.1)4TK and expression plasmids of PU.1, c-Jun, AML1, and core-binding factor β (CBFβ). Results (Figure 1C) show that AML1 did not affect the PU.1 or PU.1/c-Jun transactivation capacity. In the same experiment AML1 could transactivate the M-CSF receptor promoter 4-fold in the presence of CBFβ (data not shown). AML1, PU.1, c-Jun, and CBFβ had no effects on control vectors (p(mutPU.1)4TK and pXP2) in the experiments above (data not shown).
AML1-ETO binds to PU.1 and down-regulates transactivation capacity of PU.1.
(A) AML1-ETO binds to PU.1 in vivo. (i) Kasumi-1 cell nuclear extracts (200 μg) were immunoprecipitated with rabbit IgG (lane 1), anti-AML1 antibody (lane 2), goat IgG (lane 3), or anti-ETO antibody (lane 4). The immunoprecipitates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) along with in vitro translated PU.1 (lane 6) and nuclear extracts (NE; lane 7) and further subjected to immunoblotting with PU.1 antibody. (ii) Kasumi-1 nuclear extracts were immunoprecipitated with anti-PU.1 (lane 2) or IgG (lane 3) and subjected to SDS-PAGE along with nuclear extracts of Kasumi-1 cells (lane 4) and blotted with anti-ETO antibody. (B) AML1-ETO inhibits transactivation capacity of PU.1. (i) 293T cells were transiently transfected with human monocyte-specific M-CSF receptor promoter or promoterless vector pXP2 or pSRE (serum response element) and with expression plasmids of PU.1 (100 ng), c-Jun (50 ng), AML1-ETO (20 ng), and activated Ras (50 ng). Promoter activities (fold) were determined 24 hours after transfection and normalized to the activities of the internal control plasmid pRL0. Data represent mean values of 3 independent experiments. Error bars represent +SEM. (ii) AML1-ETO does not change the expression of cotransfected PU.1. The 293T cells were transfected as shown in Figure 1Bi, and whole cell lysates were subjected to SDS-PAGE followed by immunoblot assay with PU.1-specific antibody. (C) AML1 does not affect transactivation capacity of PU.1. The 293T cells transfected with p(PU.1)4TK-luc and expression plasmids of PU.1 (100 ng), c-Jun (50 ng), AML1 (50 ng), or CBFβ (50 ng), PU.1, c-Jun, AML1, and CBFβ had no effects on negative control p(mut.PU.1)4TK (data not shown).
AML1-ETO binds to PU.1 and down-regulates transactivation capacity of PU.1.
(A) AML1-ETO binds to PU.1 in vivo. (i) Kasumi-1 cell nuclear extracts (200 μg) were immunoprecipitated with rabbit IgG (lane 1), anti-AML1 antibody (lane 2), goat IgG (lane 3), or anti-ETO antibody (lane 4). The immunoprecipitates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) along with in vitro translated PU.1 (lane 6) and nuclear extracts (NE; lane 7) and further subjected to immunoblotting with PU.1 antibody. (ii) Kasumi-1 nuclear extracts were immunoprecipitated with anti-PU.1 (lane 2) or IgG (lane 3) and subjected to SDS-PAGE along with nuclear extracts of Kasumi-1 cells (lane 4) and blotted with anti-ETO antibody. (B) AML1-ETO inhibits transactivation capacity of PU.1. (i) 293T cells were transiently transfected with human monocyte-specific M-CSF receptor promoter or promoterless vector pXP2 or pSRE (serum response element) and with expression plasmids of PU.1 (100 ng), c-Jun (50 ng), AML1-ETO (20 ng), and activated Ras (50 ng). Promoter activities (fold) were determined 24 hours after transfection and normalized to the activities of the internal control plasmid pRL0. Data represent mean values of 3 independent experiments. Error bars represent +SEM. (ii) AML1-ETO does not change the expression of cotransfected PU.1. The 293T cells were transfected as shown in Figure 1Bi, and whole cell lysates were subjected to SDS-PAGE followed by immunoblot assay with PU.1-specific antibody. (C) AML1 does not affect transactivation capacity of PU.1. The 293T cells transfected with p(PU.1)4TK-luc and expression plasmids of PU.1 (100 ng), c-Jun (50 ng), AML1 (50 ng), or CBFβ (50 ng), PU.1, c-Jun, AML1, and CBFβ had no effects on negative control p(mut.PU.1)4TK (data not shown).
AML1-ETO inhibits the coactivation of PU.1 by c-Jun
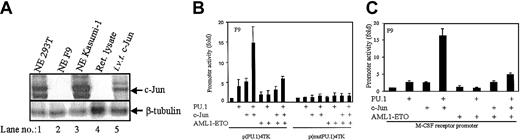
We have earlier shown that c-Jun can coactivate transactivation of PU.1 in a JNK-independent manner.46 PU.1 induced strong transactivation of p(PU.1)4TK in 293T cells (Figure 1C). This is possibly due to high expression of its coactivator c-Jun in these cells. Immunoblot assay for c-Jun indicated that 293T cells have high amounts of c-Jun (Figure 2A lane 1) comparable to Kasumi-1 cells (Figure 2A lane 3). However, F9 cells had no detectable c-Jun protein (Figure 2A lane 2). Therefore, further experiments were carried out in F9 cells, which served as a model cell line for understanding how AML1-ETO might interfere with the capacity of c-Jun in coactivating PU.1. PU.1/c-Jun could transactivate p(PU.1)4TK (Figure 2B) and also the M-CSF receptor promoter (Figure 2C) in F9 cells as described earlier.46 In the presence of AML1-ETO, the capacity of PU.1/c-Jun in transactivating the target promoters (Figure 2B-C) was down-regulated. c-Jun up-regulated the p(PU.1)4TK promoter in 293T cells (Figure 1B) and F9 cells (Figure 2B-C), which might be due to presence of noncanonical sites in the promoter construct or unknown factors in these 2 cell lines collaborating with c-Jun. A similar effect was also reported earlier.48 However, this does not influence the final conclusion.
AML1-ETO inhibits coactivation of PU.1 by c-Jun.
(A) F9 cells do not express c-Jun. Nuclear extracts (100 μg) of 293T, F9, and Kasumi-1 cells along with in vitro–translated c-Jun were subjected to SDS-PAGE and immunoblotted for c-Jun. (B) AML1-ETO inhibits PU.1/c-Jun transactivation capacity. F9 cells were transfected with p(PU.1)4TK, a minimal TK promoter driven by PU.1 DNA-binding sites only or control vector p(mut.PU.1)4TK along with expression plasmids of PU.1 (100 ng), c-Jun (50 ng), and AML1-ETO (20 ng). (C) AML1-ETO down-regulates the PU.1-regulated M-CSF receptor promoter activity by inhibiting PU.1/c-Jun function. F9 cells were transfected with M-CSF receptor promoter and PU.1 (100 ng), c-Jun (50 ng), and AML1-ETO (20 ng). PU.1, c-Jun, and AML1-ETO had no effects on control vector pXP2 (data not shown).
AML1-ETO inhibits coactivation of PU.1 by c-Jun.
(A) F9 cells do not express c-Jun. Nuclear extracts (100 μg) of 293T, F9, and Kasumi-1 cells along with in vitro–translated c-Jun were subjected to SDS-PAGE and immunoblotted for c-Jun. (B) AML1-ETO inhibits PU.1/c-Jun transactivation capacity. F9 cells were transfected with p(PU.1)4TK, a minimal TK promoter driven by PU.1 DNA-binding sites only or control vector p(mut.PU.1)4TK along with expression plasmids of PU.1 (100 ng), c-Jun (50 ng), and AML1-ETO (20 ng). (C) AML1-ETO down-regulates the PU.1-regulated M-CSF receptor promoter activity by inhibiting PU.1/c-Jun function. F9 cells were transfected with M-CSF receptor promoter and PU.1 (100 ng), c-Jun (50 ng), and AML1-ETO (20 ng). PU.1, c-Jun, and AML1-ETO had no effects on control vector pXP2 (data not shown).
AML1-ETO displaces c-Jun by binding to the β3β4 region in PU.1
The results in F9 cells (Figure 2B-C) suggest that AML1-ETO interferes with coactivation of PU.1 by c-Jun. To investigate this, in vitro protein-protein interaction assays were performed. c-Jun and AML1-ETO bind to the full-length PU.1 fused to GST (Figure3A). c-Jun was shown to interact at the β3β4 region of the DNA-binding domain of PU.1.48 Therefore, we performed protein-protein interaction assays using GST-β3β4 and found that AML1-ETO also binds to GST-β3β4(Figure 3B). In competitive protein-protein interaction assays on increasing the AML1-ETO protein, c-Jun protein bound to GST-β3β4 was reduced (Figure 3B). These results indicate that AML1-ETO competes c-Jun away from binding to the β3β4 domain of PU.1. Thus, the c-Jun coactivation function of PU.1 is down-regulated and this in turn down-regulates transcriptional activity of PU.1.
AML1-ETO displaces the coactivator c-Jun from PU.1 by binding to the β3β4 region of PU.1.
(A) AML1-ETO physically binds to PU.1 in vitro. GST pull-down assay was performed using [35S]-methionine–labeled in vitro–translated c-Jun (lane 1) or AML1-ETO (lane 4) incubated with equal amounts of bacterially expressed GST-PU.1 (lanes 2 and 5) or GST plus beads (lanes 3 and 6). GST-PU.1 or GST was recovered using glutathione-agarose beads and separated by SDS-PAGE prior to autoradiography. (B) AML1-ETO displaces c-Jun from binding to the β3β4 domain of PU.1. Saturating amounts of in vitro–translated c-Jun (20 μL) were incubated with GST-β3β4 and increasing amounts (from lanes 7-13) of in vitro–translated AML1-ETO (7.5-12.5 μL) were incubated. Densitometric quantification was also performed (given as percent input of the proteins).
AML1-ETO displaces the coactivator c-Jun from PU.1 by binding to the β3β4 region of PU.1.
(A) AML1-ETO physically binds to PU.1 in vitro. GST pull-down assay was performed using [35S]-methionine–labeled in vitro–translated c-Jun (lane 1) or AML1-ETO (lane 4) incubated with equal amounts of bacterially expressed GST-PU.1 (lanes 2 and 5) or GST plus beads (lanes 3 and 6). GST-PU.1 or GST was recovered using glutathione-agarose beads and separated by SDS-PAGE prior to autoradiography. (B) AML1-ETO displaces c-Jun from binding to the β3β4 domain of PU.1. Saturating amounts of in vitro–translated c-Jun (20 μL) were incubated with GST-β3β4 and increasing amounts (from lanes 7-13) of in vitro–translated AML1-ETO (7.5-12.5 μL) were incubated. Densitometric quantification was also performed (given as percent input of the proteins).
AML1-ETO does not change the DNA binding of PU.1
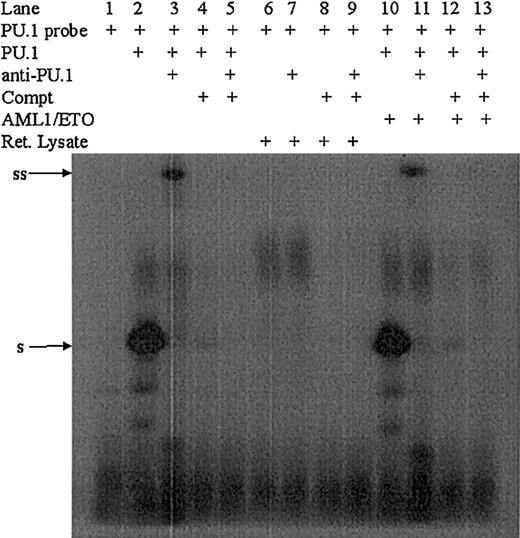
The protein-protein interactions described in Figure 3demonstrate that AML1-ETO directly interacts with PU.1. The physical interaction of AML1-ETO/PU.1 might down-regulate the DNA-binding capacity of PU.1. To address this possibility, we performed an EMSA using in vitro–translated PU.1 and AML1-ETO and oligonucleotide probes having respective DNA-binding sequences.51 52 In vitro–translated PU.1 binds specifically to the PU.1-binding oligonucleotide (Figure 4). Even in presence of AML1-ETO, no change of DNA binding of PU.1 was observed (Figure 4), indicating that AML1-ETO blocks the transactivation capacity, but not DNA binding of PU.1. In the same experiment in vitro–translated AML1-ETO was found to bind to the AML1 probe (data not shown).
AML1-ETO does not change the DNA binding of PU.1.
The PU.1-binding sequence from the CD11b promoter was chosen and labeled with γ32p-dATP (lane 1), incubated with in vitro–translated PU.1 (lane 2), or in vitro–translated PU.1 and anti-PU.1 antibody (lane 3). As a competitor, unlabeled probe was used in 100 molar excess with (lane 5) and without (lane 4) anti-PU.1 antibody. To investigate if this binding and supershift is specific for PU.1, similar experiments were performed with rabbit reticulocyte lysate (lanes 6-9). In presence of AML1-ETO, PU.1 still binds to its DNA (lane 10) and supershifts with anti-PU.1 antibody (lane 11). In presence of AML 4-ETO and competitor probe alone (lane 12) or plus anti-PU.1 antibody (lane 13), no binding of PU.1 was observed. To the left of blots, ss indicates supershift; s, shift.
AML1-ETO does not change the DNA binding of PU.1.
The PU.1-binding sequence from the CD11b promoter was chosen and labeled with γ32p-dATP (lane 1), incubated with in vitro–translated PU.1 (lane 2), or in vitro–translated PU.1 and anti-PU.1 antibody (lane 3). As a competitor, unlabeled probe was used in 100 molar excess with (lane 5) and without (lane 4) anti-PU.1 antibody. To investigate if this binding and supershift is specific for PU.1, similar experiments were performed with rabbit reticulocyte lysate (lanes 6-9). In presence of AML1-ETO, PU.1 still binds to its DNA (lane 10) and supershifts with anti-PU.1 antibody (lane 11). In presence of AML 4-ETO and competitor probe alone (lane 12) or plus anti-PU.1 antibody (lane 13), no binding of PU.1 was observed. To the left of blots, ss indicates supershift; s, shift.
AML1-ETO down-regulates PU.1 transcriptional activity in myeloid cells
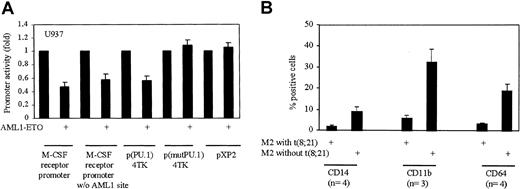
All the above transfections were performed in nonmyeloid 293T or F9 cells. We asked whether the same effects were also observed in myeloid cells. Therefore, we performed transient transfection assays in myelomonocytic U937 cells. U937 cells were transfected with wild-type M-CSF receptor promoter, M-CSF receptor promoter without AML1-binding site, minimal promoter having PU.1-binding sites (p(PU.1)4TK), minimal promoter with mutated PU.1-binding sites (p(mutPU.1)4TK) as control, and empty vector with or without AML1-ETO expression plasmid. We observed that all the promoters were down-regulated by AML1-ETO without any effect on the empty vectors (Figure5A). These data confirm that AML1-ETO down-regulates the transcriptional activity of PU.1 in myeloid cells also. U937 cells express high levels of PU.1 and C/EBPα; therefore AML1-ETO might not only down-regulate PU.1 but also C/EBPα. Therefore only 50% down-regulation of the promoters transfected into U937 cells could be seen. Furthermore, there might be proteins in myeloid cells (in contrast to 293T or F9 cells) that might interfere with the capacity of AML1-ETO to block PU.1 function.
AML1-ETO down-regulates transactivation capacity of PU.1 in myeloid cells and the expression of the PU.1 target genes in AML patients with t(8;21).
(A) AML1-ETO down-regulates transactivation of PU.1 in myeloid cells. U937 cells were electroporated with wild-type M-CSF receptor promoter, M-CSF receptor promoter without (w/o) AML1-binding site, p(PU.1)4TK, p(mutPU.1)4TK, or pXP2 with and without AML1-ETO. (B) Low expression of PU.1 target genes in patients with t(8;21). AML patients (n = number of patients) with t(8;21) have fewer positive cells for cell surface markers regulated by PU.1 as compared to patients without t(8;21). CD14 and CD64 promoters have PU.1-binding sites, but no putative C/EPBα-, AML1-, or MEF-binding sites.
AML1-ETO down-regulates transactivation capacity of PU.1 in myeloid cells and the expression of the PU.1 target genes in AML patients with t(8;21).
(A) AML1-ETO down-regulates transactivation of PU.1 in myeloid cells. U937 cells were electroporated with wild-type M-CSF receptor promoter, M-CSF receptor promoter without (w/o) AML1-binding site, p(PU.1)4TK, p(mutPU.1)4TK, or pXP2 with and without AML1-ETO. (B) Low expression of PU.1 target genes in patients with t(8;21). AML patients (n = number of patients) with t(8;21) have fewer positive cells for cell surface markers regulated by PU.1 as compared to patients without t(8;21). CD14 and CD64 promoters have PU.1-binding sites, but no putative C/EPBα-, AML1-, or MEF-binding sites.
Low expression of PU.1 target genes in patients with t(8;21)
To further understand if the down-regulation of the PU.1/c-Jun transactivation capacity by AML1-ETO leads to down-regulation of the PU.1 target genes, we performed FACS analysis of PU.1 target cell surface markers.7,51 In AML-M2 patients with t(8;21), CD14, CD11b, and CD64 were 4.6-, 5.4-, and 5.8-fold less expressed in comparison to patients with normal M2 karyotype (Figure 5B). Regulation of CD11b promoter by PU.1 has been shown51 and further analysis (by TRANSFAC analysis to identify potential transcription factor-binding sites in a promoter) of the promoter revealed potential AML1-binding sites were present (data not shown). Down-regulation of CD11b might also be due to down-regulation of AML1 in addition to PU.1's transactivation capacity by AML1-ETO. Similar analysis of CD14 and CD64 promoters showed (data not shown) that these gene promoters have PU.1-binding sites but no C/EBPα-, AML1-, or MEF-binding sites. Therefore, CD14 and CD64 down-regulation could be due to specific down-regulation of PU.1's transactivation capacity by AML1-ETO in these patients.
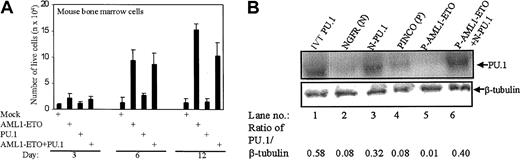
AML1-ETO causes proliferation of mouse bone marrow cells by inhibiting PU.1
To investigate the functional consequences of AML1-ETO down-regulating the transactivation capacity of PU.1, we transduced mouse bone marrow cells with PU.1 (pGsam-NGFR-PU.1) and AML1-ETO (PINCO-AML1-ETO-GFP). The cells transduced with AML1-ETO rapidly increased in number over 12 days, as did the cells overexpressed with AML1-ETO and PU.1 (Figure 6A). The cells transduced with PU.1 showed no increase in cell number (Figure 6A). Furthermore, transduction of AML1-ETO blocks PU.1-induced monocytic differentiation in mouse bone marrow cells (data not shown). The expression of transduced genes is shown in Figure 6 B-C. Densitometric quantification of the PU.1 protein expression in the same experiment revealed down-regulation of endogenous PU.1 expression on overexpression of AML1-ETO (Figure 6B). This could be due to AML1-ETO preventing the autoregulation of PU.1.54 The expression of AML1-ETO was also quantified (data not shown).
The antiproliferative effect of PU.1 is down-regulated by AML1-ETO in mouse bone marrow cells.
(A) AML1-ETO causes proliferation in mouse bone marrow cells. Live transduced mouse bone marrow cells with PU.1, AML1-ETO, or PU.1 and AML1-ETO were counted on days 3, 6, and 12 after trypan blue staining. Because both the empty vectors gave the same cell count, only one vector (PINCO) has been represented as mock. (B) Expression of PU.1 in mouse bone marrow cells. The cells of the transduction described in the legend to Panel A were lysed and immunoblot assays were performed for PU.1 and β-tubulin. NGFR (N; lysate of empty vector of PU.1), N-PU.1 (NGFR-PU.1–transduced cells), PINCO (P; lysate of empty vector of AML1-ETO–transduced cells), P-AML1-ETO (lysate of PINCO-AML1-ETO–transduced cells), and P-AML1-ETO+N-PU.1 (lysate of PINCO-AML1-ETO– and NGFR-PU.1–transduced cells) were analyzed. The ratio of PU.1/β-tubulin was calculated after densitometric quantification of the bands.
The antiproliferative effect of PU.1 is down-regulated by AML1-ETO in mouse bone marrow cells.
(A) AML1-ETO causes proliferation in mouse bone marrow cells. Live transduced mouse bone marrow cells with PU.1, AML1-ETO, or PU.1 and AML1-ETO were counted on days 3, 6, and 12 after trypan blue staining. Because both the empty vectors gave the same cell count, only one vector (PINCO) has been represented as mock. (B) Expression of PU.1 in mouse bone marrow cells. The cells of the transduction described in the legend to Panel A were lysed and immunoblot assays were performed for PU.1 and β-tubulin. NGFR (N; lysate of empty vector of PU.1), N-PU.1 (NGFR-PU.1–transduced cells), PINCO (P; lysate of empty vector of AML1-ETO–transduced cells), P-AML1-ETO (lysate of PINCO-AML1-ETO–transduced cells), and P-AML1-ETO+N-PU.1 (lysate of PINCO-AML1-ETO– and NGFR-PU.1–transduced cells) were analyzed. The ratio of PU.1/β-tubulin was calculated after densitometric quantification of the bands.
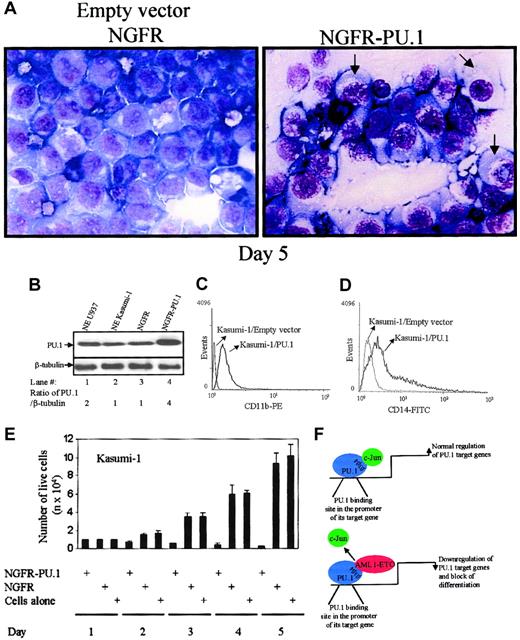
Overexpression of PU.1 initiates differentiation in t(8;21)+ Kasumi-1 cells
Our data so far shows that AML1-ETO interacts with PU.1 at the β3β4 region in the DNA-binding domain of PU.1 and displaces c-Jun from binding and coactivating PU.1 (Figures2-3). Moreover, overexpression of AML1-ETO down-regulated the PU.1 expression in mouse bone marrow cells (Figure 6B). It is important to note that Kasumi-1 cells shows high levels of c-Jun protein expression (Figure 2A). Hence, we asked whether overexpression of PU.1 could overcome the functional block of PU.1 by AML1-ETO. Transient overexpression of PU.1 (pGsam-NGFR-PU.1) in t(8;21)–bearing Kasumi-1 cells was performed. FACS sorting (for NGFR) of the transfected cells showed the PU.1 expression, which was further shown by immunoblot analysis of sorted cells for PU.1 expression (Figure7B). Fourfold overexpression was observed after transfection (Figure 7B).
Transient overexpression of PU.1 induces differentiation toward the monocytic lineage in AML1-ETO+ Kasumi-1 cells.
(A) PU.1 induces differentiation in t(8;21)+ Kasumi-1 cells. Kasumi-1 cells were transiently transfected with PU.1 (pGsam-PU.1-ires-NGFR) or the empty vector (pGsam-ires-NGFR) and morphologic changes were observed on day 5 (original magnification, × 63). Arrows indicate the differentiating cells. (B) PU.1 overexpression in Kasumi-1 cells. Western blot shows PU.1 expression and β-tubulin in transfected Kasumi-1 cells after day 5. (C) PU.1 induces CD11b expression in Kasumi-1 cells. FACS analysis was performed for the cell surface expression of CD11b in Kasumi-1 cells transfected with empty vector or PU.1. (D) PU.1 induces CD14 expression in Kasumi-1 cells. In the same experiment FACS analysis was performed for the cell surface expression of CD14 in Kasumi-1 cells transfected with empty vector or PU.1. (E) Kasumi-1 cell number decreases in PU.1-transfected cells. The transfected cells described in the legend to Panel D were counted by trypan blue staining on days 1, 2, 3, 4, and 5 after transfection. (F) Model of AML1-ETO blocking PU.1 function. Model is of AML1-ETO interacting with PU.1 and displacing its coactivator c-Jun. This down-regulation of the PU.1 transcriptional activity by AML1-ETO results in down-regulation of PU.1 target genes important for myeloid differentiation.
Transient overexpression of PU.1 induces differentiation toward the monocytic lineage in AML1-ETO+ Kasumi-1 cells.
(A) PU.1 induces differentiation in t(8;21)+ Kasumi-1 cells. Kasumi-1 cells were transiently transfected with PU.1 (pGsam-PU.1-ires-NGFR) or the empty vector (pGsam-ires-NGFR) and morphologic changes were observed on day 5 (original magnification, × 63). Arrows indicate the differentiating cells. (B) PU.1 overexpression in Kasumi-1 cells. Western blot shows PU.1 expression and β-tubulin in transfected Kasumi-1 cells after day 5. (C) PU.1 induces CD11b expression in Kasumi-1 cells. FACS analysis was performed for the cell surface expression of CD11b in Kasumi-1 cells transfected with empty vector or PU.1. (D) PU.1 induces CD14 expression in Kasumi-1 cells. In the same experiment FACS analysis was performed for the cell surface expression of CD14 in Kasumi-1 cells transfected with empty vector or PU.1. (E) Kasumi-1 cell number decreases in PU.1-transfected cells. The transfected cells described in the legend to Panel D were counted by trypan blue staining on days 1, 2, 3, 4, and 5 after transfection. (F) Model of AML1-ETO blocking PU.1 function. Model is of AML1-ETO interacting with PU.1 and displacing its coactivator c-Jun. This down-regulation of the PU.1 transcriptional activity by AML1-ETO results in down-regulation of PU.1 target genes important for myeloid differentiation.
Five days after transfection of PU.1, morphologic changes (Figure 7A) were observed by Wright-Giemsa staining of cells. PU.1-transfected cells differentiated to the monocyte like cells, whereas the empty vector (pGsam-NGFR) transfected cells showed no morphologic change. The PU.1-transfected Kasumi-1 cells also showed an increase in cell surface markers CD11b (Figure 7C; marker for myeloid differentiation) and CD14 (Figure 7D; marker for the monocytic lineage). At 24 hours after transfection, the NGFR-sorted cells were further plated and counted for live cells every 24 hours. In PU.1-transfected cells a decrease in cell number was observed (Figure 7E).
Discussion
The importance of PU.1 in myeloid differentiation is well established. Recently we have reported that PU.1 is mutated in patients with AML13 similar to C/EBPα,55 suggesting that PU.1 also plays a major role in leukemogenesis. However, PU.1 was not found to be mutated in AML patients with t(8;21), which suggests that distinct pathways of inactivation of PU.1 might be occurring in t(8;21) leukemia. We show here that PU.1 plays a major role in leukemogenesis in t(8;21) leukemia because it interacts with fusion protein AML1-ETO (Figure 1A). We have previously reported a similar phenomenon for C/EBPα,36 an important transcription factor in granulocytic differentiation. The physical interaction of PU.1 and AML1-ETO results in inactivation of the transactivation activity of PU.1 by displacing PU.1's coactivator c-Jun (Figures 1B,2B, 2C, and 5A). AML1B was shown to interact with PU.1 and synergize on M-CSF receptor promoter.10 56 In contrast, AML1B does not influence PU.1's transactivation capacity on a promoter driven by PU.1-binding sites only (Figure 1C). These data taken together suggest that AML1B and PU.1 synergy is possible in the promoters having the respective binding sites in near proximity for physical interaction, like the M-CSF receptor promoter.
We observed that AML1-ETO down-regulates the transcriptional activity of PU.1 in myeloid cells (Figure 5A) and physically interacts at the β3β4 region in the DNA-binding domain of PU.1 (Figure 3B). Our earlier data show that c-Jun, an AP-1 transcription factor complex member, binds to the β3β4 region and coactivates PU.1 in a JNK-independent manner.46 The competitive protein-protein interaction experiments with in vitro–translated proteins indicate that AML1-ETO disrupted PU.1/c-Jun interaction in a competitive manner (Figure 3B), thus blocking c-Jun from coactivating PU.1. We have described a similar mechanism for GATA-150 and C/EBPα.48 The role of c-Jun in myeloid differentiation was shown to be rather important, because it could enhance the extent of differentiation in U937 cells.57 In other studies AP-1 and C/EBPβ were shown to cooperate in regulation of common target genes, including the human TSG-6,58 collagenase-1 gene,59 and tumor necrosis factor α (TNF-α).60 These data suggest that the capacity of c-Jun to coactivate PU.1 is also a very important mechanism, which is down-regulated by AML1-ETO.
We observed that physical interaction between AML1-ETO and PU.1 did not abolish the DNA- binding capacity of PU.1 (Figure 4A), although AML1-ETO interacted with the PU.1 DNA-binding domain. Interestingly, in PU.1's crystal structure,1 the β3β4 domain does not interact with DNA, but is exposed to the solvent. This structural ability allows PU.1 to retain its DNA binding though being functionally repressed. We show here that the normal interaction between coactivators and transcription factors are altered in presence of AML1-ETO, which could be one of the important mechanisms for disrupted myelopoiesis in t(8;21)+leukemia (Figure 7F).
AML1B and AML1-ETO have been shown to transactivate the M-CSF receptor,42 suggesting that interaction between AML1B and AML1-ETO could be important for leukemogenesis. To investigate the importance of AML1-ETO/PU.1 interaction in leukemogenesis, transactivation, proliferation, and differentiation assays were performed in cells expressing wild-type AML1B protein. In the presence of AML1-ETO, the M-CSF receptor promoter was down-regulated and similarly the AML1 site mutated M-CSF receptor promoter and minimal promoter containing only PU.1-binding sites in U937 cells (Figure 5A). This could be explained by a dual function of AML1-ETO in regulation of the M-CSF receptor expression. During normal myeloid differentiation, M-CSF receptor expression is required for G1-to-S phase transition, which could be down-regulated by AML1-ETO through the functional interaction with PU.1, and then AML1-ETO cooperates with AML1B to up-regulate the M-CSF receptor expression for transformation and proliferation of abnormal progenitor cells.
In patients with t(8;21), expression of the cell surface markers CD11b, CD14, and CD64 was less in comparison to patients without t(8;21) (Figure 5B). CD14 and CD64 promoters have putative PU.1 binding sites but not AML1-, C/EBPα-, or MEF-binding sites suggesting that down-regulation of the function of PU.1 by AML1-ETO could possibly be an important step in progression toward leukemia. CD11b, another marker for differentiation, was also less expressed in patients with t(8;21) in comparison to patients without t(8;21) (Figure 5B). CD11b is regulated by PU.1 and its promoter contains putative binding sites of AML1. In this case AML1-ETO interaction and down-regulation of important myeloid transcription factors like PU.1 and AML1 could explain the lower CD11b expression. The phenotype of PU.1−/− mice suggests that PU.1 is critical for myeloid differentiation and development.4,11,12Interaction of PU.1 with AML1-ETO and subsequent suppression of PU.1 target genes (Figure 5B) might contribute to the phenotypic changes seen in t(8;21). Furthermore, PU.1 is a self-regulatory protein54 and AML1-ETO overexpression in mouse bone marrow cells down-regulated the endogenous PU.1 expression (Figure 6B).
Because AML1−/− mice lack PU.1 expression,61AML1-ETO could down-regulate PU.1 expression through repressing AML1 function. However, in a diseased condition or in presence of AML1-ETO, like in Kasumi-1 cells, the expression of AML1 and PU.1 genes was still observed. Furthermore, AML1 does not have a down-regulatory effect on PU.1 (Figure 1C). Therefore, the presence of AML1-ETO does not completely repress the expression levels of these genes, but may block their functions by protein-protein interactions. To analyze the functional impact of AML1-ETO on PU.1 in the presence of wild-type AML1B protein, we performed experiments in cells expressing endogenous AML1B protein. Overexpression of PU.1 in mouse bone marrow cells leads to a block in proliferation, but in presence of AML1-ETO this function of PU.1 was abrogated (Figure 6A). Recently, it was shown that AML1-ETO expression in human progenitor cells leads to expansion of human hematopoietic stem cells.62 Therefore, the block of differentiation and increase in abnormal proliferation of hematopoietic stem cells could be due to down-regulation of the activity of PU.1 in t(8;21) leukemia.
Overexpression of PU.1 in t(8;21)+ Kasumi-1 cells differentiates them toward the monocytic lineage (Figure 7). Morphologically, cells did not appear to be terminally differentiated even though the cell surface markers CD11b and CD14 were increased in expression. It has been earlier shown that short-term activation of PU.1 in multipotent hematopoietic cells leads to immature eosinophils.63 However, stable overexpression of PU.1 could lead to myeloid lineage in hematopoietic progenitor cells.63 Therefore, higher and stable expression of PU.1 in Kasumi-1 cells might be needed to terminally differentiate toward the monocytic lineage. The cell number of PU.1-transfected Kasumi-1 cells decreased over a course of time (Figure 7E) showing that PU.1 functions as an antiproliferative factor on overexpression in Kasumi-1 cells. However, this mechanism needs to be further elucidated. Our data suggest that the ectopic expression of PU.1 in Kasumi-1 cells overcomes the functional block by AML1-ETO. PU.1 and C/EBPα are important factors for myeloid differentiation and AML1-ETO down-regulating these 2 factors could be an important step toward leukemia. This also suggests the possibility of using these 2 factors independently or in combination for therapy of t(8;21) myeloid leukemias.
We thank Dr Atsushi Iwama, University of Tsukuba, Japan for providing pGsam-PU.1-ires-NGFR and PGsam-ires-NGFR retroviral vectors.
Prepublished online as Blood First Edition Paper, August 29, 2002; DOI 10.1182/blood-2002-04-1288.
Supported by a DFG (German research foundation) grant to G.B. (no. 2042/2-1).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Gerhard Behre, Department of Internal Medicine III, University Hospital Grosshadern, Ludwig-Maximilians-University Munich, Marchioninistr 15, D-81377 Munich, Germany; e-mail: gerdbehre@aol.com.

![Fig. 3. AML1-ETO displaces the coactivator c-Jun from PU.1 by binding to the β3β4 region of PU.1. / (A) AML1-ETO physically binds to PU.1 in vitro. GST pull-down assay was performed using [35S]-methionine–labeled in vitro–translated c-Jun (lane 1) or AML1-ETO (lane 4) incubated with equal amounts of bacterially expressed GST-PU.1 (lanes 2 and 5) or GST plus beads (lanes 3 and 6). GST-PU.1 or GST was recovered using glutathione-agarose beads and separated by SDS-PAGE prior to autoradiography. (B) AML1-ETO displaces c-Jun from binding to the β3β4 domain of PU.1. Saturating amounts of in vitro–translated c-Jun (20 μL) were incubated with GST-β3β4 and increasing amounts (from lanes 7-13) of in vitro–translated AML1-ETO (7.5-12.5 μL) were incubated. Densitometric quantification was also performed (given as percent input of the proteins).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/1/10.1182_blood-2002-04-1288/6/m_h80133591003.jpeg?Expires=1769145442&Signature=jQWLs~CMyEAMIdkDkPLDfjlkzkMJjOVo5iJe6~n7Ucpg-Cpqm8kkBKoLRdQru10gBkNL6SDgkNOTRxinQcjwAaZ8Q3xFuL4V44s1dRdOAHJ1gd2HQVoswuU7GwQZmEm2hCwpkl3AdZ8XiLqs8F43Z-42FB55tCnqA9GWd5VsySZiWoeKLRkUgsSiz4Vb4lNxrmpctFFsKBtLcOSw0~hIzQSpOsodr6cxlkViwscjEtIMu6R8IZV~lr5NSwi1dWBtGhKzlMrkA7PXhTPoVf~Qs5Gaa2Pz1zxLVC6rJp1EyvTG0QW9SjLvIY574vBcnhtvRUtAc-6fSUyVAjYsrG0Yeg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal