Abstract
Locus control regions (LCRs) are operationally defined by their ability to enhance the expression of linked genes to physiological levels in a tissue-specific and copy number–dependent manner at ectopic chromatin sites. Although their composition and locations relative to their cognate genes are different, LCRs have been described in a broad spectrum of mammalian gene systems, suggesting that they play an important role in the control of eukaryotic gene expression. The discovery of the LCR in the β-globin locus and the characterization of LCRs in other loci reinforces the concept that developmental and cell lineage–specific regulation of gene expression relies not on gene-proximal elements such as promoters, enhancers, and silencers exclusively, but also on long-range interactions of variouscis regulatory elements and dynamic chromatin alterations.
Introduction
Locus control regions (LCRs) are operationally defined by their ability to enhance the expression of linked genes to physiological levels in a tissue-specific and copy number–dependent manner at ectopic chromatin sites. The components of an LCR commonly colocalize to sites of DNAse I hypersensitivity (HS) in the chromatin of expressing cells. The core determinants at individual HSs are composed of arrays of multiple ubiquitous and lineage-specific transcription factor–binding sites.
The LCR was first identified in the human β-globin locus.1 (For a review, see Stamatoyannopoulos and Grosveld,2 Fraser and Grosveld,3 and Li et al.4) Early studies showed that a 5-kilobase (kb) β-globin gene segment, including a 1.5-kb promoter region, was expressed in erythroleukemia cell lines, implying that this fragment contains all the regulatory elements necessary for proper expression. However, this fragment did not uniformly promote gene expression in transgenic mice.5-7 The gene was expressed in only a small proportion of transgenic mice, but expression was far below physiologically significant levels and was variable between lines. These findings suggested that a major regulatory element required for reproducible, high-level expression in vivo was missing in this construct. Clues regarding the nature of the missing element came from several observations. For example, in some forms of β-thalassemia the genes of the β-globin locus are intact but not expressed.8,9 A defect common to the loci underlying these conditions was a large deletion upstream of the β-like globin genes. This deletion results in a closed chromatin conformation spanning the whole locus and leads to suppression of gene expression.8,10 Thus, these data suggested that the deleted DNA segment contained an indispensable cis-acting regulatory element required for β-globin expression in vivo. The existence of such a regulatory element was also implied by the presence of developmentally stable, erythroid-specific HSs 6 to 20 kb 5′ to the ε-globin gene.11,12 Definitive evidence for the presence of the LCR came from transgenic mouse studies.1 Linkage of this region to a β-globin gene resulted in expression of the gene at a level comparable to the endogenous mouse β-globin genes in a position-independent, copy number–dependent manner. LCRs have been described in a broad spectrum of mammalian gene systems, suggesting that they play an important role in the control of eukaryotic gene expression.
Properties of LCRs
Transcriptional enhancer activity
The most prominent property of the LCRs is their strong, transcription-enhancing activity. The β-globin LCR is located 6 to 22 kb 5′ to the first (embryonic) globin gene in the locus (Figure1). It consists of 5 DNAse I–hypersensitive sites, 5′HSs 1 to 5. HSs 1 to 4 are formed only in erythroid cells, while 5′HS5 is found in multiple lineages of cells, but it is not constitutive.13 When the LCR is absent, transcription of the human β-globin gene is usually less than 1% of the endogenous murine β-globin mRNA in transgenic mice, if it is expressed at all.5-7 Inclusion of the LCR increases β-globin gene expression to a level comparable to that of the mouse β-globin genes in all transgenic animals, indicating that the LCR has strong enhancer activity.1 LCR enhancer activity is also significant at its endogenous location, as demonstrated by LCR-deletion experiments.14-16 These deletions in the native chromosomes of mouse or human cell lines severely reduce the expression of globin genes.
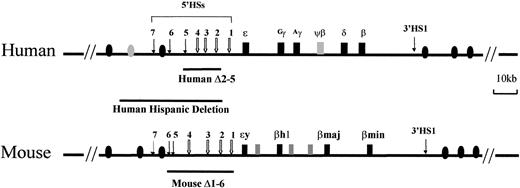
The human and mouse β-globin loci.
The human locus consists of 5 functional genes, indicated as dark boxes, arrayed in their order of developmental expression, 5′-ε-Gγ-Aγ-δ-β-3′. There are 2 developmental switches in globin chain synthesis coincident with changes in site and type of erythropoiesis. During primitive erythropoiesis, the ε-globin gene is expressed in the embryonic yolk sac. The first switch occurs at approximately 8 weeks' gestation; the ε-globin gene is silenced and the Gγ- andAγ-globin genes are expressed during definitive erythropoiesis in the fetal liver. The second switch occurs shortly after birth; the γ-globin genes are silenced and the β-globin gene and, to a lesser extent, the δ-globin gene are activated in the bone marrow. The HSs 5′HS1 through 5′HS7 are located −6, −11, −15, −18, −22, −28, and −35 kb relative to the ε-globin gene, respectively, and are indicated by arrows. 5′HS1 through 5′HS4 are erythroid specific, but 5′HS5 through 5′HS 7 are not. Another HS (3′HS1) is located 20 kb downstream of the β-globin gene; 3′HS1 is found only in erythroid cells. Boxes represent globin genes and ovals represent olfactor receptor genes; filled ones represent the productive genes and shaded ones the pseudogenes. The lines below the diagram of the locus indicate deletions of the LCR discussed in “In vivo function of LCRs.” The Hispanic deletion, which causes (γδβ)0 thalassemia in humans, extends an additional 20 kb 5′ of the LCR 5′HS5.
The human and mouse β-globin loci.
The human locus consists of 5 functional genes, indicated as dark boxes, arrayed in their order of developmental expression, 5′-ε-Gγ-Aγ-δ-β-3′. There are 2 developmental switches in globin chain synthesis coincident with changes in site and type of erythropoiesis. During primitive erythropoiesis, the ε-globin gene is expressed in the embryonic yolk sac. The first switch occurs at approximately 8 weeks' gestation; the ε-globin gene is silenced and the Gγ- andAγ-globin genes are expressed during definitive erythropoiesis in the fetal liver. The second switch occurs shortly after birth; the γ-globin genes are silenced and the β-globin gene and, to a lesser extent, the δ-globin gene are activated in the bone marrow. The HSs 5′HS1 through 5′HS7 are located −6, −11, −15, −18, −22, −28, and −35 kb relative to the ε-globin gene, respectively, and are indicated by arrows. 5′HS1 through 5′HS4 are erythroid specific, but 5′HS5 through 5′HS 7 are not. Another HS (3′HS1) is located 20 kb downstream of the β-globin gene; 3′HS1 is found only in erythroid cells. Boxes represent globin genes and ovals represent olfactor receptor genes; filled ones represent the productive genes and shaded ones the pseudogenes. The lines below the diagram of the locus indicate deletions of the LCR discussed in “In vivo function of LCRs.” The Hispanic deletion, which causes (γδβ)0 thalassemia in humans, extends an additional 20 kb 5′ of the LCR 5′HS5.
The enhancer activity of the β-globin LCR resides in 5′HS2, 3, and 4, but not in 5′HS1 or 5 (for a review, see Stamatoyannopoulos and Grosveld,2 Fraser and Grosveld,3 Li et al,4 and Hardison et al17). 5′HS2 behaves as a classical enhancer; that is, its activity can be detected in transient transfection assays. Enhancer activity in 5′HS3 or 4 can be detected only when they are integrated into chromatin (for a review, see Hardison et al17 and references therein). A requirement for chromosomal integration suggests that alteration of chromatin structure may be involved in propagating the enhancer activity of these 2 HSs. 5′HS5 functions as a chromatin insulator.18-20 The function of 5′HS1 remains to be defined.
The enhancer activity of the β-globin LCR is tissue specific; that is, the expression of globin genes is confined to erythroid cells when linked to the β-globin LCR.1,21 In addition, the LCR is able to enhance expression of linked heterogeneous nonglobin gene promoters in erythrocytes. However, when a nonglobin gene was coupled to the LCR, ectopic expression was observed in some transgenic mice.22 In these instances, although the LCR conferred erythroid-specific gene expression on the heterogeneous gene, the natural function of the linked promoter allowed expression outside the erythroid compartment. Thus, tissue-specific control of basal transcription may reside in the promoter, as is the case for the globin genes, whereas tissue-specific enhancement of gene expression may be a property of the LCR. These data suggest that tissue specificity is not really an intrinsic property of the LCR but depends on both the LCR and the promoter that it interacts with.
Central to understanding the enhancer functions of β-globin LCR is the identification of the transcription factors mediating enhancer activity. Enhancer activity of 5′HSs 2-4 resides in a 200-bp to 300-bp core, which contains an array of binding sites for ubiquitous and erythroid-specific transfactors. A conserved sequence within 5′HS2, TGCTGA(C/G)TCA(T/C), is critical for strong enhancer activity.23,24 This Maf recognition element (MARE) is bound by multiple homodimeric and heterodimeric transcription factors in vitro.25 These factors include Maf homodimers, heterodimers containing a Maf subunit and another bZIP protein (NF-E2, Nrf1, Nrf2, Bach1, Bach2), and heterodimers lacking a Maf subunit (AP1).26-31 NF-E2 is the major protein found in nuclear extracts from murine erythroleukemia (MEL) cells that binds the tandem MAREs of 5′HS2, and globin gene expression closely parallels the level of NF-E2 binding activity.32 The MEL cell line CB3, which lacks p45, is severely impaired in globin gene expression, and transcription can be rescued by expression of NF-E2.32,33The erythroid-specific transactivator p45/NF-E2 binds directly and specifically to 5′HS2 in erythroleukemia cells and mouse fetal liver. Chromatin immunoprecipitation (ChIP) assay showed that specific recovery of the 5′HS2 sequences was dependent upon the presence of p45 and intact MARE sites within 5′HS2.34 Investigation of the binding of the p45/p18 (MafK) heterodimer or other small Maf proteins within the globin locus showed that prior to induction of MEL cell differentiation, the LCR was occupied by small Maf proteins, and that during erythroid maturation, the NF-E2 complex was recruited to the LCR and the active globin promoters, even though the promoters do not contain MAREs. This differentiation-coupled recruitment of the NF-E2 complex correlates with a more than 100-fold increase in β-major globin transcription, but is not associated with a significant change in locus-wide histone H3 acetylation. Thus, the β-globin gene locus may exist in a constitutively open chromatin conformation before terminal differentiation, and the recruitment of the NF-E2 complex to the LCR and active promoters may be a rate-limiting step in the activation of β-globin gene expression.35 While the in vivo association of NF-E2 and HS2 of the βLCR is confirmed by ChIP assay, a knockout of p45 gene does not inhibit globin gene expression.36 The absence of phenotype in the p45 knockout mice is not due to the result of compensation by Nrf-2, a factor closely related to p45, as demonstrated by the study of a double knockout of p45 and Nrf-2, which also fails to interfere with expression of the α- and β-globin genes.37 These observations suggest an interchangeable function between members of the cap'n collar (CNC) subfamily of bZIP transcription factors.
LCR functions may affect the basic transcription machinery directly. RNA polymerase II (pol II), one of the essential components of the eukaryotic transcription apparatus, was found to be associated with the β-globin LCR in a p45/NF-E2–independent manner, whereas its recruitment to the promoter required p45/NF-E2. These data suggest that pol II accesses the LCR and p45/NF-E2 induces long-range transfer of pol II to the promoter, resulting in transcriptional activation.38
Copy number–dependent gene expression and chromatin domain-opening activity
Another property of the LCRs is their ability to confer position-independent, copy number–dependent expression on a linked gene.1 Copy number–dependent expression is widely considered to be indicative of open chromatin structure, that is, DNA that is accessible to transcription factors. Involvement of the β-globin LCR in creating open chromatin was suggested from analysis of β-thalassemia mutants with deletion of the LCR.8,10In the Hispanic form of β-thalassemia, an approximately 35-kb region upstream of 5′HS1 is deleted, but the remainder of the globin locus is intact. However, none of the globin genes is expressed. The deletion produces a closed chromatin conformation that spans the entire locus.10 Consistent with this, only the intact LCR (5′HS1-5) can provide position-independent chromatin-opening activity in single-copy transgenic mice carrying the entire β-globin locus.39 When one of the HSs was deleted from the LCR, expression of the β-globin genes appeared to be sensitive to the position of integration. In transgenic mice carrying single copies of small, recombinant 5′HS-globin gene constructs, only 5′HS3 is able to confer copy number–dependent gene expression. This observation led to the conclusion that 5′HS3 possesses the dominant chromatin-opening activity of the β-globin LCR.40 However, 5′HS3 chromatin-opening activity may not be dominant, since it appears to be dependent upon the constitution of the constructs.41Formation of hypersensitivity is a result of interaction of multiple ubiquitous and erythroid-specific transacting factors in the HS regions.42
Recent studies have established that the human CD2 LCR achieves position-independent expression in the T cells of transgenic mice by overcoming heterochromatin-mediated position effect variegation (PEV).43 Fluorescence in situ hybridization (FISH) was used to identify the sites of transgene integration in individual mouse lines and allowed a correlation between the type of position effects induced by such chromosomal locations and the DNA sequences required to overcome them. Transgenic mice carrying a CD2 minigene attached only to the 3′ CD2 transcriptional enhancer (the CD2 HSs 1 and 2) exhibited variegated expression when the transgene integrated in the centromere. In contrast, mice carrying a transgene with additional 3′ sequences (the CD2 HS3) showed no variegation even when the latter integrated in centromeric positions. This indicates that the CD2 HS3 functions in the establishment and/or maintenance of an open chromatin domain and that human CD2 LCR is able to overcome the gene repression imposed by constitutive centromeric heterochromatin.
In conclusion, the ability to confer copy number–dependent expression of a transgene is used to distinguish a DNA fragment functioning as an LCR rather than a transcriptional enhancer. This criterion has been employed in identification of all LCR or LCR-like elements.
Timing and origin of DNA replication
The mammalian genome is made up of defined zones that undergo DNA replication in a programmed manner during the S phase of the cell cycle. Studies of individual genes have demonstrated that there is a correlation between replication timing and gene expression.44,45 The human β-globin locus replicates late in most cell types, but replicates early in erythroid cells.46 47
Data generated from transgenic mice by FISH analysis mapped and characterized the replication zone surrounding the human globin locus on chromosome 11. These results showed that the β-globin LCR region (5′HSs 1-5) was sufficient for directing replication timing in a developmentally specific manner in vivo.48 The LCR (5′HSs 1-5) also plays a role in setting up regional erythroid-specific, open chromatin structure in transgenic mice, and this function is likely intertwined with the ability to direct early replication timing.49 Although early replication is generally correlated with gene expression, it has not been possible to decipher the cause-and-effect relationship between these 2 parameters.48 Other results using targeted deletion of the LCR (5′HSs 1-5) showed that early replication timing and an open chromatin structure do not, by themselves, guarantee high levels of globin transcription in erythroid cells.50 Therefore, an as yet undefined class of cis-acting elements may play a role in mediating control of replication timing, independent of transcription.
Many studies have emphasized the relationship between early replication and globin transcription in erythroid cells. However, these replication elements within the LCR also function in nonexpressing cell types. Thus, one of the major roles for replication timing control at the globin locus may be to set up late replication with its accompanying inactive chromatin structure in nonerythroid cells. In this manner, repression of background transcription may be achieved.48Perhaps this is accomplished by restricting the exposure of newly assembled nucleosomes to histone deacetylases, specifically during replication in late S phase.49 Recent evidence supporting this hypothesis suggests that HDAC2 is preferentially associated with late replication foci.51 Further data are required to determine whether the effects on replication are general features of LCRs and whether these effects influence transcription or are secondary to it.
Histone modification and heterochromatin
Despite numerous studies on the role of the LCR in controlling β-globin gene expression, the mechanism of long-range transactivation by the LCRs is poorly understood. Several models (including looping, tracking, linking, topologic alterations, and modification of proteins associated with chromatin) have been invoked to explain the functions of LCR.4 52-55 All the models, directly or indirectly, implicate the ability of LCRs to alter chromatin configuration and conformation.
The effects of LCRs on chromatin acetylation have been studied in different model systems. Function of the human growth hormone (hGH) LCR has been linked to specific patterns of core histone acetylation. The hGH locus consists of 5 genes expressed in either the pituitary or the placenta.56 This LCR consists of 5 HSs: 2 pituitary-specific (HSI, HSII), 1 placenta-specific (HSIV), and 2 shared (HSIII, HSV). In the pituitary, the LCR is encompassed in a somatotrope-specific domain of hyperacetylated chromatin that extends from the most 5′ LCR component to the hGH-N promoter. Further analysis shows that the hGH LCR, located 14.5 kb upstream from the hGH-N promoter, plays a critical, specific, and nonredundant role in facilitating promoter transacting factor binding and activation of hGH-N transcription. It also plays an essential role in establishing a 32-kb acetylated region that encompasses the entire hGH LCR contiguous with the hGH-N promoter. Separate positive elements in the LCR (HSI, HSII) for pituitary-expressed genes, or in gene-proximal sequences (P-elements) for placenta-expressed genes, activate their respective target genes by tissue-specific recruitment of different histone acetyltransferase activities, resulting in distinct patterns of acetylation across the locus.57 These data support a model for long-range gene activation via LCR-mediated targeting and extensive spreading of core histone acetylation.58
The functions of the LCR in the β-globin locus appear to be different from those of the hGH gene cluster. Deletion of 5′HS2-5 of the human β-globin LCR did not affect the general pattern of histone H4 acetylation of a β-globin locus transgene.59 Other studies reported that although deletion of the murine β-globin LCR decreased the rate of β-globin transcription, it did not alter the acetylation status of histone H3 or H4 within the promoter region.60 Thus, histone H3 or H4 acetylation at the β-globin promoter may be independent of LCR function.
NF-E2 is required for histone hyperacetylation at the adult β-globin promoter, but not at the LCR.38,61 Other data demonstrated that the β-globin LCR and transcriptionally active promoters were enriched in acetylated histones in fetal liver relative to fetal brain, whereas the inactive promoters were hypoacetylated. In contrast, the LCR and both active and inactive promoters were hyperacetylated in yolk sac. 5′HS2 was also hyperacetylated in murine ES cells, whereas β-globin promoters were hypoacetylated. Thus, the acetylation pattern varied at different developmental stages. Histone deacetylase inhibition selectively increased acetylation at a hypoacetylated promoter in fetal liver, suggesting that active deacetylation contributes to silencing of promoters. Therefore, dynamic histone acetylation and deacetylation activities may play an important role in the developmental control of β-globin gene expression.61
DNA methylation is important in mammalian development because it controls gene expression through chromatin closure and gene silencing. During development, gene loci expressed in a tissue-specific manner become selectively demethylated in the appropriate cell types by poorly understood processes. The LCRs may play a role in tissue-specific DNA demethylation. Studies of the methylation status of the LCR for the mouse T-cell receptor (TCR) α/δ locus support such a role. Tissue-specific functions of this LCR depend largely on 2 HSs, HS1 (T-cell receptor α enhancer) and HS1′. These HSs induce lymphoid organ–specific DNA demethylation in a region located 3.8 kb away, with little effect on intervening methylated DNA. Demethylation is impaired in mice with a germ line deletion of the HS1/HS1′ clusters. Using 5′-deletion mutants of a transgenic LCR reporter gene construct, HS1′ can act in the absence of HS1 to direct this tissue-specific DNA demethylation event. Therefore, elements of an LCR may control tissue-specific DNA methylation patterns both in transgenes and in native loci.62
In vivo function of LCRs
As discussed above, LCRs possess all the properties necessary for opening a chromosome domain and preventing heterochromatinization at ectopic sites. This property of the LCR most prominently distinguishes it from enhancers. Thus, a broadly accepted model for the major role of the β-globin LCR in vivo is to open and/or maintain a permissive chromatin conformation within the β-globin locus in erythroid cells, although enhancement of transcription is also an essential function. Surprisingly, when the entire mouse β-globin LCR (5′HS1-6) was deleted by homologous recombination, the formation of the general DNAse I-sensitivity associated with the β-globin locus domain was not affected; however, transcription of all β-like globin genes was strikingly reduced.14 63 These observations raise several questions regarding the real in vivo function of the β-globin LCR. Is this LCR simply another enhancer in the β-globin locus? If the β-globin LCR functions only as an enhancer within endogenous β-globin loci, how can this fact be reconciled with observations from transgenic mouse studies in which chromatin-opening activity is characteristic?
Understanding the in vivo function of the LCRs is associated with our knowledge of the process of gene activation. A prevailing model for gene activation is that it is a stepwise process. The first step is chromatin opening. Opening allows transacting transcription factors and cofactors to access chromatin and assemble a functional transcription apparatus. Genes in open chromatin domains are poised for expression. When protein activators are present, transcription commences and high-level gene expression is achieved. Chromatin opening is manifested by an increase in its sensitivity to DNAse I or other nucleases. General DNAse I sensitivity represents a level of sensitivity on the order of one magnitude greater than bulk chromatin. General sensitivity may stretch over regions of several hundred kilobases. Within these regions of general sensitivity, small regions (< 300 bp) of DNAse I sensitivity may be 2 orders of magnitude more sensitive than bulk chromatin; these regions are termed DNAse I-hypersensitive sites. Although increased DNAse I sensitivity may be due to improved accessibility of DNA packed in chromatin, DNAse I sensitivity indeed is an ambiguous indicative of chromatin structure. The precise molecular nature of the alterations underlying accessibility has not been delineated. Questions persist as to whether the changes occur at the 30-nm fiber level or at the nucleosome level and whether all histone tails in the general DNAse I sensitive regions are modified (acetylated, methylated, or phosphorylated) in the same fashion. Although general DNAse I sensitivity was detected in both normal and β-globin LCR knockout mice in erythroid cells, previous data do not indicate whether the general DNAse I sensitivity detected in LCR knockout mice and that detected in normal mice represent identical or different chromatin configurations.
Regardless of what the chromatin configuration may be, chromatin of the globin locus is more sensitive than bulk DNA in LCR knockout mice. In the absence of the LCR, an alternate pathway for establishment and maintenance of open chromatin must exist. Chromatin is not a structurally inert entity. Most likely it undergoes many dynamic conformational transitions that may be important in facilitating interactions between transacting factors and DNA. DNA probably unwraps from the edge of the nucleosome, since sites within nucleosomal DNA are transiently separated from histones with a probability of 1 in 103 to 105 moving from the periphery of the nucleosome toward the center.64,65 Thus, given the dynamic nature of this system, factors present at sufficient concentrations and having high affinities for naked DNA may be able to compete efficiently with histone proteins for binding, thereby ensuring significant loading of these proteins at their cognate DNA elements in chromatin. Moreover, some transacting factors, such as GATA-4, are able to bind to compacted chromatin and open up a local chromatin.66 Other transacting factors then attain an opportunity to access enhancer or promoter elements and further remodel chromatin by recruiting and targeting chromatin modifying and remodeling machinery. Since a large number of factor-binding sites are scattered throughout the β-globin locus, particularly at promoters, they are able to recruit various proteins and cofactors in the erythroid environment. Accumulation of a large amount of small, qualitative changes may finally lead to a major change in chromatin structure. Such a synergistic mechanism could result in an open chromatin at low level in the absence of the LCR. Synergistic mechanisms have been postulated for transcription activation via cooperation of multiple transactivators67and for heterochromatin formation in a mass action model.68 Based on this model, the LCR does not necessarily possess a specified chromatin opening activity.
The LCR chromatin-opening activity manifested in transgenic studies indeed results from the unique feature of the LCR that numerous binding sites clustered in the region induce an exponent (synergistic) effect on chromatin structure. Closed chromatin is considered the default status and is found in the vast majority of the chromosome. Thus, most transgenes are integrated in sites of closed chromatin. When transgene expression is detected, the chromatin region surrounding the transgene is invariably open. Although it is easy to surmise that chromatin opening is a prerequisite of gene transcription, this conclusion is not definitive. The 2 processes may be separated during in vitro assays, but more likely these 2 events are mutually interdependent in vivo. Thus, the chromatin-opening activity of the LCR manifested in transgenic studies must also function at the endogenous locus in vivo in a similar manner. Experimental description of chromatin opening indeed includes multiple distinctive states in chromatin conformation and configuration. Chromatin-opening activity has to be considered as an integrated but not necessarily linear event in gene activation.
Mechanisms of globin gene activation by the LCR
Clearly, the LCR has a role in enhancement of globin gene expression, although some uncertainty exists regarding the direct effects of the LCR on chromatin conformation. Analysis of LCR function at its endogenous location in cell lines suggests that it is limited to globin gene transcription activation,14 15 whereas transgenic experiments suggest that it is also necessary for the establishment and maintenance of an open β-globin chromatin domain. Regardless of whether the LCR functions in one or both of these processes, it does so over a long distance. Studies using both native loci and constructs in transgenic mice offer insights as to how the β-globin LCR accomplishes transcriptional activation. Four models of LCR function have been proposed: looping, tracking, facilitated tracking, and linking (Figure 2). Available data neither strongly support nor preclude any of them.
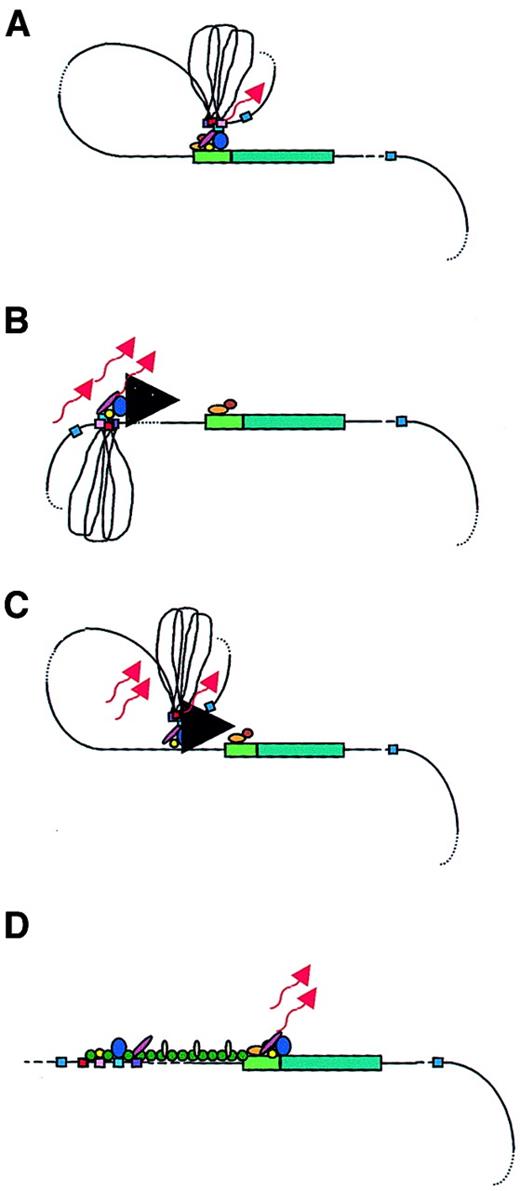
Models of LCR function.
A globin gene is denoted by a green rectangular box with the promoter region indicated in a lighter green. Transcription factors are shown as colored ovals and circles. The 4 erythroid-specific hypersensitive site cores (HSs) are indicated by small red boxes. Blue boxes are the positions of 5′HS5 and 3′HS1, representing likely insulator elements. The flanking DNA sequences of the HSs are depicted as loops between the HS cores. Transcripts are denoted by wavy arrows. (A) Looping model. Transcription factors bind to the LCR HSs and the gene promoter. The LCR directly interacts with the gene promoter by looping out the intervening DNA, thus forming an active transcription complex at the gene promoter. (B) Tracking model. Sequence-specific transcription factors bind to the LCR, forming a complex that tracks down the DNA sequence, as depicted by the large black arrowhead, until encountering transcription factors bound to the appropriate gene promoter, initiating high-level gene expression. (C) Facilitated tracking model. Aspects of both looping and tracking models are combined. Sequence-specific transcription factors bind the LCR; looping then occurs to deliver the bound transcription factors proximal to the gene promoter, followed by tracking, until they encounter transcription factors bound to the appropriate gene promoter. (D) Linking model. Sequential binding of transcription factors along the DNA directs changes in chromatin conformation and defines the transcriptional domain. The transcription factors are linked to one another from the LCR to the gene promoter by non–DNA-binding proteins and chromatin modifiers (shown as small colored circles).
Models of LCR function.
A globin gene is denoted by a green rectangular box with the promoter region indicated in a lighter green. Transcription factors are shown as colored ovals and circles. The 4 erythroid-specific hypersensitive site cores (HSs) are indicated by small red boxes. Blue boxes are the positions of 5′HS5 and 3′HS1, representing likely insulator elements. The flanking DNA sequences of the HSs are depicted as loops between the HS cores. Transcripts are denoted by wavy arrows. (A) Looping model. Transcription factors bind to the LCR HSs and the gene promoter. The LCR directly interacts with the gene promoter by looping out the intervening DNA, thus forming an active transcription complex at the gene promoter. (B) Tracking model. Sequence-specific transcription factors bind to the LCR, forming a complex that tracks down the DNA sequence, as depicted by the large black arrowhead, until encountering transcription factors bound to the appropriate gene promoter, initiating high-level gene expression. (C) Facilitated tracking model. Aspects of both looping and tracking models are combined. Sequence-specific transcription factors bind the LCR; looping then occurs to deliver the bound transcription factors proximal to the gene promoter, followed by tracking, until they encounter transcription factors bound to the appropriate gene promoter. (D) Linking model. Sequential binding of transcription factors along the DNA directs changes in chromatin conformation and defines the transcriptional domain. The transcription factors are linked to one another from the LCR to the gene promoter by non–DNA-binding proteins and chromatin modifiers (shown as small colored circles).
The looping model suggests that the 5′HSs of the β-globin LCR fold to form a holocomplex, with the HS core elements forming an active site that binds transcription factors and the core-flanking sequences constraining the holocomplex in the proper conformation (Figure 2A). This structure physically “loops,” so that the LCR comes in close proximity to the appropriate promoter. Close association with gene-proximal promoter and enhancer elements allows the delivery of LCR-bound transcription proteins and other coactivators that interact with the basal transcription apparatus, already bound at the promoter to form a stable transcription complex, thus enhancing globin gene expression.39,52,69,70 A variation of this model suggests that the LCR initially serves as a multiple element receptor that acts as a hub for factor binding to direct chromatin remodeling.71 Once chromatin-remodeling activity has been completed, the LCR directly interacts with downstream genes to facilitate their expression.
Several data support the looping model. Deletion of the 5′HS2 core abolished expression of the ε-, γ-, and β-globin genes.70 However, when the entire 5′HS2 region of conserved sequence similarity (core and flanking sequences) was removed, the ε-, γ- and β-globin genes were expressed in the correct temporal order, although the levels of each were decreased severalfold.72 These data suggested that, in the case of the core deletion, the 5′HS2 flanking regions were able to interact with the flanking sequences of the remaining intact 5′HSs to form the normal holocomplex conformation. Removal of only the 5′HS2 core, in effect, destroyed the active site of the holocomplex, resulting in a dominant-negative mutation that crippled LCR function. In contrast, when the entire 5′HS2 region was deleted, the remaining 5′HS sites were able to adapt an alternate holocomplex conformation with a slightly less effective active site consisting of the remaining 5′HS cores and constrained in form by the remaining 5′HS flanking sequences.72 Similar results were found with 5′HS3 core deletions versus complete deletion of 5′HS3.72-74
Further evidence supporting the notion of a holocomplex suggests that the LCR interacts with only 1 globin gene promoter at a time and that it may “flip-flop” between 2 or more promoters, depending on the stage of development.39,52 In this model, the LCR holocomplex is free to move from gene to gene. A parameter relevant to holocomplex function is the distance between the LCR and its target gene, which has been shown to affect the probability that these 2 elements will interact.52,75-77 This probability is constant for a gene at a specific stage of development. As development proceeds, the LCR has increasingly stable interactions with more distant globin genes, which is largely a function of the changing transcription factor milieu. Thus, it is mainly the availability of specific transcription factors and distance of a gene from the LCR that constrain the frequency of LCR-gene interactions during development.75-77
In the tracking, or scanning, model, erythroid-specific and ubiquitous transcription factors and cofactors bind recognition sequences in the LCR sequences, forming an activation complex that migrates, or tracks, linearly along the DNA helix of the locus (Figure2B).53,78 When this transcription complex encounters the basal transcription machinery located at the correct (according to the developmental stage) promoter, the complete transcriptional apparatus is assembled and transcription of that gene is initiated. If this model is valid, the expectation would be that some aberrant transcripts would arise from cryptic start sites along the locus. In fact, transcripts were detected across the LCR and intergenic regions in erythroid cells, but not in nonerythroid cells.53,79 However, these transcripts were nuclear-specific; they were not found in the cytoplasm, suggesting that they were not processed into mature messenger RNAs. The function of such intergenic transcription may be to deliver transcription complex proteins to the globin gene promoters via the tracking mechanism. Alternatively, it is possible that the function of these transcripts is to establish and maintain an open chromatin conformation permissive for gene transcription, although the persistence of DNAse I sensitivity following deletion of the LCR in cell lines argues against this role.14 15 Deacetylases and methylases within the complex may reorganize chromatin after the transcription complex activates transcription, possibly to limit activation to a particular developmental stage.
The facilitated-tracking model incorporates aspects of both the looping and tracking models (Figure 2C).78 An LCR bound-transcription factor and coactivator complex loops to contact downstream DNA in promoter-distal regions, where the transcription factor complex is released. This complex then tracks in small steps along the chromatin until it encounters the appropriate promoter with its associated bound proteins. A stable loop structure is established and gene expression proceeds.
The linking model proposes that chromatin facilitator proteins bound throughout the locus define the domain to be transcribed and mediate the sequential stage-specific binding of transcription factors (Figure2D).55 Non–DNA-binding facilitator proteins form a continuous protein chain from the LCR to the globin gene to be transcribed, linking proteins bound at a transcriptionally primed gene to one another.54 Support for this model comes from theDrosophila Chip protein complex.80 Chip protein complexes interact with transcription factors bound at a promoter region at a specific developmental time point. The Chip-tagged promoter is targeted for transcriptional activation. It was speculated that a homologous mammalian protein complex may act as the facilitating guide for transcription initiation, associating with transcription factors in the globin gene promoter regions and aiding gene activation.55 This Chip-like protein complex may allow transcriptional activation of one gene at a time, while simultaneously blocking transcription outside of the region, accounting for the developmental stage-specific expression of the β-like globin genes. The β-globin locus may have several transcription factor–bound promoter regions linked in a chainlike fashion. Chip-like proteins then dissociate and move to another promoter link to target that promoter for LCR interaction. Thus, globin gene switching proceeds.
LCRs in other systems
Several elements have been characterized in mammals (including humans, mice, rats, chickens, rabbits, sheep, and goats) that meet the criteria for LCR function (Table1). Several other elements have been identified that likely will be confirmed as LCRs, including one in the medaka fish tyrosinase gene.81-84 Structurally, these LCRs are composed of varying numbers of tissue-specific DNAse I–hypersensitive sites. The HSs of nonglobin LCRs have been extensively characterized and consist of a 150- to 300-bp central core containing a high density of transcription factor binding sites.85-87 Although the β-globin LCR consists of 5 HSs clustered on one contiguous piece of DNA, the sequences that embody a complete LCR do not have to be located together, whether upstream of, downstream of, or within the genes they control. Other LCRs are a collection of elements with different numbers of HSs spread over large distances. The relative simplicity of the β-globin LCR with regard to its single group of HSs may have contributed to its early discovery. Identification of LCRs in complex multigene loci, where the elements are interspersed among the genes, is a difficult task. Functionally, they all exhibit some or all of the properties associated with the β-globin LCR, most commonly the hallmark of copy number–dependent, site-of-integration–independent expression of their cognate loci or linked transgenes.
Mammalian LCR and LCR-like elements
| Human: |
| 1. β-globin locus1 |
| 2. Adenosine deaminase gene, human86 |
| 3. Apolipoprotein E/C-1 gene locus, human105 |
| 4. T cell receptor α/δ locus, human96 |
| 5. CD2 gene, human95 |
| 6. S100 β gene, human106 |
| 7. Growth hormone gene, human107 |
| 8. Apolipoprotein B gene, human108 |
| 9. β myosin heavy chain gene, human109 |
| 10. MHC class I HLA-B7 gene, human87 |
| 11. Immunoglobulin heavy chain locus, human110 |
| 12. Immunoglobulin C alpha 1 & 2 genes, human111 |
| 13. Keratin 18 gene, human112 |
| 14. MHC class I HLA G gene, human113 |
| 15. Complement component C4A & B genes, human114 |
| 16. Red and green visual pigment genes, human115 |
| 17. CD4 gene, human116 |
| 18. α-lactalbumin, human117 |
| 19. Desmin gene, human118 |
| 20. CYP19 (aromatase) gene, human119 |
| 21. c-fesproto-oncogene, human120 |
| Mouse: |
| 1. β-globin locus121 |
| 2. MHC Class II Ea gene, mouse122 |
| 3. Tyrosinase gene, mouse123 |
| 4. α-fetoprotein gene, mouse124 |
| 5. Immunoglobulin μ heavy chain, mouse125 |
| 6. CD34 gene, mouse126 |
| 7. Metallothionein II gene, mouse127 |
| 8. Kallikrein genes, mouse and rat128 |
| 9. Glycophorin gene, mouse129 |
| 10. γ1 heavy chain gene, mouse130 |
| 11. λ5-VpreB1 locus, mouse131 |
| 12. granzyme B gene, mouse132 |
| 13. T cell receptor γ locus, murine102 |
| 14. Interleukin-2 gene, mouse84 |
| Rat: |
| 1. Aldolase C gene, rat133 |
| 2. Whey acidic protein (WAP) gene, rat134 |
| 3. Kallikrein genes, mouse and rat128 |
| 4. LAP (C/EBP β) gene, rat135 |
| Other: |
| 1. β-globin locus, goat, rabbit136 137 |
| 2. β-lactoglobulin gene, ovine138 |
| Human: |
| 1. β-globin locus1 |
| 2. Adenosine deaminase gene, human86 |
| 3. Apolipoprotein E/C-1 gene locus, human105 |
| 4. T cell receptor α/δ locus, human96 |
| 5. CD2 gene, human95 |
| 6. S100 β gene, human106 |
| 7. Growth hormone gene, human107 |
| 8. Apolipoprotein B gene, human108 |
| 9. β myosin heavy chain gene, human109 |
| 10. MHC class I HLA-B7 gene, human87 |
| 11. Immunoglobulin heavy chain locus, human110 |
| 12. Immunoglobulin C alpha 1 & 2 genes, human111 |
| 13. Keratin 18 gene, human112 |
| 14. MHC class I HLA G gene, human113 |
| 15. Complement component C4A & B genes, human114 |
| 16. Red and green visual pigment genes, human115 |
| 17. CD4 gene, human116 |
| 18. α-lactalbumin, human117 |
| 19. Desmin gene, human118 |
| 20. CYP19 (aromatase) gene, human119 |
| 21. c-fesproto-oncogene, human120 |
| Mouse: |
| 1. β-globin locus121 |
| 2. MHC Class II Ea gene, mouse122 |
| 3. Tyrosinase gene, mouse123 |
| 4. α-fetoprotein gene, mouse124 |
| 5. Immunoglobulin μ heavy chain, mouse125 |
| 6. CD34 gene, mouse126 |
| 7. Metallothionein II gene, mouse127 |
| 8. Kallikrein genes, mouse and rat128 |
| 9. Glycophorin gene, mouse129 |
| 10. γ1 heavy chain gene, mouse130 |
| 11. λ5-VpreB1 locus, mouse131 |
| 12. granzyme B gene, mouse132 |
| 13. T cell receptor γ locus, murine102 |
| 14. Interleukin-2 gene, mouse84 |
| Rat: |
| 1. Aldolase C gene, rat133 |
| 2. Whey acidic protein (WAP) gene, rat134 |
| 3. Kallikrein genes, mouse and rat128 |
| 4. LAP (C/EBP β) gene, rat135 |
| Other: |
| 1. β-globin locus, goat, rabbit136 137 |
| 2. β-lactoglobulin gene, ovine138 |
Most of the data regarding LCR function have come from studies of the human and murine β-globin LCRs. Several insights have been gained with studies of the chicken LCR. Organization of the chicken LCR is similar to its human counterpart except that one of the LCR elements is located between the adult βA and embryonic ɛ genes (the βA/ɛ enhancer).88 This enhancer is able to confer site-independent expression to the chicken βA-globin gene in transgenic mice.89Chromatin unfolding of the chicken β-locus requires the presence of both the LCR and the promoter.90 Chicken HS4 demarcates the 5′ border of the locus, which functions as a powerful chromatin insulator.91 The insulating function of chicken HS4 is manifested by enhancer blocking activity and position-effect protection. These two activities are separable:92 the former is mediated by a transacting factor CTCF93 and the latter function may be achieved by highly efficient recruitment of histone acetyltransferase by the HS4 element.94
Some novel data regarding LCR function have come from studies of nonglobin LCRs. As previously discussed, the human CD2 LCR was shown to be essential for establishing an open chromatin configuration, even in the absence of enhancer activity.43,95 Thus, LCRs appear to operate by ensuring an open chromatin configuration. As discussed earlier, the T cell–specific TCR α/δ (TCRα) LCR has been implicated in tissue-specific DNA demethylation, an important role for LCRs, since DNA methylation may cause chromatin closure and gene silencing. Additional information regarding LCR function was obtained from studies of this LCR. The TCRα LCR consists of 8 HSs located downstream of the T-cell receptor (TCR) gene.96 It is a bifunctional element, regulating both the TCR gene and the adjacent, ubiquitously expressed Dad1 antiapoptosis gene. Two subregions of the TCRα LCR were identified: one that constituted a novel non–tissue-restricted chromatin-opening element and one an immediate upstream sequence comprising the 4 proximal HSs that restored tissue specificity to the downstream chromatin-opening element.97 The HSs of this tissue-specificity region map near 2 transcriptional silencers, the TCRα enhancer (HS1) and a region of unknown function; the region between the enhancer and the unknown HS appears to be responsible for the tissue specificity.98 The proximal tissue-specific element may insulate the TCR gene from the LCR in other tissues, without affecting the TCRα LCR-Dad1 interaction. The occurrence of activators and insulators in LCRs appears to be a common theme, suggesting that the interaction of these elements may modulate LCR function. In fact, the tissue-unrestricted HS element suppressed PEV of a linked transgene in a wide variety of tissues and was bound by several ubiquitously expressed transcription factors.99However, when the full-length LCR was present, tissue-specific binding of tissue-restricted proteins was observed, demonstrating that a widely active LCR element can interact synergistically with other LCR elements to produce tissue-specific LCR activity via differential protein binding.
Other results suggest that LCRs may, in some instances, activate gene expression through a mechanism that includes increased histone acetylation. A cassette derived from 4 HSs of the 3′ murine immunoglobulin heavy chain (IgH) locus LCR was linked to the c-myc gene.100 This LCR mediated a widespread increase in acetylation, not only within the promoter region of the c-myc gene, but also over substantial distances upstream and downstream of the transcription site. Studies of the hGH LCR described earlier suggest that LCRs may increase histone acetylation by targeted recruitment and subsequent spreading of histone acetyltransferase activity to encompass and activate remote target genes.101
Two elements of the T-cell receptor γ (TCRγ) locus, a 3′ enhancer (3′E (Cγ1)) and a region called HsA located between the Vγ5 and Vγ2 genes, constitute an LCR.102 HsA alone supported position-independent transcription in mature, but not immature, T cells; 3′E (Cγ1) alone supported position-dependent expression in both immature and mature cells. Copy number–dependent, position-independent expression was obtained at both stages of development, suggesting that HsA provides chromatin-opening activity. In addition, HsA was required for rearrangement of transgenic recombination substrates, an essential component of TCR loci.
Another interesting variant of the LCR theme is the human keratin-18 (K18) gene, which contains a 323-bp fragment that confers position-independent, copy number–dependent expression upon heterologous transgenes.103 This fragment is composed primarily of an Alu repetitive element that was partly responsible for the protective effects of sequences flanking K18, perhaps through its pol III transcriptional potential and inhibition of transcriptional interference from neighboring genes. Thus, Alu elements may function as regulatory domains within LCRs.
A pair of DNAse I hypersensitive sites (site II) of the murine γ1 heavy-chain gene lie approximately 2 kb 3′ of the γ1 promoter and exon 1 and just 5′ of the γ1 switch region. Site II functions as an LCR, conferring insertion site independence and copy number dependence on linked transgene expression, the hallmark of LCRs.104Messenger RNA is induced from γ1 transgenes lacking site II by interleukin 4 (IL-4) and by CD40 ligation (CD40 ligand–CD40 interaction). However, in the absence of site II, the induction of transgenic RNA expression by CD40 ligation was greater than expected, suggesting that the elements within site II also participate in negative regulation of the number of germ line transcripts after CD40 ligation, an effect opposite to the enhancement of transcription observed with most LCRs.
Conclusions
LCRs have been identified in various loci of vertebrates. While their composition and location relative to their cognate genes are different, they share the common property of maintaining physiological levels of gene expression, either at their natural position or at ectopic sites. This feature highlights the complexity of gene regulation. Developmental and cell lineage–specific regulation of gene expression relies not upon gene-proximal elements such as promoters, enhancers, and silencers exclusively, but also upon long-range interactions of various cis regulatory elements and dynamic chromatin alterations. The discovery of the LCR in the β-globin locus and the characterization of LCRs in other loci reinforces the need to study in vivo transcriptional regulation in the context of whole loci, so that essential regulatory elements are not excluded or overlooked.
Prepublished online as Blood First Edition Paper, June 21, 2002; DOI 10.1182/blood-2002-04-1104.
Supported by National Institutes of Health grants DK53510, HL67336, HL20899, DK61805, and DK61804 and a Faculty Scholar Award from the Madison and Lila Self Graduate Fellowship awarded to K.R.P.
References
Author notes
George Stamatoyannopoulos, Department of Genome Sciences, University of Washington, Box 357730,1705 NE Pacific St, Health Sciences K-357, Seattle WA 98195; e-mail:gstam@u.washington.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal