We have identified and cloned a novel human cytokine with homology to cytokines of the interleukin-17 (IL-17) family, which we have termed human IL-17E (hIL-17E). With the identification of several IL-17 family members, it is critical to understand the in vivo function of these molecules. We have generated transgenic mice overexpressing hIL-17E using an apolipoprotein E (ApoE) hepatic promoter. These mice displayed changes in the peripheral blood, particularly, a 3-fold increase in total leukocytes consisting of increases in eosinophils, lymphocytes, and neutrophils. Splenomegaly and lymphoadenopathy were predominant and included marked eosinophil infiltrates and lymphoid hyperplasia. CCR3+ eosinophils increased in the blood and lymph nodes of the transgenic mice by 50- and 300-fold, respectively. Eosinophils also increased 8- to 18-fold in the bone marrow and spleen, respectively. In the bone marrow, most of the eosinophils had an immature appearance. CD19+ B cells increased 2- to 5-fold in the peripheral blood, 2-fold in the spleen, and 10-fold in the lymph nodes of transgenic mice, whereas CD4+ T lymphocytes increased 2-fold in both blood and spleen. High serum levels of the cytokines IL-2, IL-4, IL-5, granulocyte colony-stimulating factor, eotaxin, and interferon γ were observed. Consistent with B-lymphocyte increases, serum immunoglobulin (Ig) M, IgG, and IgE were significantly elevated. Antigenic challenge of the transgenic mice with keyhole limpet hemocyanin (KLH) resulted in a decrease in anti-KLH IgG accompanied by increases of anti-KLH IgA and IgE. In situ hybridization of transgenic tissues revealed that IL-17Rh1 (IL-17BR/Evi27), a receptor that binds IL-17E, is up-regulated. Taken together, these data indicate that IL-17E regulates hematopoietic and immune functions, stimulating the development of eosinophils and B lymphocytes. The fact that hIL-17E overexpression results in high levels of circulating eosinophils, IL-4, IL-5, eotaxin, and IgE suggests that IL-17E may be a proinflammatory cytokine favoring Th2-type immune responses.
Introduction
Recently, the cytokine interleukin 17 (IL-17) has been recognized as a key contributor to inflammation and numerous diseases. Elevated expression levels of IL-17 have been demonstrated in tissues from patients with inflammatory diseases, including synovium of patients with rheumatoid arthritis, mononuclear cells from patients with multiple sclerosis, and T cells from psoriatic lesions.1-5 IL-17 is associated with organ allograft rejection, as IL-17–producing cells were identified in rejected human kidney allografts, and an IL-17 receptor antagonist interfered with cardiac allograft rejection and acute vascular rejection in mice.6-8
Initially identified due to its homology to an open reading frame (ORF) of T lymphotrophic herpesvirus saimiri, IL-17 has been shown to be produced only by activated CD4+ cells and to exert numerous proinflammatory responses including the induction of IL-1β, IL-6, IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), prostaglandin E2 (PGE2), leukocyte inhibitory factor (LIF), oncogene-α, and intercellular adhesion molecule 1 (ICAM-1) up-regulation in synoviocytes, keratinocytes, fibroblasts, epithelial cells, endothelial cells, and macrophages.1,3,4,9,10,12-16 IL-17 increased IL-8 release from human bronchial cells in vitro and recruited neutrophils into rat airways after intratracheal instillation.17 Arthritis associations of IL-17 were demonstrated by collagen type I degradation in human rheumatoid arthritis synovium ex vivo samples treated with IL-17 as well as cartilage degradation in mice on intra-articular administration of IL-17.18 The maturation of CD34+ progenitor cells into neutrophils was sustained when cocultured with fibroblasts and IL-17.10 Furthermore, by the use of adenoviral-mediated gene transfer, overexpression of IL-17 stimulates granulopoiesis in mice.19
Recently, cytokines homologous to IL-17, which form a distinct family, have been identified. Four new family members have been identified, namely, IL-17B, IL-17C, IL-17E/IL-25, and IL-17F.20-25These molecules possess approximately 25% to 30% homology to IL-17, consist of 155 to 197 amino acids with a molecular mass of 20 to 30 kDa, and bind to specific cognate IL-17 receptors with little cross-reactivity. To date, 2 IL-17 receptors have been identified, IL-17R, which binds IL-17, and IL-17Rh1 (IL-17BR/Evi27), which binds both IL-17B and IL-17E.21,22 26-28 The binding specificity of the IL-17 ligands suggests that other IL-17 receptors may exist. Differential expression of the ligands by activated T cells has been observed. Unlike IL-17 and IL-17F, IL-17B and IL-17C are not expressed by activated T cells but are expressed in other tissues. These differences in receptor utilization and tissue expression may account for differences in target populations and function.
In this report, we have identified a novel human cytokine with homology to IL-17 as seen by conservation of critical cysteine residues. We term this particular molecule human (h) IL-17E. We report here the identification, cloning, and functional characterization of hIL-17E and show its role in the regulation of hematopoietic and immune function.
Materials and methods
Antibodies
Antimurine monoclonal antibodies, purified or fluorochrome-labeled, were purchased from BD Pharmingen (San Diego, CA), Chemicon (Temecula, CA), Roche Molecular Biochemicals (Indianapolis, IN), Vector Laboratories (Burlingame, CA), Zymed Laboratories (San Francisco, CA), and R & D Systems (Minneapolis, MN) and used for enzyme-linked immunosorbent assays (ELISAs), immunophenotyping, and immunohistochemistry of the transgenic mouse tissues.
Cloning of hIL-17E
An IL-8 family profile search of the Amgen and Genbank dbEST database was performed, resulting in the identification of the mouse expressed sequence tag (EST), zmgb-ai430337. The overall homology between the zmgb-ai430337 predicted amino acid sequence and human IL-8 was low. The conservation of cysteines and other key residues suggested that zmgb-ai430337 was a novel member of the IL-17 family. The clone corresponding to this EST was obtained from the National Institutes of Health IMAGE Consortium via Research Genetics (an Invitrogen Company, Huntsville, AL) and was fully sequenced. A BLAST search revealed that the mouse EST sequence corresponded to regions of the GenBank BAC clone sequence CNS0000B, which is a genomic DNA (gDNA) sequence derived from human chromosome 14.
The GenBank matches supported the existence of a human homologue to the novel mouse IL-17–like protein, which we have termed hIL-17E. Polymerase chain reaction (PCR) primers based on the hIL-17–related sequence were designed to screen various sources for the IL-17–related cDNA. The forward primer, designated 2392-73 (5′-AGAGTCCTGTAGGGCCAGTGAAGATGG-3′) and the reverse primer, designated 2374-88 (5′-TACAGCCTGCGCTCCAGGCAGTAGCC-3′), were used to screen human cDNA samples (Clontech Laboratories, Palo Alto, CA) to identify the IL-17–related cDNA. Found most abundantly expressed in the testis, the full-length hIL-17E cDNA clone was obtained using the Rapid Screen human testis cDNA library (Origene Technologies, Rockville, MD). Standard PCR conditions were used for all of the cDNA library screening, and positive clones requiring further PCR of secondary subplates containing original clones identified 4 positive clones, numbers 70, 78, 85, and 89. The plasmid DNAs corresponding to these colonies were sequenced using a combination of vector primers and gene-specific primers. Clone 89 was fully sequenced and the other clones were sequenced at the ends and in the coding sequence, revealing that all 4 clones had identical coding sequences (GenBank accession no.AF461739).
Construction of hIL-17E transgenic expression vector and generation of transgenic mice
To generate transgenic mice overexpressing hIL-17E, the cDNA encoding full-length hIL-17E cDNA of 483 bp was cloned into the transgenic HE8 vector as previously described, which contains the human liver-specific apolipoprotein E (ApoE) promoter, hepatic control region (HCR), and a simian virus 40 (SV40) polyadenylation site.29 Specifically, an SpeI/NotI fragment containing the hIL-17E coding sequence and immediate upstream altered Kozac sequence CCACC was subcloned into the corresponding sites of the HE8 transgenic construct, yielding ApoE–hIL-17E. Microinjection fragments were obtained by XhoI, ScaI, andAflII restriction digestion of ApoE–hIL-17E and purified as previously described, yielding a 2.7-kb transgene insert. Microinjection of single-cell embryos from BDF1 × BDF1-bred mice with the hIL-17E transgene insert was performed as previously described.29,30 Embryos were cultured overnight in a 37°C and 5% CO2 incubator and 15 to 20 2-cell embryos were transferred to the oviducts of 13 pseudopregnant CD1 female mice. Transgenic offspring were identified by PCR screening with primers that amplify a 368-bp fragment of the human ApoE first intron from DNA prepared from ear biopsies as described.29
Taqman mRNA quantitative tissue expression analysis
The tissue distribution of hIL-17E was determined by real-time quantitative Taqman PCR using a fluorogenic probe and PCR primers complementary to hIL-17E. Human tissue cDNA was generated from 2 μg human total RNA (Clontech) by first-strand cDNA synthesis using Superscript II (Invitrogen Life Technologies, Carlsbad, CA) and following PCR conditions provided in the protocols. Taqman PCR reactions were performed using standard protocol on an ABI PRISM 7700 instrument, and the data were analyzed by Sequence Detection System software (Applied Biosystems, Foster City, CA). The tissue expression levels of IL-17E were normalized to the levels of a common cellular housekeeping gene, cyclophilin.
Animals and tissue sample preparation
Two independent full-necropsy experiments were performed from both founder (F0) and 2 high-expressing F1 lines of transgenic mice. All organs and tissues from a minimum of 5 transgenic mice and 5 negative littermates, aged 6 to 8 weeks and 8 to 10 weeks, respectively, were harvested for histologic analysis or flow ctyometry or both. Blood was collected by cardiac puncture and transferred to EDTA (ethylenediaminetetraacetic acid)–containing and serum-separation tubes. Spleens, femurs, mesenteric lymph nodes, and thymus were collected in Hanks balanced salt solution containing 2% heat-inactivated fetal bovine serum (FBS), 2 mMl-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin for flow cytometric analysis. Tissues were homogenized using standard methods and femurs were flushed, washed, and resuspended in phosphate-buffered saline plus 0.5% bovine serum albumin. Cells were counted using a Coulter counter.
Immunophenotyping and cell sorting
Cells from murine spleen, whole blood, mesenteric lymph nodes, femurs, and thymi were incubated at 0.5 to 1 × 106 cells with 1 μg monoclonal antibody (BD Pharmingen or R & D Systems) conjugated with either fluorescein isothiocyanate (FITC), phycoerythrin (PE), or Cychrome fluorochromes. The following inclusive hematopoietic panel was used: CD3, CD4, CD5, CD8, CD11b, CD11c, CD19, CD23, CD44, CD45, B220, CD54, CD69, c-kit, GR-1, and CCR3 (R & D Systems). Following antibody incubations and washes, red blood cells (RBCs) were lysed with fluorescence-activated cell sorting (FACS) lysing buffer (BD Pharmingen) and read on Becton Dickinson FACScan cytometer (Franklin Lakes, NJ). Femoral transgenic cell populations were sorted on Becton Dickinson Vantage SE cell sorter.
Histology and immunohistochemistry
Sections of all major organs and tissues were fixed in 10% neutral buffered zinc formalin, paraffin embedded, sectioned at 3 μm, and stained with hematoxylin and eosin for routine histologic examination. Immunohistochemical staining of 4-μm thick paraffin-embedded sections were deparaffinized, blocked with CAS BLOCK (Zymed Laboratories), and incubated with a rat antimouse CD45R/B220 monoclonal antibody (BD Pharmingen) or a rat antimouse immunoglobulin (Ig) G1 monoclonal antibody (BD Pharmingen) antibody. The primary antibody was detected using a biotinylated rabbit or goat antirat Ig secondary (Vector Laboratories). Standard quenching and detection procedures were performed.
Riboprobe preparation and in situ hybridization of IL-17Rh1
A 380-bp fragment of the IL-17 receptor homologue (IL-17Rh1) was amplified by PCR using mouse gDNA as a template using the following primers: sense primer (5′-GTACAGTGGCTG ACCACTCAGAAG) and the antisense primer (5′-GGTGGACTACAAGGGTGAACA GC-3′).28 PCR conditions were as follows: 35 cycles of 94°C for 15 seconds, 65°C for 30 seconds, and 72°C for 30 seconds. This 380-bp fragment was then subcloned into pGEMT vector (Promega, Madison, WI). All plasmids were linearized with NcoI restriction enzyme and the antisense33P-labeled RNA probe was synthesized with Sp6 RNA polymerase (Invitrogen Life Technologies).
Tissue sections of 5 μm from 2 expressing hIL-17E founder (F0) transgenic mice and one nonexpressing littermate control were immersion fixed, paraffin embedded on slides. Standard in situ hybridization protocol was followed involving overnight hybridization at 60°C followed by RNase digestion and a high-stringency wash in 0.1 times standard sodium citrate (SSC) at 55°C for 30 minutes. Slides were dipped in emulsion and allowed to expose for 3 weeks.
Cytokine and Ig production assays
Murine IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, granulocyte colony-stimulating factor (G-CSF), GM-CSF, and eotaxin levels in serum were detected using ELISA kits (Pierce Endogen, Rockford, IL) or the mouse Th1/Th2 cytokine bead assay detection kit (BD Pharmingen) using protocols provided.
Serum Ig ELISAs were done, as previously described, using the following antibodies: primary antibodies (at 5 μg/mL) rat antimouse IgE, IgA, IgG2a, IgG2b, IgG3 (BD Pharmingen), goat antimouse IgM (Chemicon), and goat anti-IgG1 (Roche Molecular Biochemicals); secondary antispecies horseradish peroxidase (HRP)–linked antibodies (at 0.25 μg/mL from BD Pharmingen, Roche Molecular Biochemicals, or Zymed Laboratories).31
KLH antigen challenge and antibody production
Eight nontransgenic and 6 hIL-17E transgenic animals (14-15 weeks old) were immunized subcutaneously with 100 μg KLH (Pierce, Rockford, IL) in complete Freund adjuvant on day 0 as previously described.32 Blood was drawn on days 7, 14, and 21 and analyzed for serum anti-KLH Igs. All classes and subclasses of Igs were examined, that is, IgM, IgE, IgA, and IgG, composed of IgG1, IgG2a, IgG2b, and IgG3.
Results
Identification of the hIL-17E gene
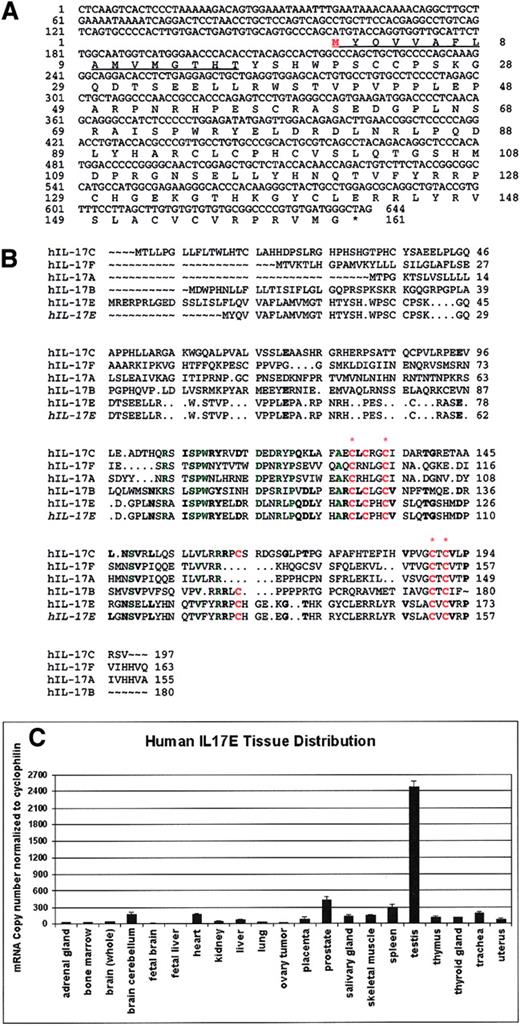
The hIL-17E gene is 3987 bp in length containing a 483 bp ORF encoding 161 amino acids. This gene product possesses a putative hydrophobic signal peptide of 16 amino acids and a mature protein of 145 amino acids (Figure 1A). Like other IL-17 family members, IL-17E is predicted to have a molecular weight of 16 to 17 kDa. Flanking the ORF exist large 5′- and 3′-untranslated sequences (data not shown). A FASTA search of the SwissProt database with our predicted IL-17E protein sequence revealed a 25.0% identity over 160 amino acids overlap with IL-17, a 35.6% identity over 90 amino acids overlap with IL-17B, and a 34.5% identity over 171 amino acids overlap with IL-17C. Alignment of the IL-17 family member protein sequences reveals that 4 cysteine residues located in the C-terminus are conserved, as well as 11 additional residues (Figure 1B). Greater conservation of residues with IL-17B and IL-17C largely in the C-terminus exists as well, including 2 additional cysteines. Like other IL-17 family members, this novel human IL-17E is predicted to be a secreted protein and a cytokine.
Sequence of hIL-17E, protein alignment with IL-17 cytokine family members, and human tissue expression.
(A) Nucleotide sequence and predicted protein sequence of hIL-17E molecule (GenBank accession number AF461739). The 3′ untranslated sequence upstream with start Met is indicated in red. Underlined sequence is the predicted signal sequence. (B) Alignment of hIL-17 protein family members with hIL-17E molecule reported herein (indicated in italics). FASTA searches of the Swiss Protein Database revealed hIL-17E conserved cysteine residues (indicated in red; complete conservation among all members indicated by red asterisk) and approximately 25% to 35% homology with all other IL-17 proteins. Other important conserved residues are highlighted in green. Residues conserved with IL-17B and IL-17C are indicated in bold. (C) IL-17E expression in human tissues (RNA purchased from Clontech) byTaqman quantitative analysis.
Sequence of hIL-17E, protein alignment with IL-17 cytokine family members, and human tissue expression.
(A) Nucleotide sequence and predicted protein sequence of hIL-17E molecule (GenBank accession number AF461739). The 3′ untranslated sequence upstream with start Met is indicated in red. Underlined sequence is the predicted signal sequence. (B) Alignment of hIL-17 protein family members with hIL-17E molecule reported herein (indicated in italics). FASTA searches of the Swiss Protein Database revealed hIL-17E conserved cysteine residues (indicated in red; complete conservation among all members indicated by red asterisk) and approximately 25% to 35% homology with all other IL-17 proteins. Other important conserved residues are highlighted in green. Residues conserved with IL-17B and IL-17C are indicated in bold. (C) IL-17E expression in human tissues (RNA purchased from Clontech) byTaqman quantitative analysis.
Initial reverse transcription–PCR (RT-PCR) analysis of human cDNA samples indicated that although IL-17E is expressed at low levels in a wide variety of tissues, strong expression was identified in human testis (data not shown). Normal hIL-17E tissue expression was examined by Taqman quantitative RT-PCR of human tissues, revealing very high expression in the testis, with moderate expression in the prostate and spleen (Figure 1C). Low expression was observed in the brain cerebellum, heart, liver, placenta, salivary gland, skeletal muscle, thymus, thyroid gland, and trachea. IL-17E expression was not detectable by human tissue Northern blot analysis (data not shown).
Overexpression of hIL-17E results in leukocytosis, eosinophilia, and enlarged secondary lymphoid organs
To investigate the biologic effect of IL-17E, we generated transgenic mice overexpressing hIL-17E using a previously described transgenic HE-vector driven by the ApoE hepatic promoter.29 With this construct, hIL-17E is secreted from the liver where the transgene is expressed. The cDNA sequence encoding the full-length hIL-17E sequence was subcloned into the HE-vector (Figure 2A). Resultant founders were identified by genomic PCR analysis using primers against the HE and SV40 vector sequences. Northern blots of hepatic mRNA revealed 16 transgenic founders expressing the transgene. Four positive male founders were bred with wild-type BDF1 females generating F1 progeny. Western blot analysis of the F1 progeny showed very high expression of IL-17E for one founder line (Figure 2B). Molecular mass of hIL-17E was shown to be about 24.7 kDa, suggesting it to be highly glycosylated. No behavioral or health abnormalities were observed in any of the hIL-17E transgenic mice.
Human IL-17E transgenic construct and protein expression in mice.
(A) Transgenic IL-17E overexpression construct, ApoE–hIL-17E, indicating ApoE promoter and HCR upstream of subcloned hIL-17E cDNA sequence. Restriction sites used are indicated. (B) Mouse sera (5 transgenics, T1-T5, and 3 littermate controls, C1-C3) were run in reducing conditions on a 14% Tris-Glycine gel, probed with conditioned media containing anti–hIL-17E, followed by incubation with HRP-linked secondary antibody (1:4000) and detected by chemiluminescence; 10-second exposure.
Human IL-17E transgenic construct and protein expression in mice.
(A) Transgenic IL-17E overexpression construct, ApoE–hIL-17E, indicating ApoE promoter and HCR upstream of subcloned hIL-17E cDNA sequence. Restriction sites used are indicated. (B) Mouse sera (5 transgenics, T1-T5, and 3 littermate controls, C1-C3) were run in reducing conditions on a 14% Tris-Glycine gel, probed with conditioned media containing anti–hIL-17E, followed by incubation with HRP-linked secondary antibody (1:4000) and detected by chemiluminescence; 10-second exposure.
Six- to 8-week-old mice were analyzed by extensive histologic and immunophenotypic analysis. The hIL-17E founders and F1 mice revealed striking hematopoietic changes compared with nonexpressors. Splenomegaly and lymphadenopathy were prevalent in the high-expressing transgenic mice (Table 1). Splenic hematopoietic cellularity increased on averages by 2- to 3-fold. Mesenteric lymph nodes were significantly enlarged, and mild to moderate inflammation was observed throughout the intestines. Peribronchial inflammation was observed as well in the lungs of some higher expressing transgenic mice (data not shown). The peripheral blood revealed a significant 3-fold increase in total leukocytes. The most striking observation was a 50-fold increase in eosinophils, from 0.03 × 106 to 1.99 × 106 cells/mL. Along with eosinophils, a 3-fold increase in lymphocytes (from 3.09 × 106 to 11.93 × 106 cells/mL blood), a 2-fold increase in neutrophils (from 0.92 × 106 to 2.29 × 106 cells/mL), and a 13-fold increase in a large unstained cell population contributed to the remaining white blood cell (WBC) expansion.
Spleen weights and peripheral blood leukocyte counts in IL-17E transgenic mice*
| . | Spleen wt.† (% body weight, g) . | Total WBC (103cells/μL) . | Neutrophils (103 cells/μL) . | Eosinophils (103 cells/μL) . | Lymphocytes (103cells/μL) . | Lg unstained‡ (103 cells/μL) . |
|---|---|---|---|---|---|---|
| Transgenics | 1.08 ± 0.27 | 11.93 ± 4.47 | 2.29 ± 0.67 | 1.99 ± 0.38 | 6.76 ± 2.32 | 1.41 ± 1.11 |
| Nontransgenics | 0.37 ± 0.33 | 3.09 ± 0.79 | 0.92 ± 0.53 | 0.03 ± 0.01 | 1.99 ± 0.38 | 0.10 ± 0.05 |
| P | .0007 | .003 | .032 | .017 | .0025 | .031 |
| . | Spleen wt.† (% body weight, g) . | Total WBC (103cells/μL) . | Neutrophils (103 cells/μL) . | Eosinophils (103 cells/μL) . | Lymphocytes (103cells/μL) . | Lg unstained‡ (103 cells/μL) . |
|---|---|---|---|---|---|---|
| Transgenics | 1.08 ± 0.27 | 11.93 ± 4.47 | 2.29 ± 0.67 | 1.99 ± 0.38 | 6.76 ± 2.32 | 1.41 ± 1.11 |
| Nontransgenics | 0.37 ± 0.33 | 3.09 ± 0.79 | 0.92 ± 0.53 | 0.03 ± 0.01 | 1.99 ± 0.38 | 0.10 ± 0.05 |
| P | .0007 | .003 | .032 | .017 | .0025 | .031 |
Values for IL-17E founders (n = 4, for each set of nontransgenic and transgenic mice).
Transgenic spleens, n = 5.
Large unstained cells, possibly large granular lymphocytes.
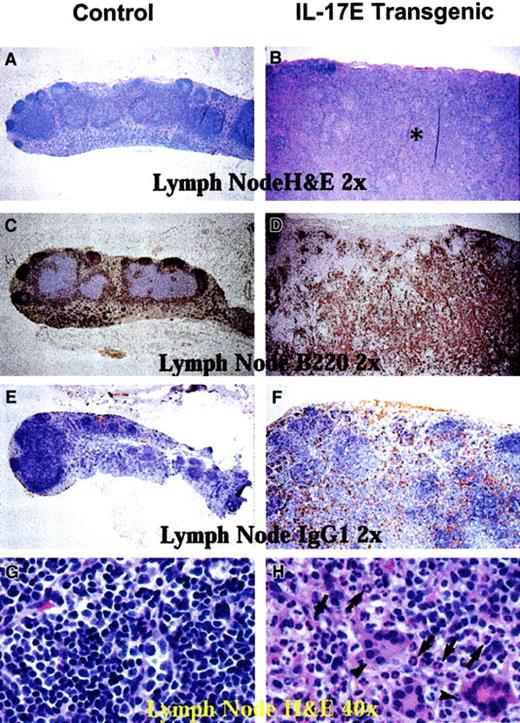
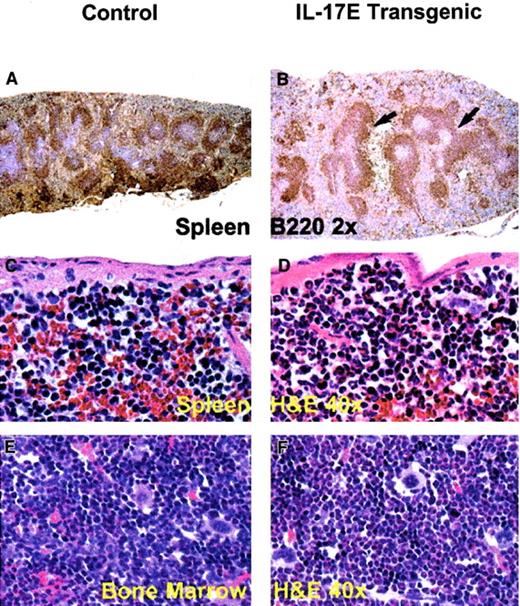
Histologic analysis of transgenic mice lymph nodes revealed marked lymphadenopathy with a predominant eosinophilic infiltration and a distinct loss in normal nodal architecture (Figure3A,B). Immunostaining for B220 and mouse IgG1 revealed lymph node medullary expansion due to increases in reactive B cells and plasma cells, respectively (Figure 3C-F). Immunostaining for F4/80 revealed an increase in macrophages (data not shown). Consistent with changes in the lymph nodes, the transgenic mouse spleen and bone marrow displayed eosinophilic myeloid hyperplasia as well as lymphoid hyperplasia with a predominance of B220+ cells (Figure 4). These findings suggest a role of IL-17E in inflammation of lymphoid and intestinal tissues, as well as a role in myelopoiesis and lymphopoiesis in the blood and bone marrow.
Human IL-17E transgenic lymph nodes revealed lymphadenopathy.
Histologic sections of nontransgenic control littermates (left panels) and transgenic (right panels) mesenteric lymph nodes. Panels A, B, G, and H are stained with hematoxylin and eosin; panels C and D, B220-stained sections; E and F, mouse IgG1-stained sections. All panels illustrate markedly enlarged transgenic lymph nodes disrupted of their normal architecture by the infiltration of eosinophils (B, asterisk; H, arrows), B220+ B lymphocytes (D), and mouse IgG1+ plasma cells (F) as compared to the nontransgenic controls. Panel H also illustrates that transgenic lymph nodes had a substantial infiltration of multinucleated inflammatory giant cells (arrowheads). Magnification of × 2 or × 40 as indicated.
Human IL-17E transgenic lymph nodes revealed lymphadenopathy.
Histologic sections of nontransgenic control littermates (left panels) and transgenic (right panels) mesenteric lymph nodes. Panels A, B, G, and H are stained with hematoxylin and eosin; panels C and D, B220-stained sections; E and F, mouse IgG1-stained sections. All panels illustrate markedly enlarged transgenic lymph nodes disrupted of their normal architecture by the infiltration of eosinophils (B, asterisk; H, arrows), B220+ B lymphocytes (D), and mouse IgG1+ plasma cells (F) as compared to the nontransgenic controls. Panel H also illustrates that transgenic lymph nodes had a substantial infiltration of multinucleated inflammatory giant cells (arrowheads). Magnification of × 2 or × 40 as indicated.
Splenomegaly and bone marrow myeloid hyperplasia in the hIL-17E transgenic mice.
Transgenic spleen (B,D) and bone marrow (F) illustrate B220+ lymphoid hyperplasia (arrows) and marked eosinophilic myeloid hyperplasia, respectively, compared to the nontransgenic controls (A,C,E). The transgenic spleen also exhibited eosinophilic hyperplasia in the red pulp (D). Tissues are stained with hematoxylin and eosin (H&E) or B220 as indicated; magnification of × 2 or × 40 as indicated.
Splenomegaly and bone marrow myeloid hyperplasia in the hIL-17E transgenic mice.
Transgenic spleen (B,D) and bone marrow (F) illustrate B220+ lymphoid hyperplasia (arrows) and marked eosinophilic myeloid hyperplasia, respectively, compared to the nontransgenic controls (A,C,E). The transgenic spleen also exhibited eosinophilic hyperplasia in the red pulp (D). Tissues are stained with hematoxylin and eosin (H&E) or B220 as indicated; magnification of × 2 or × 40 as indicated.
To examine leukocyte populations in the IL-17E transgenic mice, cells from peripheral blood, mesenteric lymph nodes, spleen, bone marrow, and thymus were analyzed by flow cytometry using antibodies against the following lineage-specific surface markers: CD3, CD4, CD5, CD8, CD11b, CD11c, CD19, CD23, CD44, CD45, B220, CD54, CD69, CCR3, c-kit, and GR-1. In all secondary hematopoietic tissues, B cells, as indicated by CD19 expression, increased significantly (Table2). Specifically, there was a 2- to 5-fold increase of CD19+ B cells in the peripheral blood and a 2-fold increase in the spleen, whereas in the lymph nodes, this population increased up to 10-fold. CD4+ T cells were similarly increased by 2-fold in both the spleen and blood. Interestingly, CD19+ B cells in the bone marrow decreased by about 2-fold. These results suggest a role for IL-17E in lymphocyte development and in transferring B cells from the bone marrow to the peripheral compartments.
Inflammatory infiltrate populations in hIL-17E transgenic tissues
| . | CD19+ . | CD4+ . | CD3+ . | CD5+CD19+ . | CD34+CD19+ . | CCR3+ . |
|---|---|---|---|---|---|---|
| Blood (× 106cells)* | ||||||
| Nontransgenic (NTg) | 1.49 ± 0.39 | 0.78 ± 0.26 | 1.85 ± 0.55 | ND | ND | ND2-153 |
| Transgenic (Tg) | 6.17 ± 4.81 | 2.27 ± 0.78 | 3.43 ± 1.41 | ND | ND | ND2-153 |
| Spleen (× 107cells)* | ||||||
| Nontransgenic | 3.88 ± 0.94 | 1.49 ± 0.12 | 2.71 ± 0.55 | ND | ND | 0.34 ± 0.12 |
| Transgenic | 9.80 ± 0.33 | 2.72 ± 0.81 | 4.03 ± 1.40 | ND | ND | 6.78 ± 5.82 |
| Lymph node (× 106cells)†,2-155 | ||||||
| Nontransgenic | 4.20 ± 0.21 | 4.94 ± 0.14 | 8.0 ± 2.7 | 0.31 ± 0.112-155 | 0.10 ± 0.032-155 | 0.09 ± 0.062-155 |
| Transgenic | 64.12 ± 5.21 | 28.43 ± 27.37 | 43.0 ± 35.4 | 6.66 ± 0.842-155 | 4.78 ± 1.932-155 | 28.67 ± 18.482-155 |
| Bone marrow (× 106cells)‡ | ||||||
| Nontransgenic | 3.02 ± 1.61 | ND | ND | 0.08 ± 0.04 | 0.13 ± 0.06 | 0.08 ± 0.03 |
| Transgenic | 1.90 ± 1.17 | ND | ND | 0.04 ± 0.03 | 0.05 ± 0.03 | 0.74 ± 0.40 |
| . | CD19+ . | CD4+ . | CD3+ . | CD5+CD19+ . | CD34+CD19+ . | CCR3+ . |
|---|---|---|---|---|---|---|
| Blood (× 106cells)* | ||||||
| Nontransgenic (NTg) | 1.49 ± 0.39 | 0.78 ± 0.26 | 1.85 ± 0.55 | ND | ND | ND2-153 |
| Transgenic (Tg) | 6.17 ± 4.81 | 2.27 ± 0.78 | 3.43 ± 1.41 | ND | ND | ND2-153 |
| Spleen (× 107cells)* | ||||||
| Nontransgenic | 3.88 ± 0.94 | 1.49 ± 0.12 | 2.71 ± 0.55 | ND | ND | 0.34 ± 0.12 |
| Transgenic | 9.80 ± 0.33 | 2.72 ± 0.81 | 4.03 ± 1.40 | ND | ND | 6.78 ± 5.82 |
| Lymph node (× 106cells)†,2-155 | ||||||
| Nontransgenic | 4.20 ± 0.21 | 4.94 ± 0.14 | 8.0 ± 2.7 | 0.31 ± 0.112-155 | 0.10 ± 0.032-155 | 0.09 ± 0.062-155 |
| Transgenic | 64.12 ± 5.21 | 28.43 ± 27.37 | 43.0 ± 35.4 | 6.66 ± 0.842-155 | 4.78 ± 1.932-155 | 28.67 ± 18.482-155 |
| Bone marrow (× 106cells)‡ | ||||||
| Nontransgenic | 3.02 ± 1.61 | ND | ND | 0.08 ± 0.04 | 0.13 ± 0.06 | 0.08 ± 0.03 |
| Transgenic | 1.90 ± 1.17 | ND | ND | 0.04 ± 0.03 | 0.05 ± 0.03 | 0.74 ± 0.40 |
Populations are representative of transgenic founders or F1 progeny and were determined by FACS analysis using conjugated antibodies against indicated markers. Values reported as mean ± SEM (absolute number of cells); ND indicates not determined.
NTg, n = 5; Tg, n = 6.
NTg, n = 2; Tg, n = 4.
NTg, n = 5; Tg, n = 10.
Indicates eosinophil values reported in Table 1 by CBC analysis.
CD5+CD19+ and CD34+CD19+, NTg, n = 3; Tg, n = 3. CCR3+, NTg, n = 4; Tg, n = 6; all values are × 106cells.
Immunophenotyping of eosinophils by expression of the eotaxin receptor, CCR3, revealed significant increases in eosinophils in all of the lymphohematopoietic tissues. A striking 300-fold increase in CCR3+ eosinophils was identified in the transgenic lymph nodes. An 8-fold increase in CCR3+ eosinophils in the bone marrow and an 18-fold increase in the spleen was also observed.
Overexpression of hIL-17E results in marked bone marrow eosinophilia and increase in CD5+CD19+ and CD34+CD19+ B lymphocytes
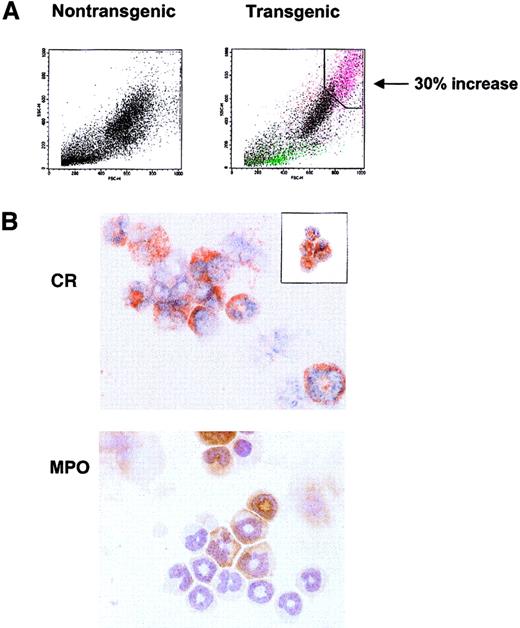
The histology and complete blood count (CBC) studies revealed a large, highly granular CCR3+ population in the spleen, blood, lymph node, and bone marrow that could not be singularly identified. To characterize these cells, flow cytometric analysis of bone marrow revealed a novel cell population observed only in the transgenic mice. This population constituting up to 30% of the total transgenic marrow nucleated cells possessed high side-scatter (high granularity) properties that did not correlate with scatter characteristics typical of normal hematopoietic cells (Figure5A). To identify the specific leukocytes (or precursors) comprising this increased population, FACS sorts gated on this high scatter population in the transgenic marrow revealed a mixed population of eosinophilic and neutrophilic cells, as seen by Congo Red (CR) and myeloperoxidase (MPO) staining, respectively (Figure5A,B). This population consisted largely of immature, band eosinophils with a smaller percentage of immature neutrophils. An additional sort of this marrow population successfully identified eosinophils (Figure5B, inset).
Distinct granular populations arise in the hIL-17E transgenic bone marrow.
(A) FACS light scatter dot plots of nontransgenic control (left plot) and transgenic (right plot) bone marrow cells, identifying novel highly granular, increased population in the transgenic animal. This population is indicated in pink, correlating to a 30% increase over the control. Plots represent one transgenic animal (Tg, n = 5; all displayed similar phenotype). (B) Sort of the transgenic granulocytic population reveals immature eosinophils and neutrophils comprise this population. Bone marrow from one high-expressing hIL-17E transgenic line was sorted based on high scatter properties identified in panel A and cytospots of sorted cells were stained with either CR (upper panel) or MPO (lower panel). Inset of CR staining represents transgenic bone marrow sort using CD11c-FITC and CD45-PE marker as well as light scatter plots.
Distinct granular populations arise in the hIL-17E transgenic bone marrow.
(A) FACS light scatter dot plots of nontransgenic control (left plot) and transgenic (right plot) bone marrow cells, identifying novel highly granular, increased population in the transgenic animal. This population is indicated in pink, correlating to a 30% increase over the control. Plots represent one transgenic animal (Tg, n = 5; all displayed similar phenotype). (B) Sort of the transgenic granulocytic population reveals immature eosinophils and neutrophils comprise this population. Bone marrow from one high-expressing hIL-17E transgenic line was sorted based on high scatter properties identified in panel A and cytospots of sorted cells were stained with either CR (upper panel) or MPO (lower panel). Inset of CR staining represents transgenic bone marrow sort using CD11c-FITC and CD45-PE marker as well as light scatter plots.
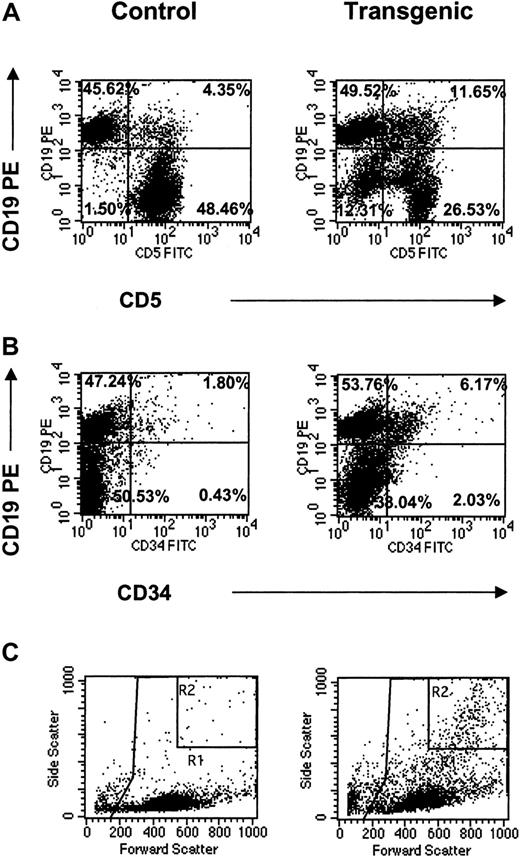
The appearance of these distinct myeloid populations in the bone marrow suggested abnormal hematopoiesis in the transgenic animals. In all lymphoid tissues, these cell populations were analyzed for the coexpression of surface antigens identified in leukemic transformation. These markers included CD5, CD34, and CD19.33-36 The most striking observation was increased numbers of CD19+ cells expressing either CD5 or CD34 in the lymph nodes (Figure6). Specifically, CD5+CD19+ cells increased more than 20-fold and CD34+CD19+ cells increased more than 30-fold compared to the littermate controls (Figure 6A,B; Table 2). Furthermore, light scatter plots of the transgenic lymph nodes revealed the presence of the highly granular (eosinophilic) population consistently seen by FACS analysis in the transgenic bone marrow as well as other hematopoietic tissues (Figure 6C). Gating indicated that this granular population correlated with both CD5+CD19+- and CD34+CD19+-coexpressing myeloid and lymphoid cells (data not shown). Along with the CD5+CD19+ double-positive population, this granular population accounted for a CD5−CD19−double-negative population, which increased 10-fold over the controls (Figure 6A). Given these data, it is suggested that IL-17E acts pleiotropically to increase various myeloid and lymphoid populations that coexpress progenitor markers, which appear to originate in the bone marrow and are identified in the periphery.
CD19+ populations in transgenic lymph nodes.
FACS analysis of CD19+ populations in mesenteric lymph nodes from control and transgenic mice analyzed for coexpression of CD5 (A) or CD34 (B) surface antigens. Light scatter plots (C) are also shown with important gates indicated (R1 = total leukocyte gate, on which the fluorescent plots were gated; R2 = increased granular population identified in the transgenic mice). Percentages for all fluorescent populations are indicated. Data are representative of one animal per plot.
CD19+ populations in transgenic lymph nodes.
FACS analysis of CD19+ populations in mesenteric lymph nodes from control and transgenic mice analyzed for coexpression of CD5 (A) or CD34 (B) surface antigens. Light scatter plots (C) are also shown with important gates indicated (R1 = total leukocyte gate, on which the fluorescent plots were gated; R2 = increased granular population identified in the transgenic mice). Percentages for all fluorescent populations are indicated. Data are representative of one animal per plot.
Overexpression of hIL-17E results in high serum levels of proinflammatory cytokines and Igs
To determine the factors responsible for these increases in leukocytes, that is, direct IL-17E activation of these cell populations or indirectly through increased cytokine production that can induce these cells to proliferate, the blood sera of the transgenic mice were examined for changes in levels of various cytokines. The Th1 cytokines (IL-2, IFN-γ, and TNF-α), Th2 cytokines (IL-4, IL-5, IL-6, IL-10), and other cytokines (IL-1α, IL-1β, IL-3, G-CSF, GM-CSF, and eotaxin) were examined in the mice.37 These cytokines are known to activate T and B cells, eosinophils, and other cell populations. IL-5 levels increased 60- to 80-fold in 4 of the transgenic animals (Table 3). Northern analysis indicated that these 4 transgenic animals expressed high levels of IL-17E mRNA, whereas the remaining 3 transgenic mice had undetectable transcript levels (data not shown). IL-2 and IL-4 serum concentrations increased in 3 of the transgenic animals, at levels 20- to 150-fold and 180- to 500-fold, respectively; and IL-1β and GM-CSF increased in one animal each, both expressing high levels of the transgenic construct. G-CSF serum levels increased from 2- to 8-fold above negative control levels, whereas the eosinophil-specific chemokine, eotaxin, increased similarly 2- to 5-fold above controls. The only increased Th1 cytokine was IFN-γ, at levels ranging from 12- to 2500-fold higher than control levels. IL-3, IL-6, and IL-10 were unchanged. Thus increased cytokine levels due to overexpressed hIL-17E may be responsible for increases in cellularity in the transgenic mice.
Increased cytokine and immunoglobulin levels in transgenic blood serum
| Cytokines (pg/mL) . | IL-2 . | IL-4 . | IL-5 . | G-CSF . | Eotaxin . | IFN-γ . |
|---|---|---|---|---|---|---|
| Nontransgenic | ND | ND | ND | 87.5 | 226.2 | 3.32 |
| Transgenic | 65.263-151 | 346.633-151 | 97.623-150 | 328.03-151 | 721.2 | 2264.9 |
| Fold increase (range) | 20-150× | 180-500× | 60-80× | 2-8× | 2-5× | 12-2500× |
| Cytokines (pg/mL) . | IL-2 . | IL-4 . | IL-5 . | G-CSF . | Eotaxin . | IFN-γ . |
|---|---|---|---|---|---|---|
| Nontransgenic | ND | ND | ND | 87.5 | 226.2 | 3.32 |
| Transgenic | 65.263-151 | 346.633-151 | 97.623-150 | 328.03-151 | 721.2 | 2264.9 |
| Fold increase (range) | 20-150× | 180-500× | 60-80× | 2-8× | 2-5× | 12-2500× |
Nontransgenic and transgenic sera were measured by ELISA for the indicated cytokines and immunoglobulins. Values are reported as the mean for NTg, n = 5 animals and Tg, n = 4 animals, unless otherwise indicated for cytokines.
ND indicates not detectable.
Tg, n = 7.
Tg, n = 3.
Because B cells were increased in the blood, changes in serum Ig levels were examined by ELISA. IgE, IgM, and IgG1 were increased 40- to 80-fold, 10- to 30-fold, and 8- to 20-fold, respectively (Table4). Similar to the cytokines, the increases in most of the Igs are indicative of a Th2 response. All levels of other classes and subclasses of Ig were unchanged in the transgenic mice (data not shown).
Increased immunoglobulin levels in transgenic blood serum
| Immunoglobulins . | IgE (μg/mL) . | IgM (μg/mL) . | IgG1 (μg/mL) . | IgG2A (μg/mL) . |
|---|---|---|---|---|
| Nontransgenic | 0.7 | 213.0 | 164.2 | 83.0 |
| Transgenic | 49.3 | 3824.4 | 1852.6 | 47.0 |
| Fold increase (range) | 40-80× | 10-30× | 8-20× | — |
| Immunoglobulins . | IgE (μg/mL) . | IgM (μg/mL) . | IgG1 (μg/mL) . | IgG2A (μg/mL) . |
|---|---|---|---|---|
| Nontransgenic | 0.7 | 213.0 | 164.2 | 83.0 |
| Transgenic | 49.3 | 3824.4 | 1852.6 | 47.0 |
| Fold increase (range) | 40-80× | 10-30× | 8-20× | — |
Nontransgenic and transgenic sera were measured by ELISA for the indicated immunoglobulins. Values are reported as the mean for NTg, n = 4 animals and Tg, n = 5 animals.
Altered production of antigen-specific antibodies in hIL-17E transgenic mice
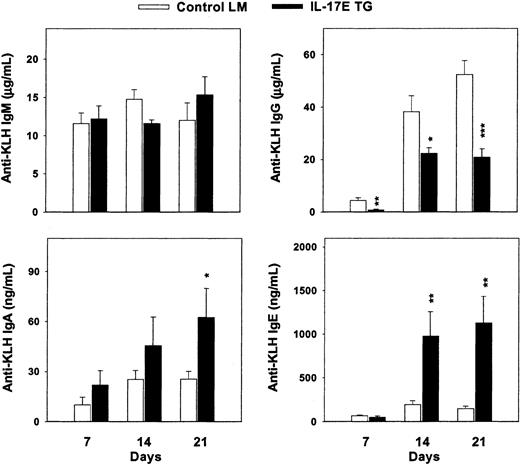
With the increased production of Th2-type cytokines and Igs in hIL-17E transgenic mice, the immunologic response of these animals on antigen challenge was studied. Exposure to antigen may result in antibody production that corroborates the serum Ig levels. Both hIL-17E transgenic mice and nontransgenic littermate controls were injected subcutaneously with 100 μg KLH antigen. Serum anti-KLH Ig levels were analyzed 7, 14, and 21 days after injection. Total anti-KLH IgG levels decreased, with a maximum decrease of 2-fold by day 21 compared to the controls (Figure 7). In contrast, anti-KLH IgE and IgA production increased significantly in the transgenic mice, at 6-fold and 2-fold, respectively, over the control Ig production. IgM levels were similarly increased between the 2 groups. These results indicate that in the presence of overexpressed hIL-17E, antigenic challenge alters antibody production, possibly by increased B-cell production of IgA and IgE or by increased Ig switching of IgG to IgE and IgA cells.
Increased immunoglobulin production on antigenic challenge.
Human IL-17E transgenic mice and nontransgenic littermate controls were immunized with 100 μg KLH on day 0. Blood draws on days 7, 14, and 21 were analyzed by ELISA for anti-KLH antibody production in the serum. *P < 0.05; **P < .01; ***P < .001.
Increased immunoglobulin production on antigenic challenge.
Human IL-17E transgenic mice and nontransgenic littermate controls were immunized with 100 μg KLH on day 0. Blood draws on days 7, 14, and 21 were analyzed by ELISA for anti-KLH antibody production in the serum. *P < 0.05; **P < .01; ***P < .001.
Overexpression of hIL-17E is associated with up-regulated expression of IL-17Rh1
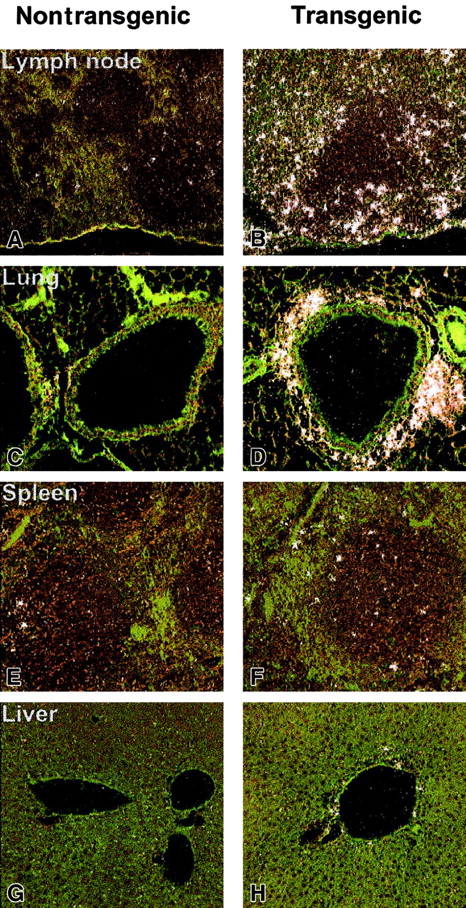
To further assess a direct mechanism by which hIL-17E affects hematopoietic cell development in the transgenic mice, expression levels of the IL-17 family of receptors were examined by in situ hybridization. IL-17E has previously been shown to bind to IL-17Rh1 (IL-17BR/Evi27), which had initially been reported as a binding receptor for the IL-17B ligand.21 22 In a full tissue panel examined, liver, lung, spleen, and lymph node, IL-17Rh1 expression was strikingly higher in the transgenic tissues (Figure8). Strongest expression was present in the lymph node and lung, specifically in areas of inflammatory cellular infiltration (Figure 8B,D). The increased expression of IL-17Rh1 in the lymph node appeared to occur in the perifollicular zones. In the lung, a strong cluster of IL-17Rh1+ cells was present around the airways (Figure 8C,D). A moderate increase in IL-17Rh1 expression was seen in the hIL-17E transgenic spleen and liver. In all of the nontransgenic littermate tissues examined, very little receptor expression was observed, indicating that increased levels of hIL-17E resulted in increased receptor expression. A sense control probe hybridized to normal mouse tissue did not produce a detectable, nonspecific binding signal.
In situ hybridization of IL-17Rh1 of IL-17E transgenic tissues.
Tissues from nontransgenic (A,C,E,G) and hIL-17E transgenic (B,D,F,H) mice. Lymph node, lung, spleen, and liver samples were hybridized in situ with a 33P-labeled anti–IL-17Rh1 riboprobe as described in “Materials and methods.” Highly increased IL-17Rh1expression in lymph nodes and areas of inflammation in the lung are indicated. Increased expression is also present in spleen and liver.
In situ hybridization of IL-17Rh1 of IL-17E transgenic tissues.
Tissues from nontransgenic (A,C,E,G) and hIL-17E transgenic (B,D,F,H) mice. Lymph node, lung, spleen, and liver samples were hybridized in situ with a 33P-labeled anti–IL-17Rh1 riboprobe as described in “Materials and methods.” Highly increased IL-17Rh1expression in lymph nodes and areas of inflammation in the lung are indicated. Increased expression is also present in spleen and liver.
Discussion
We have identified and cloned a novel cytokine that is a member of the IL-17 family. Identification of this molecule was made through EST searches of GenBank and Amgen proprietary databases using amino acid motifs centered on conserved cysteine residues. This molecule, hIL-17E, is the fifth member of the IL-17 family to be described, possessing 25% to 35% homology to other IL-17 family members and a conserved secondary protein structure of a distinct N-terminal signal sequence followed by a mature protein. The full-length sequence of IL-17E consists of 161 amino acid residues with a predicted monomeric mass of 17 kDa. Western blot analysis of IL-17E, however, reveals a molecular mass of 24.7 kDa, suggesting posttranslational modification.
Recently, 2 other IL-17E molecules, IL-17E and IL-25, have been reported.22,25 Although possessing more than 95% homology to our molecule, significant differences exist in both the N- and C-termini among the proteins. Specifically, the IL-17E reported by Lee et al22 possesses a longer N-terminal signal sequence, and both IL-17E and IL-25, reported by Fort et al,25 possess amino acid differences in the C-terminus. These differences could be the result of polymorphisms among individuals, molecular isoform distinctions, or simply different genes encoding distinct molecules in the IL-17 family. Moreover, differences in expression patterns were observed among these molecules. A generally low expression pattern has been reported for the IL-17E molecule described by Lee et al,22 whereas we observe high expression in the testis as well as moderate expression in spleen and prostate. IL-25 mRNA expression also differed with highest expression seen in the gastrointestinal tract and uterus with lower levels in the kidney and lungs.25
Overexpression of hIL-17E in mice reported herein was used to understand the biologic significance of this molecule. All transgenic animals were healthy, viable, and reproductively normal. Species cross-reactivity between the human ligand with the murine receptor is apparent, as striking phenotypes were observed. The predominant phenotype identified in the transgenic mice was displayed in organs of the hematopoietic and lymphoid systems and included remarkable inflammatory infiltrates. Two particularly interesting and distinct populations of hematopoietic cells, eosinophils and CD19+ B lymphoid cells, were significantly elevated in the overexpressing mice. A striking, ubiquitous eosinophilia was detected in most primary and secondary sites of hematopoiesis. In the bone marrow, these eosinophils were predominantly immature band cells, suggesting that there was an increase in newly developed eosinophils as a result of IL-17E overexpression. In the secondary tissues, eosinophils were largely mature bilobed cells. Also identified in this increased population in the bone marrow was the presence of immature band neutrophils, corresponding to increased circulating levels of neutrophils in the blood. CD19+ B lymphoid cells were increased in the spleen, lymph nodes, and other secondary lymphohematopoietic tissues. However, CD19+ cells decreased in the bone marrow of the transgenic mice, indicating that these cells may be prematurely released to the periphery and may account for the CD19+ cell increases observed there.
Two potential mechanisms might explain the distinct phenotypic changes observed in the hIL-17E transgenic mice. One mechanism would be through direct stimulation of eosinophils or B cells (or their precursors) by IL-17E. To directly stimulate these cells, the receptor for IL-17E necessarily needs to be expressed on their cell surfaces. We show that IL-17Rh1, a binding receptor for IL-17E, is up-regulated in inflammatory tissues but is not present in high copy number on either murine eosinophils or B cells (data not shown). Preliminary studies of direct treatment of murine B and T cells with hIL-17E does not induce proliferation of these cells (data not shown).
Although these results indicate that IL-17E does not directly induce proliferation of mature lymphocytes, IL-17E may act on precursors of eosinophils or B cells, which may express the receptor. FACS analysis revealed up-regulation of the IL-17E binding receptor on the expanded immature myeloid population in marrow described above (data not shown). This supports the possibility that IL-17E may directly stimulate a progenitor cell population that may undergo subsequent differentiation and migration to peripheral tissues. Lastly, another IL-17E receptor may exist on either mature or progenitor cell populations, which could induce proliferation or maturation of these cells.
A second mechanism is the indirect role of IL-17E on the production of secondary cytokines, specifically those known to induce eosinophil or lymphocyte production. We report that hIL-17E overexpression in vivo or stimulation in vitro leads to the induction of numerous cytokines, which is consistent with the ability of other IL-17 family members to induce cytokine production from various stromal cells.3,4,9,10,12,14-18,20-22,24,38 In particular, we show that overexpression of hIL-17E resulted in elevated levels of IL-5 and eotaxin in mouse serum. Furthermore, IL-17E stimulated IL-5 production in conditioned medium of treated whole human marrow cultures (data not shown). Induction of IL-5 could result in the development of eosinophils observed in the periphery and marrow of the hIL-17E transgenic mice, because IL-5 is a primary cytokine responsible for eosinophilopoiesis.39-42 Both IL-5 and the chemokine eotaxin have been implicated in the production and recruitment of eosinophils to sites of allergic or asthmatic reactions, particularly in the lungs.42 43 With increased IL-17Rh1 expression on cells in the transgenic lungs, IL-17E may have a direct stimulatory effect on these cells resulting in inflammation, as well as in inducing IL-5 and eotaxin production to recruit eosinophils to the lungs.
Transgenic overexpression of IL-5 leads to significant eosinophilia, the production of autoantibodies, and abnormal and ectopic bone formation.39-41 Although similar phenotypic characteristics were identified between IL-17E and IL-5 transgenic mice, including eosinophilia, increased WBC counts, and splenomegaly, we did not observe any abnormal bone development. Autoantibody production in hIL-17E transgenic mice may occur, because increased Igs were observed. This suggests that although IL-17E promotes eosinophilia similarly to IL-5, likely through the induction of IL-5 production, IL-17E displays distinct biologic functions differing from IL-5.
In addition to increased IL-5 levels in hIL-17E overexpressing mice, we detected increases in other cytokines including IL-2, IL-4, G-CSF, and eotaxin. These cytokines promote T-cell activation (IL-2), B-cell proliferation and differentiation (IL-4 and IL-5), and hematopoiesis, particularly of progenitor cells (G-CSF).44-53 IL-4 and IL-5 are produced by CD4+ Th2 helper cells. Interestingly, we observed an increase in CD4+ T cells in the transgenic mice, which may include increased Th2 differentiated cells. As stated, the direct mechanism of cytokine production by hIL-17E is unclear. However, it is likely that other cells including stromal cells, dendritic cells, macrophages, or other cell populations (rather than the Th2 cytokine-producing T cells) are induced by IL-17E to produce these secondary cytokines. In further support of this indirect mechanism, we have observed that recombinant hIL-17E protein can bind to and stimulate cytokine release from the leukemic monocytic cell line THP-1 (data not shown). Lee et al reported induction of IL-8 on IL-17E treatment of renal carcinoma cell lines as well.22Additional studies are needed to further understand the role of IL-17E in inducing these cytokines to promote all these effects.
Altered antibody production, both in the transgenic serum and antigen-challenged mice, was observed, demonstrating a function of hIL-17E in regulating humoral immune responses. The increased production of IgE and IgA in response to KLH antigen suggested that overexpressed hIL-17E interfered with normal Ig production. The increases in both total and anti-KLH IgE in the transgenic mice correlate with the eosinophilia, and along with the increases in IL-5, these data taken together may suggest a role for IL-17E to mount a Th2-type response in the animals.54,55 A more extensive correlation of these phenotypes of increased Igs, cytokines (particularly IL-5 and eotaxin), and inflammation observed in the lungs may further suggest a role for IL-17E in the pathogenesis of allergic diseases, especially asthma.55 56 Further studies to define the role of IL-17E in regulating antibody production, particularly in the pathogenesis of asthma, will need to be performed.
The increased coexpression of CD5 and CD34 on CD19+ cells, observed most significantly in the transgenic lymph nodes, suggested that chronic overexpression of hIL-17E may be related to certain proleukemic transformation events during hematopoietic and lymphopoietic development in the transgenic mice. These increased double-positive populations consisted of lymphocytes and novel myeloid populations produced in the transgenic mice. The coexpression of these markers characterizes many leukemias, including B-cell chronic lymphocytic leukemia (B-CLL), CLL, and acute myeloid leukemia (AML).33-36,57,58 These increased myeloid populations expressing CD5 and CD34 suggests a possible preleukemic state in the hIL-17E transgenic animals, possibly due to a loss of lineage commitment among these myeloidlike cells. In support of this, the myelocytic leukemia, AML M4Eo, is associated with marrow eosinophilia.33,55 More directly associated with IL-17E, Evi27 (IL-17Rh1) was found to be up-regulated in numerous B, T, and myeloid leukemic cell lines, including M1 (myeloid leukemia), EL-4 (thymoma), and WEHI231 (pre–B-cell lymphoma).28 To determine the possible leukemic state of the transgenic mice, an age-onset condition due to overexpression of IL-17E, and to correlate IL-17E expression and myeloid transformation events, aging studies of the transgenic mice are presently underway.
Concurrent with our study on overexpression of hIL-17E in mice, 2 studies recently reported the results of either overexpression of mIL-17E in mice or of exogenous treatment of mice with mIL-25.25,59 Several consistent observations were reported across the 3 studies including a dominant eosinophilia, inflammatory cell infiltrates in lung, liver, spleen and lymph nodes, and increases in serum levels of the cytokines IL-4 and IL-5 and the immunoglobulin IgE. However, there were certain distinct differences noted among the studies. In particular, Pan et al reported that overexpression of mIL-17E resulted in growth retardation and increased serum bilirubin levels.59 Neither Fort et al25 nor our own study reported these pathologic observations. Secondly, in contrast to the other published observations, we detected increases in B- and T-lymphoid cells, abnormal surface marker expression on these cells, increases in several Ig classes, and changes in Ig levels as a result of antigenic challenge. The distinct differences among the 3 studies could result from differences in gene promoter usage (ie, tissue nonspecific versus tissue specific), the gene sequences used (Pan et al59 and Fort et al25 used murine IL-17E versus our study, which used human IL-17E), as well as the genetic background on which the transgenic animals were produced or were treated with exogenous IL-17E protein. A more generic function of the IL-17 family to stimulate granulopoiesis is further supported by our study as well.19 Thus our study provides significant corroboration of the biologic consequence of IL-17E overexpression and provides new and exciting insights into the critical role that IL-17E plays in inflammation and possibly in leukemic transformation.
We have shown that IL-17E plays a role in the proinflammatory response, similar to its related family members. This response results in increases in myeloid and lymphoid cell populations, induction of proinflammatory cytokine production or release, and infiltration of eosinophilic myeloid cells throughout the tissues in transgenic overexpressing mice. Moreover, overexpression of IL-17E may be correlated with certain oncogenic transformation events in hematopoietic cells. The direct association of IL-17E with inflammatory or oncogenic diseases requires further investigation and may define IL-17E as not only a key regulatory cytokine but as a potential therapeutic target directed against such diseases including arthritis, asthma, allergy, and leukemias.
We thank James McCabe, Kathy Christensen, Carl Whitley, Laura Chiu, Diane Duryea, and the Amgen Transgenics and Histology Laboratories for extensive assistance with animal production and maintenance, flow cytometry, and tissue preparation. We thank Jenny Hu for preparation of the riboprobe used for in situ hybridization.
Prepublished online as Blood First Edition Paper, May 31, 2002; DOI 10.1182/blood-2002-01-0012.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mee Rhan Kim, One Amgen Center Dr, MS 35-1-B, Thousand Oaks, CA 91302; e-mail: mkim@amgen.com.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal