The cyclin-dependent kinase inhibitorp57KIP2 is thought to be a potential tumor suppressor gene (TSG). The present study examines this possibility. We found that the expression ofp57KIP2 gene is absent in various hematological cell lines. Exposing cell lines to the DNA demethylating agent 5-aza-2′-deoxycytidine restoredp57KIP2 gene expression. Bisulfite sequencing analysis of its promoter region showed thatp57KIP2 DNA was completely methylated in cell lines that did not express thep57KIP2 gene. Thus, DNA methylation of its promoter might lead to inactivation of thep57KIP2 gene. DNA methylation of this region is thought to be an aberrant alteration, since DNA was not methylated in normal peripheral blood mononuclear cells or in reactive lymphadenitis. Methylation-specific polymerase chain reaction analysis found frequent DNA methylation of thep57KIP2 gene in primary diffuse large B-cell lymphoma (54.9%) and in follicular lymphoma (44.0%), but methylation was infrequent in myelodysplastic syndrome and adult T-cell leukemia (3.0% and 2.0%, respectively). These findings directly indicate that the profile of the p57KIP2gene corresponds to that of a TSG.
Introduction
Cell cycle progression is promoted by cyclins and cyclin-dependent kinases (CDKs) and is counterbalanced by CDK inhibitors.1 These components are regulated in the normal cell cycle, and disrupted regulation is closely related to tumorigenesis. For example, activation of cyclin D1 by bcl-1 translocation (t(11;14)(q13;q23)) is a critical genetic alteration in mantle cell lymphoma,2,3 and the CDK inhibitorp16INK4A is genetically lost or mutated in many malignancies.4 CDK inhibitors have been categorized into 2 families. One family, inhibitors of CDK4 (INK4s), includes p16 and p15, has repeated ankyrinlike sequences, and is a specific inhibitor of CDK4 and CDK6, resulting in competing cyclin D.5,6The other family, kinase inhibitor proteins (KIPs), includes p21CIP1, p27KIP1, and p57KIP2,7-10 and this family potently inhibits the binding of several cyclin/CDK complexes. The presence of mutations and deletions in INK4s indicates that they are tumor suppressor genes (TSGs)4 and that their function as negative regulators of cell proliferation fits the profile of TSGs. Because the KIP family also consists of CDK inhibitors, genetic alterations of the p21CIP1gene11 and p57KIP2genes12-14 have been extensively studied. In particular, the p57KIP2 gene is located at chromosome 11p15.5, a region implicated in sporadic cancers, including those of the breast,15 liver,16 and bladder.17 Beckwith-Wiedemann syndrome (BWS),18 which is characterized by the somatic overgrowth of various tissues, including the kidneys, liver, and skeletal muscle, and by a predisposition to embryonal tumors of these organs (Wilms tumor, hepatoblastoma, and embryonal rhabdomyosarcoma), is linked to this region.19 Although germ line point mutations of the p57KIP2 gene resulting in protein truncation have been identified in some patients with BWS,20,21 somatic mutations have not been found in sporadic cancers, including Wilms tumor.12-14,21 Thep57KIP2 gene is also an imprinted gene that preferentially expresses the maternal allele. In mice, genomic imprinting of the p57KIP2 gene is absolute and is regulated by DNA methylation,22 but DNA methylation has not been detected in healthy humans.23 24 The maternal monoallelic expression of thep57KIP2 gene might be regulated by a mechanism other than DNA methylation in humans.
DNA methylation of CpG islands in the promoter region leads to epigenetic gene inactivation by transcriptional silencing. Recently, aberrant DNA methylation of candidate TSGs have been found in various tumors,25-27 and this mechanism might be an alternative pathway of TSG inactivation. DNA hypermethylation of the INK4 family is frequent in lymphoid malignancies,25 26 and this mechanism is responsible for the inactivation of these genes.
Here we demonstrate that the aberrant DNA methylation of the promoter region of p57KIP2 gene is found in hematological malignancies, and we examine the relationship between DNA methylation of the p57KIP2 gene and its gene expression.
Materials and methods
Samples and DNA/RNA preparation
DNA was isolated from biopsied lymph nodes of 71 patients with diffuse large B-cell lymphomas (DLBCLs), 18 patients with follicular lymphomas (FLs), and 6 patients with reactive lymphadenitis. We also isolated DNA from the mononuclear cells of peripheral blood or bone marrow of 18 patients with chronic lymphocytic leukemias (CLLs), 26 with multiple myeloma (MM), 51 with adult T-cell leukemia (ATL), 52 with myelodysplastic syndrome (MDS), 14 with acute myeloid leukemia (AML), 6 with acute lymphocytic leukemias (ALL), and from 7 healthy volunteers as described.28 All patients were informed and consented to participate in the study. We also examined 17 hematological cell lines (NAMALWA, RAMOS, DAUDI, RAJI, RPMI1778, RPMI8226, KMS-12-PE, HS-SULTAN, IM-9, NALL-1, KOPT-K1, NALM-17, THP-6, CMK11-5, HL60, KU812F, and CMK86). RNA was extracted by means of the RNeasy Kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions.
Reverse transcriptase–polymerase chain reaction
First-strand cDNA was synthesized from 1 μg total RNA with the use of random hexamers as primers and Moloney murine leukemia virus–H reverse transcriptase (Gibco-BRL, Rockville, MD). Reverse transcriptase–polymerase chain reaction (RT-PCR) proceeded as described, with the use of first-strand cDNA as a template.28 In brief, 35 PCR cycles proceeded at an annealing temperature of 56°C with the use of the primers KIP2-1 (5′-CTGATCTCCGATTTCTTCGC-3′) and KIP2-2 (5′-TCTTTGGGCTCTAAATTGG-3′) to generate 162-bp products (Figure 1). The PCR products were separated on 2% agarose gels and were visualized by ethidium bromide staining. The control was glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA-amplified by RT-PCR (25 cycles) in separate tubes from p57KIP2amplification with the use of the primers GAPDH1 (5′-CCATGGAGAAGGCTGGGG-3′) and GAPDH2 (5′-CAAAGTTGTCATGGATGACC-3′) to generate 225-bp products. The KIP2-1, KIP2-2, and GAPDH primers were designed to not amplify the genome sequences.
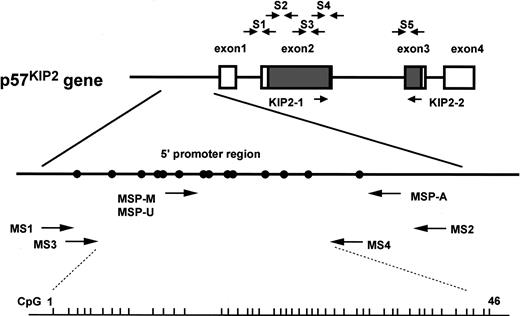
Genomic structure ofp57KIP2 gene and location of primers used in this study.
Short vertical bars on bottom line show sites of CpG(1-46) analyzed by bisulfite sequencing; open rectangle, noncoding region ofp57KIP2 gene transcripts; shaded rectangle, coding region ofp57KIP2 gene transcripts; closed circle, putative transcriptional factor binding site (Hatada and Mukai22); S1-S5, primer sets for PCR–single-strand conformation polymorphism (PCR-SSCP) (Hatada and Mukai22); KIP2-1 and KIP2-2, primer set of RT-PCR (see “Materials and methods”). Methylation-specific PCR (MSP) with the use of primer sets specific for methylated DNA (MSP-M), MSP with the use of primer sets specific for unmethylated DNA (MSP-U), and MSP-A indicate primers for MSP analysis (see “Materials and methods”); MS1, MS2, MS3, and MS4, primers for bisulfite sequencing analysis (see “Materials and methods”). Locations of these primers (in relation to the transcriptional start site ofp57KIP2 gene) are as follows: MSP-M, bp −282 to −263; MSP-U, bp −289 to −263; MSP-A, bp −41 to −20; MS1, bp −460 to −441; MS2, bp +75 to +91; MS3, bp −418 to −401; MS4, bp −148 to −125.

Genomic structure ofp57KIP2 gene and location of primers used in this study.
Short vertical bars on bottom line show sites of CpG(1-46) analyzed by bisulfite sequencing; open rectangle, noncoding region ofp57KIP2 gene transcripts; shaded rectangle, coding region ofp57KIP2 gene transcripts; closed circle, putative transcriptional factor binding site (Hatada and Mukai22); S1-S5, primer sets for PCR–single-strand conformation polymorphism (PCR-SSCP) (Hatada and Mukai22); KIP2-1 and KIP2-2, primer set of RT-PCR (see “Materials and methods”). Methylation-specific PCR (MSP) with the use of primer sets specific for methylated DNA (MSP-M), MSP with the use of primer sets specific for unmethylated DNA (MSP-U), and MSP-A indicate primers for MSP analysis (see “Materials and methods”); MS1, MS2, MS3, and MS4, primers for bisulfite sequencing analysis (see “Materials and methods”). Locations of these primers (in relation to the transcriptional start site ofp57KIP2 gene) are as follows: MSP-M, bp −282 to −263; MSP-U, bp −289 to −263; MSP-A, bp −41 to −20; MS1, bp −460 to −441; MS2, bp +75 to +91; MS3, bp −418 to −401; MS4, bp −148 to −125.
Culture with 5-aza-2′-deoxycytidine
RAJI and HL60 cells (2 × 105/mL) were incubated with 5-aza-2′-deoxycytidine (0, 5, or 10 μM) for 24 hours on day 2 after seeding. The cells were washed and incubated for another 72 hours. Cells were harvested on day 5 to analyzep57KIP2 gene expression and DNA methylation.
Bisulfite sequencing
We modified 500 ng genomic DNA extract from clinical samples and cell lines with sodium bisulfite as described26 using a DNA modification kit (CpGenome DNA Modification Kit; Oncor, Gaithersburg, MD) according to the manufacturer's instructions. Modified DNA (25 ng) was amplified by nested PCR to obtain the promoter sequence of the p57KIP2 gene. First-round PCR consisted of 35 cycles at an annealing temperature of 56°C, with the use of the primers MS1 (5′-TTTCGTTTGTAGATAAAGGA-3′) and MS2 (5′-CTAACTATCCGATAATAAACTCTTCTA-3′) (Figure 1). With the use of one twentieth of the first-round PCR product as a template, second-round PCR consisted of 35 cycles at an annealing temperature of 60°C with the primers MS3 (5′-GGGGGTGGGGAGTGTTGT-3′) and MS4 (5′-ATATTTTCAATTTCAACAACACCA-3′) (Figure 1). Under these conditions, we could assess the DNA methylation status ofp57KIP2 gene at the position 400 to 149 bp upstream of the transcription initiation site. PCR products were separated on 2% low-melting agarose gels, excised, and then digested with β-agarase (New England Biolabs, Beverly, MA). The digestion products were subcloned into the PGEM-Teasy vector, and at least 10 clones of each product were subjected to cycle sequencing (PE Applied Biosystems, Warrington, United Kingdom) and were analyzed by means of an ABI 310 (Applied Biosystems, Foster City, CA).
Methylation-specific PCR (MSP)
DNA modified with sodium bisulfite was selectively amplified by PCR with the use of primers MS1 and MS2 under the conditions described above. The concentrated modified DNA was then analyzed by MSP with the use of primer sets specific for unmethylated DNA (MSP-U, 5′-TTTGTTTTGTGGTTGTTAATTAGTTGT-3′, and MSP-A, 5′-ACACAACGCACTTAACCTATAA-3′) and methylated DNA (MSP-M, 5′-CGCGGTCGTTAATTAGTCGC-3′, and MSP-A). The annealing temperatures were 56°C for MSP-U with MSP-A, and 63°C for MSP-M with MSP-A. Each PCR was “hot-started,” and the products were separated on 2% agarose gels and then visualized by ethidium bromide staining.
The sensitivity of the MSP method was confirmed with the use of a mixture of bisulfite-modified DNA of HL60 and of U937, which have completely methylated and unmethylatedp57KIP2 gene promoter sequences, respectively, at ratios of 100:0, 10:90, 1:99, 0.1:99.9, and 0:100.
PCR-SSCP
Mutations of the p57KIP2 gene were screened by PCR-SSCP analysis29 with the use of primers as listed in a previous report.12 Thirty cycles of PCR amplification proceeded at annealing temperatures ranging from 56°C to 62°C in the presence of 1 μCi (0.037 MBq) α-[32P]–deoxycytidine triphosphate (α-32P]-dCTP) per reaction. The products were separated on 0.5 × MDE gels (FMC BioProducts, Rockland, ME). Bands were exposed on phosphorimaging plates and evaluated by means of a laser image analyzer (FUJI, Kanagawa, Japan).
Results
Expression of p57KIP2gene
We examined p57KIP2 gene expression in 17 hematological cell lines, 2 lymphadenitis samples, and 1 normal peripheral blood mononuclear cell (PBMNC) sample by RT-PCR using the primer set KIP2-1 and KIP2-2 (Figure 1). Expression of the p57KIP2gene was undetectable in 5 cell lines (RAMOS, DAUDI, RAJI, HS-SULTAN, and HL60) after 35 amplification cycles (Figure2). These cell lines were derived from diverse hematological malignancies, but loss of expression tended to be more frequent in lymphoid cell lines of the B-cell lineage. PCR analysis of genomic DNA using primers for PCR-SSCP analysis showed that none of the cell lines analyzed here had genetically lost thep57KIP2 gene (data not shown).
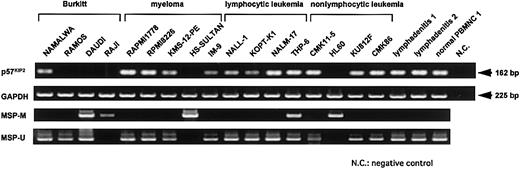
RT-PCR analysis of p57KIP2 gene expression and MSP analysis of p57KIP2gene promoter region in various cell lines.
Upper arrow shows 162-bp RT-PCR product ofp57KIP2 gene; lower arrow, 225-bp RT-PCR product of GAPDH gene; U, PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A).

RT-PCR analysis of p57KIP2 gene expression and MSP analysis of p57KIP2gene promoter region in various cell lines.
Upper arrow shows 162-bp RT-PCR product ofp57KIP2 gene; lower arrow, 225-bp RT-PCR product of GAPDH gene; U, PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A).
Restoration of p57KIP2gene expression by 5-aza-2′-deoxycytidine
We examined the mechanism of down-regulatedp57KIP2 gene expression. When 2 cell lines (HL60 and RAJI) that did not expressp57KIP2 gene were exposed to the demethylating agent 5-aza-2′-deoxycytidine (0, 5, or 10 μM),p57KIP2 gene expression was restored in both (Figure 3). The unmethylated status at the promotor region of the p57KIP2gene in both cell lines was detected after this procedure. Methylated bands remained after exposure to 5-aza-2′-deoxycytidine in RAJI cells, but the methylated bands disappeared from HL60 cells. All methylated sequences of the p57KIP2 gene in HL60 might have become unmethylated by this procedure. In RAJI cells, this conversion might not have been complete. These data suggested that DNA methylation of thep57KIP2 gene or of genes that transcriptionally regulate p57KIP2might be one mechanism of inactivation of thep57KIP2 gene expression.

Restoration of p57KIP2gene expression in RAJI and HL60 cells by 5-aza-2′-deoxycytidine (5-aza-2′C), detected by RT-PCR.
Top row of panels shows expression ofp57KIP2 gene; second row of panels, expression of GAPDH; third row of panels, detection of unmethylated status of p57KIP2 gene by MSP method; bottom row of panels, detection of methylated status ofp57KIP2 gene by MSP method. Upper arrow, 162-bp RT-PCR product ofp57KIP2 gene; lower arrow, 225-bp RT-PCR product of GAPDH gene.

Restoration of p57KIP2gene expression in RAJI and HL60 cells by 5-aza-2′-deoxycytidine (5-aza-2′C), detected by RT-PCR.
Top row of panels shows expression ofp57KIP2 gene; second row of panels, expression of GAPDH; third row of panels, detection of unmethylated status of p57KIP2 gene by MSP method; bottom row of panels, detection of methylated status ofp57KIP2 gene by MSP method. Upper arrow, 162-bp RT-PCR product ofp57KIP2 gene; lower arrow, 225-bp RT-PCR product of GAPDH gene.
Bisulfite sequence of the p57KIP2gene promoter region
We further examined the involvement of DNA methylation in relation to p57KIP2 gene expression. We performed bisulfite genomic sequencing of 46 CpG sites at the promoter region of the p57KIP2 gene (Figure 1) in 2 samples of lymphadenitis, 1 PBMNC sample from a healthy volunteer, 2 cell lines that expressed p57KIP2 gene (NAMALWA and THP-6), and 4 cell lines that did not (RAMOS, RAJI, HL60, and HS-SULTAN; Figure 4). The sequence analyzed here contained 12 putative transcriptional factor–binding sites.12 The lymphadenitis samples and normal PBMNC sample had no DNA methylation in 46 CpG sites of the promoter region. The presence of p57KIP2gene expression was confirmed in these samples by RT-PCR (Figure 2). NAMALWA expressed the p57KIP2 gene, and the sequence in this region was completely unmethylated. THP-6 also expressed the p57KIP2 gene. These sequences were methylated in 6 of 12 clones sequenced, and unmethylated in another 6. Unmethylated sequences at this region might be responsible for expression of the p57KIP2gene. All of the sequences were almost completely methylated in 3 of 4 cell lines without p57KIP2 gene expression, which was consistent with the loss of this gene expression. Although RAMOS did not express thep57KIP2 gene, its promoter region was completely unmethylated. Thus, silencing mechanisms other than DNA methylation in the promoter region might exist. The completely unmethylated p57KIP2 gene promoter in nonmalignant samples suggested thatp57KIP2 promoter DNA methylation itself is aberrant in healthy individuals.
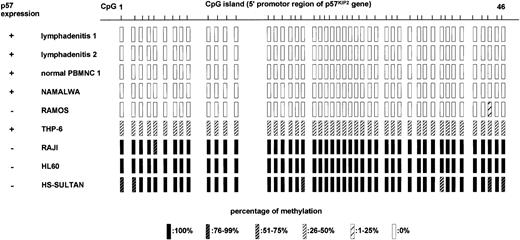
Methylation status of promoter region ofp57KIP2 gene in CpG islands in 2 samples of reactive lymphadenitis, 1 normal PBMNC sample, and 6 cell lines determined by sequencing after bisulfite modification of genomic DNA.
CpG 1 to CpG 46 correspond to what is depicted in Figure 1. Expression of p57 was detected by RT-PCR, and the percentage of methylation was determined from ratios of methylated cytosine in 10 to 12 sequenced clones.

Methylation status of promoter region ofp57KIP2 gene in CpG islands in 2 samples of reactive lymphadenitis, 1 normal PBMNC sample, and 6 cell lines determined by sequencing after bisulfite modification of genomic DNA.
CpG 1 to CpG 46 correspond to what is depicted in Figure 1. Expression of p57 was detected by RT-PCR, and the percentage of methylation was determined from ratios of methylated cytosine in 10 to 12 sequenced clones.
Methylation of p57KIP2gene detected by MSP
We screened for DNA methylation of the promoter region of thep57KIP2 gene, using the MSP method. We initially examined the sensitivity of the method using mixed samples of completely unmethylated and methylated cell lines. The results showed that the MSP method could detect one methylated cell among 1 × 102 unmethylated cells (Figure5). We then examined mRNA expression of the p57KIP2 gene in 17 hematological cell lines and compared the results of MSP analysis and bisulfite sequencing. The results of the 2 methods to detect DNA methylation were comparable (Figures 2 and 4). DAUDI cells have an unmethylated band, but mRNA was not detected. This discrepancy was also revealed in the RAMOS cell line by bisulfite sequencing, indicating an alternative mechanism of silencing the p57KIP2 gene. Six samples of lymphadenitis and 7 of normal PBMNCs were analyzed by MSP, and methylated bands were undetectable (Figure 2 and6). This finding means that nonmalignant mononuclear cells from peripheral blood and lymphadenitis tissue do not have a methylated promoter region in thep57KIP2 gene. We then screened the methylation status of the p57KIP2 gene promoter in primary hematological malignancies, including 71 DLBCL, 18 FL, 18 CLL, 26 MM, 6 ALL, 53 ATL, 14 AML, and 52 MDS samples, using the MSP method. The p57KIP2 gene promoter was methylated in 39 of 71 DLBCL samples (54.9%), in 4 of 26 MM samples (15.3%), and in 1 of 51 ATL samples (2.0%; Figure7). Table1 lists the frequencies ofp57KIP2 gene methylation in various hematological malignancies. These data suggested that aberrant DNA methylation of p57KIP2 gene is detected in some populations of hematological malignancies, and that the frequency of this methylation is higher in lymphoid malignancies of the B-cell phenotype.

MSP analysis of p57KIP2gene promoter region in mixtures of completely unmethylated and methylated cell lines.
Unmethylated (HL60) and methylated (RAMOS) cell lines were mixed at various ratios as indicated above each column. U shows PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A).

MSP analysis of p57KIP2gene promoter region in mixtures of completely unmethylated and methylated cell lines.
Unmethylated (HL60) and methylated (RAMOS) cell lines were mixed at various ratios as indicated above each column. U shows PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A).
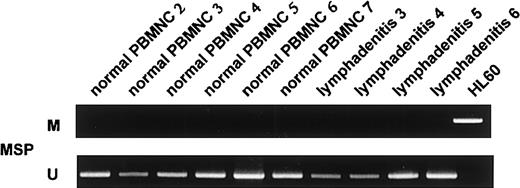
Methylation analyses ofp57KIP2 gene in 4 samples of lymphadenitis and 6 of normal PBMNCs detected by the MSP method.
U shows PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A).

Methylation analyses ofp57KIP2 gene in 4 samples of lymphadenitis and 6 of normal PBMNCs detected by the MSP method.
U shows PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A).
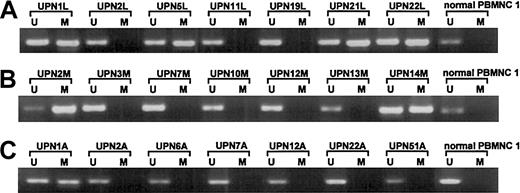
Methylation analyses ofp57KIP2 gene in primary lymphoid malignancies detected by the MSP method.
U shows PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A); and UPN, unique patient number. (A) DLBCL and normal PBMNC 1. (B) MM and normal PBMNC 1. (C) ATL and normal PBMNC 1.

Methylation analyses ofp57KIP2 gene in primary lymphoid malignancies detected by the MSP method.
U shows PCR products amplified with primers specific for unmethylated DNA (MSP-U and MSP-A); M, PCR products amplified with primers specific for methylated DNA (MSP-M and MSP-A); and UPN, unique patient number. (A) DLBCL and normal PBMNC 1. (B) MM and normal PBMNC 1. (C) ATL and normal PBMNC 1.
Frequency of p57KIP2 gene methylation in various hematological malignancies
| Malignancy . | Methylation ofp57KIP2 . | Frequency, % . |
|---|---|---|
| DLBCL | 39/71 | 54.9 |
| FL | 8/18 | 44.0 |
| CLL | 6/18 | 33.0 |
| MM | 4/26 | 15.4 |
| ALL | 1/6 | 16.7 |
| ATL | 1/51 | 2.0 |
| AML | 0/14 | 0 |
| MDS | 2/52 | 3.0 |
| Malignancy . | Methylation ofp57KIP2 . | Frequency, % . |
|---|---|---|
| DLBCL | 39/71 | 54.9 |
| FL | 8/18 | 44.0 |
| CLL | 6/18 | 33.0 |
| MM | 4/26 | 15.4 |
| ALL | 1/6 | 16.7 |
| ATL | 1/51 | 2.0 |
| AML | 0/14 | 0 |
| MDS | 2/52 | 3.0 |
Methylation was detected by MSP analysis.
Mutation of coding region ofp57KIP2gene
We screened for mutations of thep57KIP2 gene coding region in 31 of these 71 DLBCL samples by PCR-SSCP analysis. We did not find any mutations of the p57KIP2 gene (data not shown).
Discussion
The present study indicates that aberrant DNA methylation ofp57KIP2 gene at its promoter region might constitute one mechanism of silencingp57KIP2 gene expression. We also found this epigenetic alteration not only in cell lines but also in primary hematological malignancies.
In addition to point mutations or gene deletions, transcriptional repression by the hypermethylation of promoter sequences may be an alternative mechanism for TSG inactivation in cancers. Hypermethylation of CpG islands of TSG was originally detected in the retinoblastoma gene.30 The methylation of CpG sites in the p16INK4A gene has recently been recognized as a mechanism that suppresses its transcription in lymphoma, myeloma, and solid tumors.26,31,32 The major biological effect of p16INK4A is to halt cell cycle progression at the G1/S boundary, and the loss of p16INK4Afunction may result in cancer progression by allowing unregulated cellular proliferation. Because of its function as a CDK inhibitor like p16INK4A and p15INK4B, and its chromosomal location (11p15.5, which is recurrently deleted in various tumors), thep57KIP2 gene has been considered a good TSG candidate. Nevertheless, no direct evidence has confirmed this notion. Expression of the p57KIP2 gene is decreased in gastric cancer,33 bladder cancer,34 blastic crisis of chronic myelogenous leukemia,35 and Wilms tumor.24 36 These reports weakly supported one aspect ofp57KIP2 as a TSG candidate.
The p57KIP2 gene is imprinted, which means preferential maternal expression.37 The mechanism ofp57KIP2 gene imprinting is now under investigation. So far, no evidence correlates the expression ofp57KIP2 and DNA methylation at the promoter region under physiological conditions. In addition, we did not detect DNA methylation of the p57KIP2gene in the promoter region in normal blood or lymphadenitis samples. Methylation at the promoter region might not contribute to the monoallelic expression of this gene. However, the DNA might be methylated at a site other than at the promoter region, resulting inp57KIP2 gene imprinting. The chromosomal 11p15.5 region harbors several imprinted genes, and clusters of these genes might be divided into 2 separate imprinted domains.13 The centromeric domain containsp57KIP2 and the telomeric domain containsH19 and IGF2, and these regions are independently regulated.13 In the centromeric region, a paternally expressed transcript, LIT1,38 located withinKvLQT1 and transcribed in antisense orientation to it, might play a role in p57KIP2 regulation similar to that of H19 in IGF2 regulation.13The establishment of an imprint of thep57KIP2 gene might be coordinately regulated throughout the entire imprinted domain.
Shin et al33 showed promoter methylation ofp57KIP2 gene by means of a conventional PCR-based method in gastric cancer. We did not detectp57KIP2 mRNA expression in several hematological cell lines by RT-PCR. Treatment of RAJI and HL60, which do not express the p57KIP2 gene, with the demethylating agent 5-aza-2′-deoxycytidine restoredp57KIP2 gene expression. Sequence analysis of the p57KIP2 gene promoter region after bisulfite modification precisely revealed the methylation status. We found that cell lines with complete DNA methylation at promoter region of p57KIP2 gene did not express this gene. In contrast, cell lines that expressed thep57KIP2 gene had unmethylated promoter sequences. In THP-6, the p57KIP2 gene promoter was unmethylated in half of the clones analyzed by bisulfite sequencing and methylated in the other half. We could not determine which clones were of maternal origin, but these unmethylated promoter sequences might lead to expression of thep57KIP2 gene.
RAMOS and DAUDI did not express thep57KIP2 gene despite the presence of unmethylated promoter sequence. There might exist mechanisms to inactivate the p57KIP2 gene mechanisms other than DNA methylation in the promoter region, such as DNA methylation of p57KIP2 gene at other regions, deregulation of a gene transcriptionally upstream of thep57KIP2 gene, or histone deacetylation. The formation of inactive chromatin through histone deacetylation is an alternative mechanism for inactivation of thep57KIP2 gene in gastric cancer cells.33
Bisulfite sequencing revealed no methylated sequences of thep57KIP2 promoter region in nonmalignant cells, and MSP analysis did not detect methylation of this region in 6 lymphadenitis samples and 7 normal PBMNC samples. These findings suggested that complete DNA methylation of thep57KIP2 gene promoter silenced this gene, and that methylation of this region itself was an aberrant alteration in lymphoid tissues. Thus, epigenetic silencing of thep57KIP2 gene revealed that this gene might be a TSG candidate despite the absence of genetic mutations.
MSP is a simple and sensitive method of detecting DNA methylation and requires only small specimens containing limited amounts of DNA.26 We used this method to detect aberrant DNA methylation of the p57KIP2 gene promoter region at different frequencies among primary hematological malignancies. This region was 54.9% methylated in DLBCL, but 1.9% in ATL and 0% in AML. This observation suggested that the aberrant p57KIP2 gene methylation was not random among the hematological malignancies, and that the samples with the B-cell phenotype were preferentially affected. Such differences in the frequencies of methylation of TSGs among hematological malignancies have been identified in the p16INK4A and p15INK4B genes.39
An understanding of the relationship between methylation of thep57KIP2 gene and the clinical outcome of primary tumors is necessary to determine the clinical significance of such epigenetic alterations. Unfortunately, clinical information about the patients analyzed in this study is rather sparse, so we could not assess these points. To elucidate the role of aberrant DNA methylation of the p57KIP2 gene in primary hematological malignancies, the expression level of thep57KIP2 gene should be assessed in tumor specimens. RT-PCR detected p57KIP2 mRNA in primary tumors even when the p57KIP2gene was methylated (data not shown). We considered these mRNAs to be expressed by normal background cells of tumor specimens. We are now attempting to determine the protein level of KIP2 by immunohistochemical means in primary tumors.
Among several studies ofp57KIP2-deficient mice,40,41Zhang et al40 showed that a lack of kip2 causes some features of BWS. However, Takahashi et al41 reported that most BWS symptoms could not be reproduced. Both groups of investigators found that mice lacking kip2 did not develop tumors, in either the kidney or liver. These results do not substantiate the notion that the p57KIP2gene is a TSG. However, these results were obtained from mice models and cannot be completely extrapolated to humans. Because human neoplasms are thought to develop through several steps, analyses of double-knockout mice that lack p57KIP2and other candidate TSGs might be required to more precisely elucidate the effects of the p57KIP2 gene on tumor formation.
The presence of genetic mutations is thought to be a hallmark of many TSGs and TSG candidates. Recent investigations into epigenetic alterations in TSGs have highlighted DNA methylation as a powerful mechanism of TSG inactivation. Our findings indicate that methylation of the promoter region is a pathway ofp57KIP2 gene inactivation. We also found that the p57KIP2 gene is actually methylated in primary lymphoid malignancies as an aberrant and epigenetic event. This would support the notion that thep57KIP2 gene is a candidate TSG. Further study of its biological effect on tumorigenesis might further delineate the profile of p57KIP2 gene as a TSG.
We thank Ms K. Kondo for excellent technical assistance.
Prepublished online as Blood First Edition Paper, May 13, 2002; DOI 10.1182/blood-2001-11-0026.
Supported by Grants-in-Aid for Cancer Research (11S-1 and 11-8) from the Ministry of Health, Labor and Welfare, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hirokazu Nagai, Department of Hematology, Nagoya National Hospital, 4-1-1 Sannomaru, Naka-ku, Nagoya, 460-0001, Japan; e-mail: nagaih@nnh.hosp.go.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal