The transcription factor C/EBPα is crucial for differentiation of mature granulocytes. Recently, differentCEBPA gene mutations likely to induce differentiation arrest have been described in nearly 10% of patients with acute myeloid leukemia (AML). In the present study, we retrospectively analyzed the prognostic significance of CEBPA mutations in 135 AML patients (French-American-British [FAB]-M3 excluded). All patients were prospectively enrolled between 1990 and 1996 in a multicenter trial of the ALFA (Acute Leukemia French Association) Group (median age 45 years, median follow-up 5.7 years). Mutations were assessed using direct sequencing of the CEBPA gene. Twenty-two mutations were found in 15 (11%) of 135 patients tested. Twelve patients had at least one mutation located in the N-terminal part of the protein leading to the lack of expression of the full-length C/EBPα protein. CEBPA mutations were present only in patients belonging to the intermediate cytogenetic risk subgroup and associated with the FAB-M1 subtype (P = .02). FLT3 internal tandem duplication (ITD) was found in 5 of 15 CEBPA-mutated as compared with 30 of 119 CEBPA-nonmutated cases tested (P = .54). Presence of CEBPA mutations was identified as an independent good prognosis factor for outcome even after adjustment on cytogenetics and FLT3 status (estimated 5-year overall survival 53% vs 25%, P = .04).FLT3-ITD appeared to act as a major bad prognosis factor in patients with CEBPA-mutated AML. We thus propose a risk classification that includes in the favorable subgroup all patients from the intermediate subgroup displaying CEBPA mutations when not associated with FLT3-ITD.
Introduction
In acute myeloid leukemia (AML), genetic abnormalities frequently occur in transcriptional control elements, leading to uncontrolled proliferation and/or differentiation arrest. Transcription factors belonging to the CCAAT/enhancer binding protein (C/EBP) family have been demonstrated to be involved in the balance between cell proliferation and mitotic growth arrest during terminal differentiation.1,2 In the hematopoietic system, one member of this family, C/EBPα, is exclusively expressed in myelomonocytic cells and is essential for commitment to the granulocytic lineage and differentiation of mature neutrophils.3 C/EBPα possesses a bipartite DNA-binding domain (bZIP domain) composed of a positively charged basic region that contacts the DNA and a leucine zipper in the C terminus that mediates dimerization. The less-conserved N terminus contains 2 regulatory and transactivation domains, TAD1 and TAD2, both included in the full-length 42-kDa protein. A shorter 30-kDa protein (initial codon starting at codon 120) contains only the TAD2 domain. C/EBPαis specifically up-regulated during the granulocytic differentiation, and its conditional expression is sufficient to trigger neutrophilic differentiation in bipotential precursors. In this process, the negative regulation of c-Myc by C/EBPα is a critical event for allowing early precursors to enter the granulocytic differentiation pathway.4 In addition, C/EBPαtranscriptionally activates the promoters of the myeloid-specific receptors for macrophage colony-stimulating factor (M-CSF), granulocyte colony-stimulating factor (G-CSF), and granulocyte-macrophage colony-stimulating factor (GM-CSF) in myeloid cells. Finally, C/EBPα-deficient mice exhibit a blockage in the granulocytic differentiation at the myeloblast level with a complete absence of mature granulocytes occurring early in the fetal development, whereas other blood cell types are present in normal proportions.5
Recently, mutations of the gene coding for C/EBPα leading to inactivation of its transcriptional potential have been reported in hematologic malignancies with a particular incidence of 7% to 10% in AML cells.6-8 In addition, several silent CEBPApolymorphisms have been described, especially a frequent 836T>G transversion.8 To date, there is, however, no clinical study reporting the prognostic impact of CEBPA mutations in patients treated for AML.
In the present study, we evaluated the clinical features and the prognostic significance associated with CEBPA mutations in 135 patients with AML, all prospectively enrolled in a common prospective randomized trial. We confirm the prevalence of these mutations in an adult AML population with a higher incidence in patients from the intermediate risk cytogenetic subgroup. We also report a clear positive impact of these CEBPA mutations on patient outcome, leading us to propose taking into account the C/EBPα status for AML risk category definition.
Patients, materials, and methods
Patients and samples
A total of 135 peripheral blood or bone marrow samples from 135 AML adult patients from 3 institutions were retrospectively analyzed for the presence of CEBPA mutations by sequencing on genomic DNA. All these patients (aged 15-65 years) had been diagnosed with de novo AML (ie, without any antecedent myelodysplastic or myeloproliferative syndrome or prior history of exposure to radiation, chemotherapy, or carcinogens) and enrolled in the Acute Leukemia French Association (ALFA)–9000 prospective trial between April 1990 and February 1996 (median follow-up 5.7 years). Patients with AML-M3 were not considered for inclusion in this trial. Cytogenetic features were classified according to the British Medical Research Council (MRC) cytogenetic risk classification: (1) favorable: core binding factor (CBF) AML including inv(16) and t(8;21); (2) adverse: −7, −5, del(5q), 3q abnormality, and complex karyotype defined by the presence of a clone with at least 5 unrelated cytogenetic abnormalities; and (3) intermediate: normal karyotype and all other abnormalities including +8, +21, +22, del(7q), del(9q), and 11q23 abnormality.9 In addition, all the 135 patients except one had been retrospectively analyzed for the presence of FLT3 internal tandem duplication (FLT3-ITD) as described10 and classified as FLT3-wild type (FLT3-wt) orFLT3-ITD.
Treatments
Patients were randomized at diagnosis to receive 1 of the 3 following induction regimens: arm 1 (high-dose daunorubicin 3 + 7 induction) consisted of 80 mg/m2/d daunorubicin for 3 days (days 1-3) and 200 mg/m2/d cytarabine as continuous infusion for 7 days (days 1-7); arm 2 (double induction) consisted of the same regimen, followed at day 20 by a second induction course comprising 12 mg/m2/d mitoxantrone for 2 days (days 20 and 21) and 500 mg/m2/12-h cytarabine as 3-hour intravenous bolus infusion for 3 days (days 20-22); arm 3 (timed-sequential induction) consisted of 80 mg/m2/d daunorubicin for 3 days (days 1-3) and 500 mg/m2/d cytarabine as continuous infusion for 3 days (day 1-3), followed at day 8 by 12 mg/m2/d mitoxantrone for 2 days (days 8 and 9) and 500 mg/m2/12-h cytarabine as 3-hour intravenous bolus infusion for 3 days (days 8-10). Salvage therapy consisted of 3000 mg/m2/12-h cytarabine as 3-hour intravenous bolus infusion for 6 days (days 1-6) and 100 mg/m2/d amsacrine for 3 days (days 7-9). With the exception of 14 younger patients with an HLA-identical sibling who were considered by center investigators as eligible for allogeneic bone marrow transplantation in first complete remission (CR), all other CR patients then received 2 courses of consolidation. The first consolidation course was administered in outpatients and consisted of 90 mg/m2/d amsacrine for 1 day (day 1) and 60 mg/m2/12-h subcutaneous cytarabine for 5 days (days 1-5). The second consolidation course was an intensive timed-sequential etoposide, mitoxantrone, aracytine (EMA) course administered in hospitalized patients and comprising 12 mg/m2/d mitoxantrone for 3 days (days 1-3) with 500 mg/m2/d cytarabine as continuous infusion for 3 days (days 1-3), followed at day 8 by 200 mg/m2/d etoposide for 3 days (days 8-10) with 500 mg/m2/12-h cytarabine as continuous infusion for 3 days (days 8-10). Patients aged more than 50 years received an attenuated-dose EMA course with half doses of cytarabine and etoposide. All patients with AML-M4 and AML-M5 subtypes or with a white blood count (WBC) more than 100 × 109/L were also randomized at diagnosis to receive central nervous system prophylaxis (6 intrathecal injections of 15 mg methotrexate, 30 mg/m2 cytarabine, and 4 mg dexamethasone, followed at the end of the consolidation phase by a 18 Gy cranial irradiation) or not. Preliminary results of the ALFA-9000 trial have been presented.11
CEBPA mutation detection
The CEBPA mutation detection was performed by direct sequencing. According to Pabst et al,7 we used 2 overlapping primer pairs (PP1 and PP2) to amplify the entire coding region of the human CEBPA gene. Four additive primer pairs, PP3, PP4, PP5, and PP6, were used in cases of abnormal or ambiguous results. Polymerase chain reaction (PCR) was carried out in a final volume of 50 μL containing genomic DNA (100 ng), Tris(tris(hydroxymethyl)aminomethane)–HCl (40 mmol, pH 9.3), 5% dimethyl sulfoxide (DMSO), Triton X-100 (0.1%), KOAc (85 mmol), Mg(Oac)2 (1 mmol), GC melt (supplied by the manufacturer) (1 mol), 0.3 μmol of each primer, nucleotides (200 μmol of each deoxyribonucleoside triphosphate [dNTP]), and Tth DNA polymerase (0.5 units; Clontech, Palo Alto, CA). PCR conditions were as follows: 94°C for 5 minutes, 35 cycles at 94°C for 1 minute, 58°C for 1 minute, 72°C for 1 minute and 30 seconds, and then a final step for 10 minutes at 72°C. PCR products were loaded on ethidium bromide staining agarose gel, purified (Qiagen, Valencia, CA), and directly sequenced using ABIprism BigDye Terminators and AmpliTaq FS (Perkin Elmer, Foster City, CA). Abnormal sequencing results were controlled by 3 separate experiments. In cases with 2 different mutations, we amplified the coding sequence with the PP7 primer pairs corresponding to the PP1 forward primers and PP2 reverse primer, respectively. After purification, the PCR product was cloned in PCR II plasmids, and 10 clones were sequenced in each patient.
Statistical analysis
Response to initial therapy was evaluated after induction or after induction and salvage. Complete remission (CR) was defined according to the National Cancer Institute (NCI) criteria.12 Disease-free survival (DFS) was calculated as survival without relapse or death from the date of first CR. Event-free survival (EFS) was calculated from the date of up-front randomization until the date of CR induction failure, first relapse, or death in CR. Patient characteristics and CR rate comparisons were performed using the Fisher exact test for binary variables and the Mann-Whitney test for continuous variables. Data on treatment failure were estimated by the Kaplan-Meier method13 and compared using the log-rank test.14 Survival comparisons were adjusted for covariates using the Cox model15 and tested by the likelihood ratio test. For all outcome estimations, the 14 patients who received an allogeneic transplantation in first CR were censored at transplantation time. This was done to not introduce biases related to a putative better antileukemic effect and a higher treatment-related mortality associated with allogeneic transplantation. P < .05 was considered to indicate statistical significance. All calculations were performed using the STATA software, version 7.0 (Stata, College Station, TX).
Results
Incidence of CEBPA mutations
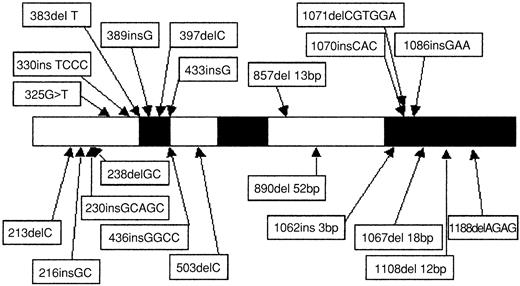
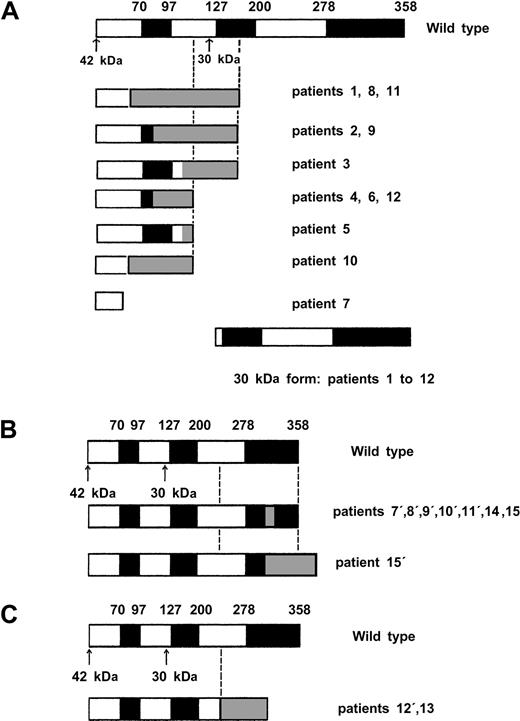
Direct sequencing showed 22 CEBPA mutations in 15 (11%) of 135 AML patients (Table 1). In addition, the 836T>G polymorphism was observed in 24 patients (17%), and 1 patient presented a 719C>T (H191H) silent mutation. Twelve of the 22 CEBPA mutations (Table 1, patients 1 to 12) were located in the N-terminal part of the protein (Figure1). As indicated in Figure2A, all these mutations induced a stop codon respectively at codon 59 (1 case), 106 (1 case), 107 (2 cases), 108 (2 cases), 159 (4 cases), 160 (1 case), and 161 (1 case). Interestingly, these 12 mutations abolished the expression of the full-length 42-kDa protein, with a normal expression of the 30-kDa protein. Eight of the 10 remaining mutations (Table 1, patients 7 to 12 and patient 15) are located in the bZip domain (Figure 1). They included duplication of Lys312 in 2 cases, addition of His at codon 307, addition of Leu at codon 304, deletion of Val-Glu307-308 with a substitution of Asn306Lys, deletion of 4 bps (1188-1191) inducing a 383 stop codon, and 2 deletions of 4 and 6 AA (Figure 2B). The 2 remaining mutations (Table 1, patients 12 and 13) were 2 large deletions of 13 and 52 bps inducing a stop codon at codon 313 and 310, respectively (Figure 2C). Overall, 7 of the 15 patients (patients 7 to 12 and patient 15) had 2 different mutations, and 1 additional patient (patient 14) had 1 in-frame deletion associated with the absence of the normal allele. The 2 alleles are implicated in all these 7 patients except one (patient 8). Finally, sequencing was performed at diagnosis and after CR achievement in 2 cases, and no mutation was detected in CR.
Summary of CEBPA mutations detected in patient samples
| Patient no. . | Age, y . | FAB . | Base pair change . | Amino acid change . |
|---|---|---|---|---|
| 1 | 46 | 1 | 213del | S18fsX159 |
| 2 | 63 | 1 | 397del | Q83fsX159 |
| 3 | 23 | 2 | 503del | A118fsX159 |
| 4 | 54 | 1 | 4-bp duplication of nt 436-439 | P97fsX108 |
| 5 | 64 | 4 | 330-331ins4bp | S61fsX108 |
| 6 | 35 | 5 | 389-390insG | D80fsX107 |
| 7 | 25 | 4 | 325G>T | Stop at Glu59 |
| 7′ | — | — | 1067-1084del | In-frame deletion of Arg-Asn-Val-Glu-Thr-Gln |
| 8* | 51 | 4 | 217-218ins | P20fsX160 |
| 8′ | — | — | 1062-1063ins3bp | In-frame addition of Leu304 |
| 9 | 42 | 1 | 383del | L78fsX159 |
| 9′ | — | — | 3-bp duplication of nt 1086-1088 | In-frame duplication of Lys312 |
| 10 | 39 | 1 | 238-239del | A30fsX106 |
| 10′ | — | — | 3-bp duplication of nt 1086-1088 | In-frame duplication of Lys312 |
| 11 | 36 | 1 | 5-bp duplication of nt 230-234 | S27fsX161 |
| 11′ | — | — | 1071-1076del | In-frame deletion of Val-Glu, Asn306Lys |
| 12 | 57 | 2 | 433-434insG | V95fsX107 |
| 12′ | — | — | 857-869del | H236fsX313 |
| 13 | 50 | 5 | 890-941del | P247fsX310 |
| 14† | 44 | 1 | 1108-1119del | In-frame deletion of Asp-Asn-Asp-Arg |
| 15 | 58 | 1 | 1070-1071ins3bp | In-frame addition of His307 |
| 15′ | — | — | 1188-1191del | E347fsX383 |
| Patient no. . | Age, y . | FAB . | Base pair change . | Amino acid change . |
|---|---|---|---|---|
| 1 | 46 | 1 | 213del | S18fsX159 |
| 2 | 63 | 1 | 397del | Q83fsX159 |
| 3 | 23 | 2 | 503del | A118fsX159 |
| 4 | 54 | 1 | 4-bp duplication of nt 436-439 | P97fsX108 |
| 5 | 64 | 4 | 330-331ins4bp | S61fsX108 |
| 6 | 35 | 5 | 389-390insG | D80fsX107 |
| 7 | 25 | 4 | 325G>T | Stop at Glu59 |
| 7′ | — | — | 1067-1084del | In-frame deletion of Arg-Asn-Val-Glu-Thr-Gln |
| 8* | 51 | 4 | 217-218ins | P20fsX160 |
| 8′ | — | — | 1062-1063ins3bp | In-frame addition of Leu304 |
| 9 | 42 | 1 | 383del | L78fsX159 |
| 9′ | — | — | 3-bp duplication of nt 1086-1088 | In-frame duplication of Lys312 |
| 10 | 39 | 1 | 238-239del | A30fsX106 |
| 10′ | — | — | 3-bp duplication of nt 1086-1088 | In-frame duplication of Lys312 |
| 11 | 36 | 1 | 5-bp duplication of nt 230-234 | S27fsX161 |
| 11′ | — | — | 1071-1076del | In-frame deletion of Val-Glu, Asn306Lys |
| 12 | 57 | 2 | 433-434insG | V95fsX107 |
| 12′ | — | — | 857-869del | H236fsX313 |
| 13 | 50 | 5 | 890-941del | P247fsX310 |
| 14† | 44 | 1 | 1108-1119del | In-frame deletion of Asp-Asn-Asp-Arg |
| 15 | 58 | 1 | 1070-1071ins3bp | In-frame addition of His307 |
| 15′ | — | — | 1188-1191del | E347fsX383 |
afsXb indicates a frameshift mutation of the amino acid at position “a” inducing a stop codon at position “b.”
Patient 8 had 2 different mutations located on the same allele.
Patient 14 had homozygous deletion.
Schematic representation and location of the 22
CEBPA mutations. Black boxes represent functional domains (TAD1, TAD2, and bZIP, respectively). The 1086insGAA mutation was found in 2 patients (patients 9 and 10).
Schematic representation and location of the 22
CEBPA mutations. Black boxes represent functional domains (TAD1, TAD2, and bZIP, respectively). The 1086insGAA mutation was found in 2 patients (patients 9 and 10).
Diagrammatic representation of proteins encoded by wild-type and mutant
CEBPA alleles. White and black boxes represent wild-type CEBPA sequence and gray boxes a shift of the reading frame encoding a novel peptide until formation of stop codon. The black boxes shows functional domains (TAD1, TAD2, bZIP), and the arrows show the 2 ATG initiation codon. (A) Proteins encoded by the 12 N-terminal mutants. (B) Proteins encoded by the 8 C-terminal mutants. Notably, mutation in patient 8′ affected only the 30-kDa form because of the presence of a N-terminal mutation (patient 8, panel A) on the same allele inducing an alteration of the 42-kDa protein. (C) Proteins encoded by the 2 middle large deletion mutants.
Diagrammatic representation of proteins encoded by wild-type and mutant
CEBPA alleles. White and black boxes represent wild-type CEBPA sequence and gray boxes a shift of the reading frame encoding a novel peptide until formation of stop codon. The black boxes shows functional domains (TAD1, TAD2, bZIP), and the arrows show the 2 ATG initiation codon. (A) Proteins encoded by the 12 N-terminal mutants. (B) Proteins encoded by the 8 C-terminal mutants. Notably, mutation in patient 8′ affected only the 30-kDa form because of the presence of a N-terminal mutation (patient 8, panel A) on the same allele inducing an alteration of the 42-kDa protein. (C) Proteins encoded by the 2 middle large deletion mutants.
As indicated in Table 2, there were no significant differences in age, sex ratio, and WBC at diagnosis betweenCEBPA-mutated and CEBPA-nonmutated patients, butCEBPA mutations were significantly associated with the M1 subtype of the French-American-British (FAB) classification (P = .02 by the Fisher exact test). Patients with or without CEBPA mutations were equally distributed among the 3 randomization arms.
Patient characteristics (N = 135 patients)
| . | Total . | CEBPA+ . | CEBPA− . | P . |
|---|---|---|---|---|
| No. of patients | 135 | 15 | 120 | |
| Median age, y (range) | 45 (15-64) | 45 (22-63) | 45 (15-64) | .59* |
| Sex (M/F) | 71/64 | 10/5 | 61/59 | .28† |
| WBC, ×109/L (range) | 14 (0.6-400) | 20 (2.2-170) | 13 (0.6-400) | .68* |
| FAB subtypes | ||||
| M0 | 1 | 0 | 1 | |
| M1 | 33 | 8 | 25 | .02‡ |
| M2 | 34 | 2 | 32 | |
| M4 | 19 | 3 | 16 | |
| M5 | 32 | 2 | 30 | |
| M6 | 5 | 0 | 5 | |
| M7 | 4 | 0 | 4 | |
| Unclassified | 7 | 0 | 7 | |
| Cytogenetics (MRC classification) | .062-153 | |||
| Favorable | 15 | 0 | 15 | |
| t(8;21) | 9 | 0 | 9 | |
| Inv(16) | 6 | 0 | 6 | |
| Intermediate | 91 | 15 | 76 | |
| Adverse | 14 | 0 | 14 | |
| Undetermined | 15 | 0 | 15 | |
| FLT3status | .54† | |||
| Wild type | 99 | 10 | 89 | |
| ITD | 35 | 5 | 30 | |
| Induction arm | .79† | |||
| Standard 3 + 7 | 45 | 6 | 39 | |
| Double induction | 47 | 4 | 43 | |
| Timed-sequential induction | 43 | 5 | 38 |
| . | Total . | CEBPA+ . | CEBPA− . | P . |
|---|---|---|---|---|
| No. of patients | 135 | 15 | 120 | |
| Median age, y (range) | 45 (15-64) | 45 (22-63) | 45 (15-64) | .59* |
| Sex (M/F) | 71/64 | 10/5 | 61/59 | .28† |
| WBC, ×109/L (range) | 14 (0.6-400) | 20 (2.2-170) | 13 (0.6-400) | .68* |
| FAB subtypes | ||||
| M0 | 1 | 0 | 1 | |
| M1 | 33 | 8 | 25 | .02‡ |
| M2 | 34 | 2 | 32 | |
| M4 | 19 | 3 | 16 | |
| M5 | 32 | 2 | 30 | |
| M6 | 5 | 0 | 5 | |
| M7 | 4 | 0 | 4 | |
| Unclassified | 7 | 0 | 7 | |
| Cytogenetics (MRC classification) | .062-153 | |||
| Favorable | 15 | 0 | 15 | |
| t(8;21) | 9 | 0 | 9 | |
| Inv(16) | 6 | 0 | 6 | |
| Intermediate | 91 | 15 | 76 | |
| Adverse | 14 | 0 | 14 | |
| Undetermined | 15 | 0 | 15 | |
| FLT3status | .54† | |||
| Wild type | 99 | 10 | 89 | |
| ITD | 35 | 5 | 30 | |
| Induction arm | .79† | |||
| Standard 3 + 7 | 45 | 6 | 39 | |
| Double induction | 47 | 4 | 43 | |
| Timed-sequential induction | 43 | 5 | 38 |
By the Mann-Whitney test.
By the Fisher exact test.
By the Fisher exact test comparing M1 subtype versus all other FAB subtypes.
By the Fisher exact test performed on the 4-subgroup repartition, including favorable, intermediate, adverse, and undetermined cytogenetics.
Outcome of patients with CEBPA mutations
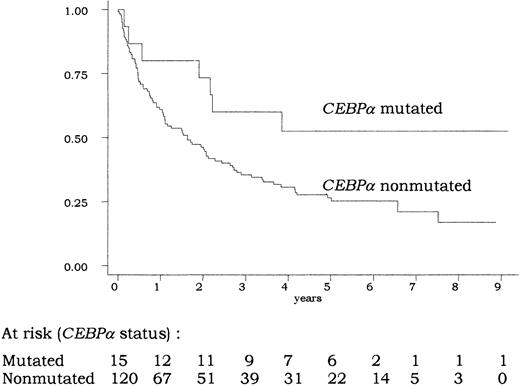
A total of 106 (79%) of the 135 patients achieved CR. In univariate analysis, the CR rate was not significantly different in patients with or without CEBPA mutations (87% vs 78%;P = .74 by the Fisher exact test). As indicated in Figure3, overall survival was longer in patients with CEBPA-mutated AML compared with those withCEBPA-nonmutated AML. At 5 years, estimated overall survival was 53% (95% confidence interval [CI], 25%-74%) in the former group and 25% (95% CI, 17%-34%) in the latter group (P = .04 by the log-rank test). This good prognosis associated with CEBPA mutations was also observed in terms of EFS and DFS (P = .04 and .05, respectively, by the log-rank test). In the CEBPA-nonmutated group, no difference in outcome was observed between patients presenting the 836T>G polymorphism or not (not shown).
Overall survival according to
CEBPA status. There was a significant difference in overall survival between patients with CEBPA-mutated and -nonmutated AML (P = .04 by the log-rank test). The 14 patients allografted in CR1 were censored at transplantation time.
Overall survival according to
CEBPA status. There was a significant difference in overall survival between patients with CEBPA-mutated and -nonmutated AML (P = .04 by the log-rank test). The 14 patients allografted in CR1 were censored at transplantation time.
Interaction with cytogenetics
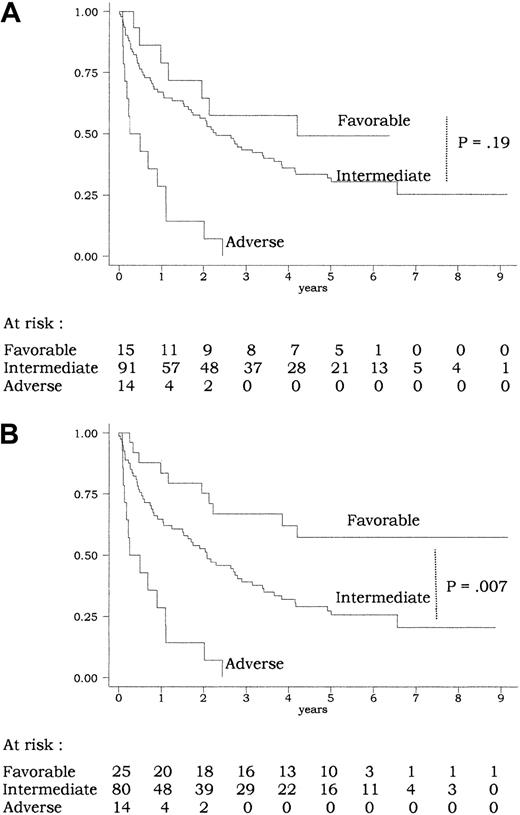
A total of 120 of the 135 patients had informative cytogenetics (Table 2). CEBPA mutations were only observed in patients from the intermediate MRC cytogenetic risk subgroup (incidence ofCEBPA mutations, 16% in the intermediate subgroup vs 0% in the favorable, adverse, and undetermined subgroups;P = .06 by the Fisher exact test). In univariate analysis, this MRC classification had a strong prognostic value for CR achievement (CR rate, 15 of 15, 74 of 91, 6 of 14, and 12 of 15 in the favorable, intermediate, adverse, and undetermined subgroup, respectively; P = .002 by the Fisher exact test) as well as for outcome (P < .001 for overall survival and EFS andP = .02 for DFS by the log-rank test). Overall survival according to the 3 subgroups of the MRC cytogenetic risk classification (favorable, intermediate, and adverse) is shown on Figure4A. However, this classification did not allow significant discrimination of patients from the favorable subgroup (with CBF-AML) from those from the intermediate subgroup (P = .32, .83, and .19 for EFS, DFS, and overall survival, respectively, by the log-rank test). This may be related to the relatively limited number of patients included in the present study and/or to the fact that survival of patients withCEBPA-mutated AML (most from the intermediate subgroup) appeared to be as good as survival of patients with CBF-AML from the favorable subgroup (Figures 3 and 4A).
Overall survival according to risk classifications.
(A) MRC classification (Table 4): using the MRC cytogenetic classification, there was a significant difference in overall survival between the 3 subgroups of patients (P < .001 by the log-rank test). No significant difference was, however, observed between the favorable and intermediate subgroups (P = .19 by the log-rank test). (B) Current classification (Table 4): using the current classification, there was a significant difference in overall survival between the 3 subgroups of patients (P < .001 by the log-rank test). Here, a significant difference was observed between the favorable and intermediate groups (P = .007 by the log-rank test). The 11 patients allografted in CR1 were censored at transplantation time.
Overall survival according to risk classifications.
(A) MRC classification (Table 4): using the MRC cytogenetic classification, there was a significant difference in overall survival between the 3 subgroups of patients (P < .001 by the log-rank test). No significant difference was, however, observed between the favorable and intermediate subgroups (P = .19 by the log-rank test). (B) Current classification (Table 4): using the current classification, there was a significant difference in overall survival between the 3 subgroups of patients (P < .001 by the log-rank test). Here, a significant difference was observed between the favorable and intermediate groups (P = .007 by the log-rank test). The 11 patients allografted in CR1 were censored at transplantation time.
Interaction with FLT3-ITD
A total of 134 of the 135 patients were tested for the presence ofFLT3-ITD. Overall, FLT3-ITD was detected in 35 patients studied (26%) and was also associated with the intermediate cytogenetic risk subgroup (ratio of FLT3-ITD cases, 0 of 15, 26 of 90, 3 of 14, and 6 of 15 in the favorable, intermediate, adverse, and undetermined cytogenetic risk subgroups, respectively;P = .04 by the Fisher exact test). The complete remission rate was identical in both FLT3-wt and FLT3-ITD subgroups (81% vs 77%, respectively; P = .63 by the Fisher exact test). In univariate analysis, the presence ofFLT3-ITD was predictive of shortened overall survival (P = .03 by the log-rank test) but not significantly shortened EFS or DFS (P = .12 and .11, respectively, by the log-rank test). As indicated in Table 2, there was no difference in the incidence of FLT3-ITD between bothCEBPA-nonmutated and CEBPA-mutated patient groups (P = .54 by the Fisher exact test). When analyzed in a 2-variable Cox model, both CEBPA mutations andFLT3-ITDs were independent factors for overall survival (P = .05 for CEBPA mutations, relative risk [RR] = 2.2 in the CEBPA-nonmutated group;P = .03 for FLT3-ITD, RR = 1.6 in theFLT3-ITD group). The 4-subgroup classification based on both C/EBPα and FLT3 status (C/EBPα+/FLT3-wt,C/EBPα+/FLT3-ITD, C/EBPα−/FLT3-wt,C/EBPα−/FLT3-ITD) significantly influenced overall survival (P = .03 by the log-rank test). Furthermore, even if numbers of patients in each subgroup were not high enough to strongly conclude, there seemed to be an interaction between these 2 prognostic factors: presence of FLT3-ITD significantly worsened overall survival in the CEBPA-mutated group (estimated 5-year overall survival, 20% vs 69% in theFLT3-ITD and the FLT3-wt groups, respectively;P = .03 by the log-rank test) but not in theCEBPA-nonmutated group (estimated 5-year overall survival, 14% vs 29% in the FLT3-ITD and the FLT3-wt groups, respectively; P = .11 by the log-rank test).
Multivariate analysis
We tested the prognostic significance of CEBPAmutations and FLT3-ITD in a multivariate analysis also including the 3-subgroup MRC cytogenetic risk classification as covariate. The lack of prognostic value of other putative prognostic factors including age (tested as continuous variable), WBC (tested as continuous variable), and treatment arm was verified in univariate analysis before performing this multivariate analysis (not shown). As shown in Table 3, the presence ofCEBPA gene mutations represented an independent good prognosis factor in this setting, either for EFS, DFS, or overall survival (P = .04, .05, and .05, respectively). In this multivariate analysis, the presence of FLT3-ITDs was only associated with a nonsignificant trend for a worse overall survival (P = .07).
Prognostic value of the MRC and current risk classifications, P (relative risk, 95% confidence interval)
| . | EFS . | DFS . | Survival . |
|---|---|---|---|
| MRC classification3-150 | |||
| Univariate analysis (n = 120 patients) | < .001 | .02 | < .001 |
| Multivariate analysis (n = 119 patients) | |||
| MRC classification | < .001 | .10 | < .001 |
| CEBPAmutation | .04 (0.47, 0.23-0.97) | .05 (0.43, 0.19-0.99) | .05 (0.45, 0.21-0.98) |
| FLT3/ITD | .32 | .50 | .07 (1.58, 0.96-2.58) |
| Current classification3-150 | |||
| Univariate analysis (n = 119 patients) | < .001 | .004 | < .001 |
| Multivariate analysis (n = 119 patients) | |||
| Current classification | < .001 | .05 | < .001 |
| CEBPAmutation | .84 | .41 | .72 |
| FLT3/ITD | .55 | .64 | .18 |
| . | EFS . | DFS . | Survival . |
|---|---|---|---|
| MRC classification3-150 | |||
| Univariate analysis (n = 120 patients) | < .001 | .02 | < .001 |
| Multivariate analysis (n = 119 patients) | |||
| MRC classification | < .001 | .10 | < .001 |
| CEBPAmutation | .04 (0.47, 0.23-0.97) | .05 (0.43, 0.19-0.99) | .05 (0.45, 0.21-0.98) |
| FLT3/ITD | .32 | .50 | .07 (1.58, 0.96-2.58) |
| Current classification3-150 | |||
| Univariate analysis (n = 119 patients) | < .001 | .004 | < .001 |
| Multivariate analysis (n = 119 patients) | |||
| Current classification | < .001 | .05 | < .001 |
| CEBPAmutation | .84 | .41 | .72 |
| FLT3/ITD | .55 | .64 | .18 |
All patients allografted in CR1 were censored at transplantation time.
Risk category definitions are given in Table 4.
We therefore tested another risk classification in which all AML cases with CEBPA mutation were considered as favorable if not associated with FLT3-ITD and/or adverse cytogenetic features (Table 4). This risk classification appeared to be more powerful than the MRC risk classification, especially in identifying a subgroup of favorable AML that would include all CBF-AMLs and all AMLs with CEBPA mutation but without FLT3-ITD and/or adverse cytogenetic features. Patients from this current favorable risk subgroup, accounting for 21% of all cases, had significantly longer EFS and overall survival than those from the intermediate risk subgroup (P = .01 and .007, respectively, by the log-rank test) (Figure 4B for overall survival). In a multivariate analysis including this current risk classification, CEBPA mutation, and FLT3-ITD as covariates, the risk classification remained the only independent prognostic factor for EFS, DFS, and overall survival (Table 3).
Risk category definitions
| Risk subgroup . | MRC coding9 . | Current study . |
|---|---|---|
| Favorable | inv(16) and t(8;21) | inv(16) and t(8;21) |
| CEBPAmutation | ||
| Without FLT3-ITD and/or adverse cytogenetic features | ||
| Intermediate | All others | CEBPAmutation |
| With FLT3-ITD | ||
| All others | ||
| With/without FLT3-ITD | ||
| Adverse | del(5q)/−5, −7, 3q abn, | del(5q)/−5, −7, 3q abn, complex |
| complex | With/without FLT3-ITD and/or CEBPA mutation |
| Risk subgroup . | MRC coding9 . | Current study . |
|---|---|---|
| Favorable | inv(16) and t(8;21) | inv(16) and t(8;21) |
| CEBPAmutation | ||
| Without FLT3-ITD and/or adverse cytogenetic features | ||
| Intermediate | All others | CEBPAmutation |
| With FLT3-ITD | ||
| All others | ||
| With/without FLT3-ITD | ||
| Adverse | del(5q)/−5, −7, 3q abn, | del(5q)/−5, −7, 3q abn, complex |
| complex | With/without FLT3-ITD and/or CEBPA mutation |
Discussion
Results of this study of CEBPA gene mutations in patients with de novo AML confirm the previously reported incidence of these mutations in AML cells of approximately 10%. These mutations may be categorized in 3 subclasses according to their effects on C/EBPα expression and structure. The first subclass comprises mutations resulting in an absence of the full-length 42-kDa protein (Figure 2A). Pabst et al have demonstrated that the transactivation potential of these C/EBPα-mutated proteins was abolished in vitro, the short 30-kDa protein being normally expressed and responsible for a dominant-negative effect.7 The second subclass comprises mutations located in the C-terminal bZIP domain (Figure 2B). These mutations, which correspond to most of the mutations reported by Gombart et al,8 also abolished the transactivation potential of C/EBPα. Interestingly, mutations from this subclass are not responsible for a dominant-negative effect of the mutated factor but only for a loss of its function.16,17The third subclass comprises 2 large deletions located in the middle part of the coding sequence but inducing termination codon before the bZip domain (Figure 2C). Three similar CEBPA alterations have been already reported,7,8 with uncertain functional consequences. A higher transactivation potential as compared with the wild-type protein has been found in vitro on G-CSF receptor promoter, while structural/function studies have shown that these mutations create polypeptides unable to localize to the nucleus, to heterodimerize, and to bind the DNA.16,17 Three of the 18 previously reported patients with CEBPA mutations had 2 different mutations, and another one had homozygous mutation.7,8 In our study, 7 of the 15 patients had 2 different mutations (biallelic in 6 cases), and another one had homozygous deletion. As previously shown by Pabst et al,7the 2 patients studied at diagnosis and after CR achievement displayed a CEBPA mutation at diagnosis and a normal sequence in CR confirming that the CEBPA mutation was acquired. In this study, we also confirmed the previous observation that CEBPAmutations are associated with the intermediate cytogenetic risk group. Regarding the FAB repartition, we confirmed the higher incidence of mutations in the AML-M1 and -M2 subtypes, even if mutations could be also observed in AML-M4 and -M5 subtypes.
Results of this first prognostic study of CEBPA gene mutations in patients with de novo AML show that CEBPAmutations are associated with better outcome. We thus propose taking into account the C/EBPα status in the risk category definition. The favorable risk subgroup (accounting for approximately 20% of the whole population) would comprise not only patients with CBF-AML but also those with CEBPA-mutated AML if not associated with other bad prognostic factors such as FLT3-ITD and/or adverse cytogenetic features. All other patients without adverse cytogenetic features (approximately 70% of the whole population) would be classified in the intermediate risk subgroup. Our results also suggest that the prognostic value of FLT3-ITDs is more marked in patients with CEBPA mutations as compared with those withoutCEBPA mutations, but these preliminary results will have to be confirmed in a larger cohort of patients. In other terms,FLT3-ITD appears to act as a major bad risk factor in patients with CEBPA-mutated AML. Even if this interaction between the prognostic impact of FLT3-ITDs and C/EBPα status has to be confirmed by further reports from other cooperative AML groups, this observation leads to caution when analyzing the results of all previously reported FLT3 studies that never referred to the C/EBPα status of patients tested. More generally, the question how to classify AML cases with FLT3-ITD is still a matter of debate. We have previously reported that the presence ofFLT3-ITD did not significantly worsen overall survival when patients were intensively treated with high cumulative doses of cytotoxic agents and timed-sequential schedules as used in our treatment protocol.10 In the present study again, theFLT3 status did not significantly influence patient outcome when analyzed in multivariate condition, especially onceCEBPA-mutated and FLT3-wt patients had been incorporated in the favorable risk subgroup. This negative observation, also reported by other cooperative groups with different intensive treatment schedules,18 might nevertheless be related to the limited population of patients studied.
The good outcome reported here in patients withCEBPA-mutated AML suggests that we define a new group of good-risk AML including PML-RARα–positive,AML1-ETO–positive, and CBFβ-MYH11–positive AML as well as CEBPA-mutated AML. The main common characteristic of this new group, which may account for 25% to 30% of all de novo AML cases, might be an acquired deficiency in normal C/EBPα function. In fact, it has been already shown that theAML1-ETO chimeric transcription factor indirectly suppresses the CEBPA gene expression.19 It has also been shown that C/EBPα homodimers interact strongly with thePML-RARα oncoprotein in a retinoic acid–dependent manner resulting in the loss of C/EBPα function inPML-RAR–positive cells.6 20 Acquired deficiency in C/EBPα function may thus be explained either throughCEBPA gene repression, C/EBPα protein binding, orCEBPA gene mutations. To date, neither the relationship between C/EBPα and the CBFβ-MYH11 fusion protein nor evidence of C/EBPα deficiency has been, however, reported inCBFβ-MYH11–positive cells.
In conclusion, while awaiting further observations emerging from the microarray technology, a simple approach based on standard karyotype associated with molecular analysis of a limited number of genes, including CBFs, FLT3, and CEBPA, provides a powerful risk classification that can be used to stratify the treatment of patients with de novo AML. This approach might still be improved through further evaluation of the prognostic significance of additional mutations of other important genes, such asp53, ras, and c-kit, in this context.
The authors are indebted to N. Philippe, M. Crepin, and the Institut Fédératif de Recherche for their excellent technical assistance and support in molecular biology and to Dr Helene Cavé and Dr Gerard Socié for helpful discussions and critical review of the manuscript.
Prepublished online as Blood First Edition Paper, June 14, 2002; DOI 10.1182/blood-2002-03-0990.
Supported by the Centre Hospitalier Universitaire of Lille (PHRC 1997), the Ligue Nationale Contre le Cancer (Comité du Nord), and the Fondation de France.
C.P. and C.S. contributed equally to the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Claude Preudhomme, Inserm U524, Laboratoire d'Hématologie, CHRU de Lille, Hôpital Calmette Bd du Pr Leclerq, 59037 Lille, France; e-mail: cpreudhomme@chru-lille.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal