Abstract
High-titer self-inactivating human immunodeficiency virus type-1 (HIV-1)–based vectors expressing the green fluorescent protein reporter gene that contained the central polypurine and termination tract and the woodchuck hepatitis virus posttranscriptional regulatory element were constructed. Transduction efficiency and biodistribution were determined, following systemic administration of these improved lentiviral vectors. In adult severe combined immunodeficiency (SCID) mice, efficient stable gene transfer was achieved in the liver (8.0% ± 6.0%) and spleen (24% ± 3%). Most transduced hepatocytes and nonhepatocytes were nondividing, thereby obviating the need to induce liver cell proliferation. In vivo gene transfer with this improved lentiviral vector was relatively safe since liver enzyme concentration in the plasma was only moderately and transiently elevated. In addition, nondividing major histocompatibility complex class II–positive splenic antigen-presenting cells (APCs) were efficiently transduced in SCID and normal mice. Furthermore, B cells were efficiently transduced, whereas T cells were refractory to lentiviral transduction in vivo. However, in neonatal recipients, lentiviral transduction was more widespread and included not only hepatocytes and splenic APCs but also cardiomyocytes. The present study suggests potential uses of improved lentiviral vectors for gene therapy of genetic blood disorders resulting from serum protein deficiencies, such as hemophilia, and hepatic disease. However, the use of liver-specific promoters may be warranted to circumvent inadvertent transgene expression in APCs. In addition, these improved lentiviral vectors could potentially be useful for genetic vaccination and treatment of perinatal cardiac disorders.
Introduction
Hepatocytes are attractive target cells for gene therapy of genetic blood diseases, particularly hemophilia, and hepatic disorders (eg, familial hypercholesterolemia). Providing a long-term cure of these diseases requires not only that the hepatocytes be efficiently transduced, but also that the vectors remain transcriptionally active and give rise to therapeutic levels of functional protein. We have recently demonstrated that hemophilia A could be cured in newborn hemophilic factor VIII (FVIII)–deficient mice by intravenous injection of Moloney murine leukemia virus (MoMLV)–based oncoretroviral vectors encoding human FVIII.1 In adult mice however, most hepatocytes are refractory to gene transfer performed with the use of MoMLV-based oncoretroviral vectors since disassembly of the nuclear membrane during cell division is required to allow the oncoretroviral preintegration complex to enter the nucleus.2 Hence, it would be desirable to develop gene delivery strategies that could overcome the requirement for hepatic cell division and be used in adults.
Macrophages and dendritic cells are also important target cells for gene therapy. By virtue of their potent antigen-presenting properties, they could be used to develop improved genetic vaccination strategies against cancer and infectious or autoimmune disease.3-6Furthermore, genetic modification of antigen-presenting cells (APCs) may have therapeutic implications for genetic (eg, lysosomal storage disease) or infectious diseases (eg, acquired acquired immunodeficiency syndrome, leishmaniasis) that are associated with APC dysfunction.3,7 However, genetic modification of APCs often relies on ex vivo gene transfer, which is relatively inefficient.8 9 Hence, the development of in vivo gene therapy approaches that could efficiently deliver potentially therapeutic transgenes into nondividing APCs in situ is warranted.
Human immunodeficiency virus type-1 (HIV-1)–based lentiviral vectors are attractive tools for gene transfer since sustained expression of the transgene can be achieved by the stable integration of the vector into the host cell genome.10,11 In addition, lentiviral vectors do not contain residual lentiviral genes precluding de novo expression of potentially immunogenic lentiviral proteins in transduced target cells. Lentiviral vectors can transduce both proliferating and quiescent cells, including neurons and including retina, muscle, and hematopoietic cells.10,12-16 However, lentiviral transduction in nondividing cells may be dependent on cell type, and the presence of HIV-1 accessory proteins (Vif, Vpr, Vpu, and Nef) in the vector particles may be required to achieve stable gene transfer in quiescent lymphocytes and hepatocytes.13-15 Cell cycle activation is required for infection with wild-type HIV although mitosis is not required for integration.16,17 Furthermore, while lentiviral transduction of quiescent hepatocytes is relatively inefficient,18,19 induction of hepatocyte proliferation by partial hepatectomy overcomes this limitation. A previous study concluded, however, that lentiviral vectors packaged with accessory proteins could transduce quiescent hepatocytes in vivo following intraparenchymal injection.13 Several viral determinants responsible for the active nuclear import of HIV-1 and HIV-1–based vectors have been identified.20 The HIV-1 Gag, Vpr, and Int proteins have been implicated in the nuclear import of the viral genome in nondividing cells.21-23 In addition, a 118–base pair (bp) sequence within the pol gene, designated as central DNA flap, is required in cis.20,24-28This sequence contains structural elements associated with the progress of reverse transcription and encompasses the central polypurine tract and central termination sequences (designated cPPT andCTS, respectively). Gene transfer by lentiviral vectors is limited by nuclear translocation but can be rescued by this so-called central DNA flap,28,29 which forms a triple-stranded HIV complementary DNA intermediate of reverse transcription.20 27 It is possible that this nuclear translocation pathway controls lentiviral transduction, irrespective of the proliferation status of the target cells.
In the present study, transduction of nondividing hepatocytes and APCs with improved self-inactivating lentiviral vectors was investigated. The vectors contained the central DNA flap and the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), which improves transgene expression.30 31 The lentiviral biodistribution profile was analyzed following systemic vector administration into adult and newborn mice.
Materials and methods
Cell lines and culture conditions
The 293T human embryonic kidney and NIH-3T3 mouse fibroblastic cell lines were cultured in Dulbecco modified Eagle medium supplemented with 2 mM l-glutamine, 100 IU/mL penicillin, 100 μg/mL, streptomycin, and 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen, Merelbeke, Belgium) (designated D10). Cells were grown in cell factories and/or single tray units (Nalge Nunc International, Roskilde, Denmark) during viral vector production in medium with 10% Nu-Serum IV instead of FBS (“serum-free” medium) (Becton Dickinson, Erembodegem, Belgium).
Animals
Severe combined immunodeficiency mice (SCID) mice were obtained from M&B (Ry, Denmark). Immunocompetent hemophilic FVIII–deficient mice containing a disruption of the murine FVIII gene in exon 16 have been described previously32 and were kindly provided by Dr H. Kazazian (University of Pennsylvania, Philadelphia). HIV–green fluorescent protein (HIV-GFP) vectors were injected via tail vein into 9-week-old adult SCID or 14-week-old adult Balb/c mice or by retro-orbital injection into 2- to 3-day-old neonatal recipient FVIII-deficient mice as described previously.1 All vector preparations were supplemented with 40 μg/mL polybrene (Sigma, Bornem, Belgium). Similarly, littermates were injected with phosphate-buffered saline (PBS) and polybrene as controls. Plasma samples were obtained from some recipient mice by retro-orbital bleeding at different intervals to assess liver toxicity by measuring serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels on a Modular System (Roche, Basel, Switzerland) according to the manufacturer's instructions. To determine whether transduced cells were cycling or quiescent, at 1 day prior to vector injection an osmotic minipump (model 2001, Alzet; Iffa Credo, Brussels, Belgium) was implanted subcutaneously into SCID and Balb/c mice in some experiments to administer bromodeoxyuridine (BrdU) (1 mg/d for 7 days) (Sigma). Animals received general anesthesia during surgical procedures and were killed prior to dissection. All animal experiments were approved by the Animal Ethical Commission of the University of Leuven.
Construction of lentiviral vectors
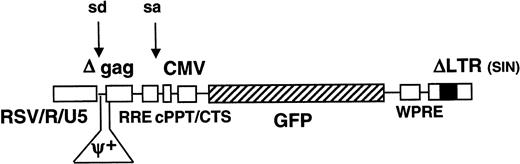
The HIV-GFP vector (aliaspRRL-SIN-18-cPPT-CMV-EGFP-WPRE) has been described previously and was similar to a self-inactivating construct,33,34 except that an additional 118-bp sequence containing the cPPT and CTS had been introduced in the vector. In addition, the WPRE backbone was included to augment transgene expression and/or viral vector titer28 30 (Figure 1).
Schematic representation of lentiviral vector.
The full-length lentiviral genomic transcript is encoded by a Rous sarcoma virus (RSV)/HIVLTR RU5 chimeric promoter. The vector contains a mutation in the 3′ long terminal repeat (LTR) that is copied onto the 5′ LTR, which results in the self-inactivation of the vector upon transduction. Consequently, only the internal CMVpromoter remains active and expresses the GFP reporter gene. The WPRE, cPPT, CTS,Rev-responsive element (RRE), splice donor and acceptor sites, and residual gag sequence (Δgag) are indicated.
Schematic representation of lentiviral vector.
The full-length lentiviral genomic transcript is encoded by a Rous sarcoma virus (RSV)/HIVLTR RU5 chimeric promoter. The vector contains a mutation in the 3′ long terminal repeat (LTR) that is copied onto the 5′ LTR, which results in the self-inactivation of the vector upon transduction. Consequently, only the internal CMVpromoter remains active and expresses the GFP reporter gene. The WPRE, cPPT, CTS,Rev-responsive element (RRE), splice donor and acceptor sites, and residual gag sequence (Δgag) are indicated.
Production and concentration of lentiviral vectors
When cells were 90% to 95% confluent, they were transfected by means of a modified calcium phosphate transfection protocol (Life Technologies, Merelbeke, Belgium). Plasmid DNA was extracted by means of Qiagen Maxi or Mega kits (Westburg, Leusden, The Netherlands). The following amount of DNA was used per cell factory: 3 mg HIV-GFP vector, 1.5 mg pMDL gag/pol RRE helper plasmid, 1.5 mg Rev-expressing plasmid, and 1.5 mgpCI-VSV-G envelope-encoding plasmid; these were previously described.34,35 At 24 hours after transfection, medium was removed, cells were washed with PBS, and 1 L cell culture medium containing 1.1 mg/mL sodium butyrate (Sigma) was added per cell factory. Conditioned medium was collected every 24 hours during the subsequent 2 days. Viral vector concentration was performed after filtration with a 0.45-μm filter (Corning, Elscolab, Kruibeke, Belgium) at 4500 rpm overnight at 4°C with an SLA1500 rotor.1 The pelleted viral vector particles were resuspended thoroughly in PBS and frozen in dry ice before being stored at −80°C. Typically, vectors were concentrated 1000-fold. Alternatively, vector concentration was performed by ultrafiltration. To achieve this, conditioned medium containing viral vectors was collected in serum-free medium. After prefiltration by means of a 0.45-μm filter, the conditioned medium was applied on a Centricon Plus-80 centrifugal filter device (Millipore, Bedford, MA) (100-kd nominal MW cutoff) and centrifuged for 100 minutes at 1500 rpm (10°C). The retentate could be retrieved by centrifuging for 2 minutes at 2000 rpm (10°C). A 200-fold concentration could be achieved with this method. Functional titer of the HIV-GFP vector was determined by transducing NIH-3T3 fibroblasts seeded in a 6-well plate (7.5 × 104 cells per well) with serially diluted unconcentrated or concentrated vector-containing supernatant supplemented with 8 μg/mL polybrene. Since GFP was used as a marker, the number of fluorescent cells per microscopic field (5 fields) were counted 48 hours after transfection, and the total number of fluorescent cells per mL viral vector-containing supernatant could be calculated on the basis of that number (titer = transducing units per milliliter [TU/mL]).
Immunohistochemistry
Mice were dissected, and organs (liver, spleen, lungs, heart, brain, stomach, gut, kidney, testes, or ovaries) were fixed overnight in 4% formaldehyde in PBS. After being washed in PBS, the organs were incubated overnight in sucrose (20% in PBS) and subsequently embedded in Tissue-Tek (Laborimpex, Brussels, Belgium) and frozen on 2-methyl butane, cooled in liquid nitrogen. Then, 7-μm sections were mounted on polylysine-coated superfrost slides. BrdU incorporation was quantified by immunohistochemistry. First, the cells were permeabilized by incubating the sections for 3 hours in Tris/borate/saline (TBS) with 0.1% Triton, and the DNA was subsequently unwound with DNAse I (Roche) (250 Kunitz units per section in PBS with Ca2+/Mg2+ for 30 minutes). After being washed in TBS and blocked for 45 minutes in preimmune goat serum diluted 5-fold in TBS with 2% bovine serum albumin (BSA), sections were incubated overnight at 4°C with rat anti–BrdU-specific antibodies (Harlan Seralab, Loughborough, United Kingdom) diluted 2-fold in TBS/BSA. Sections were rinsed in TBS and incubated for 45 minutes with goat anti–rat immunoglobulin G (IgG)–Alexa 568 (Molecular Probes, Eugene, OR) diluted 1/500 in TBS/BSA. Sections were washed and mounted with the Prolong Antifade kit (Molecular Probes). The duodenum was used as positive control for BrdU incorporation. Immunohistochemistry for laminin and smooth muscle cell actin was performed as described previously.36 37 Sections were analyzed by confocal microscopy (Axiovert, Zeiss, Zena, Germany). Quantification of the percentages of transduced hepatocytes was based on 4′6-diamidino-2-phenylindole 2HCl (DAPI)–stained (Vector Laboratories, Burlingame, CA) sections to visualize the nuclei of individual cells according to the manufacturer's instructions. A total of 30 DAPI-stained, randomly selected liver sections (from 3 different mice, 2 lobes per mice, 5 sections per lobe) were used for quantification by counting the number of transduced hepatocytes versus the total number of DAPI-stained nuclei. BrdU quantifications in liver (n = 3) and spleen (n = 2) from recipient SCID animals were based on 30 and 10 randomly selected sections, respectively. BrdU staining in the intestine of SCID mice and in the spleen of Balb/c mice (n = 3) injected with 109HIV-GFP was used as control to validate the sensitivity of the assay.
Extraction of tissue DNA and polymerase chain reaction
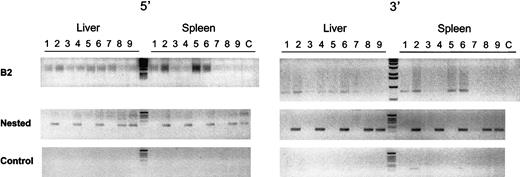
High–molecular weight DNA from liver and spleen of treated and control mice was isolated by DNeasy Kit (Qiagen, Valencia, CA). To detect integrated vector DNA in the liver and spleen of treated and control mice, a nested polymerase chain reaction (PCR) with primers specific for ubiquitous repeats found in the mouse genome (B2 family repeats38) and lentiviral vector was used. For the B2-vector PCR, 200 ng extracted genomic DNA was used as template for 2 different primers pairs: one at the 5′ end (B2 sense, 5′-GGCTGGTGAGATGGTTCAGT-3′; 5NC2 antisense, 5′-GAGTCCTGCGTCGAGAGAG-3′), and the other at the 3′ end (B2 antisense, 5′-ATATGTAAGTACACTGTAGC-3′; WPRE sense, 5′-CTTTCCATGGCTGCTCGC-3′ of the vector genome). The following amplification conditions were used for the 5′ B2-PCR: 10 minutes at 95°C once, 1 minute at 94°C, 1 minute at 57°C, 1 minute at 72°C, repeated 30 times, followed by an extra 7 minutes' extension at 72°C. The amplification conditions for the 3′ B2-PCR were similar to the 5′ B2-PCR but differed in that an annealing temperature of 49°C was used. A total volume of 50 μL was used containing 0.2 mM deoxynucleoside 5′-triphosphates, 1.5 mM MgCl2, 0.4 μM each of the primers, and 2.5 U Ampli Taq Gold (Perkin Elmer, Boston, MA). After this amplification, a nested PCR was performed with 2.5 μL B2-vector PCR product with 2 different internal primers in the vector genome. For the 5′ nested PCR, the primers used were as follows: LTR9 sense, 5′-GCCTCAATAAAGCTTGCCTTG-3′; U5PBS antisense, 5′-GGCGCCACTGCTAGAGATTTT-3′ amplifying a fragment of 121 bp. For the 3′ nested PCR, the primers used were as follows: Δnef sense, 5′-CGAGCTCGGTACCTTTAAGACC-3′; LTR8 antisense, 5′-TCCCAGGCTCAGATCTGGTCTAAC-3′ amplifying a fragment of 166 bp. The amplification conditions were similar to the B2-vector PCR but differed in that the annealing temperature was 55°C for the 5′ nested PCR and 58°C for the 3′ nested PCR. As control, a nested PCR was performed with the use of 10 ng nonamplified genomic DNA from liver and spleen of treated and control mice. The PCR mix was subsequently subjected to gel electrophoresis.
Flow cytometry
Splenocytes were isolated by repeatedly injecting D10 medium into the spleen with a 10-mL syringe and 24-gauge needle until most cells were removed. Cells were centrifuged and resuspended in lysis buffer (5 minutes followed by resuspension in D10) to eliminate erythrocytes. To reduce the aspecific background signals, splenocytes were preincubated with unconjugated anti-Fc receptor (rat anti–mouse CD16/CD32 FcγIII/II receptor) antibodies to block the Fc receptor. Then, one million cells were incubated with 5 μL phycoerythrin (PE)–labeled antibodies for at least 20 minutes. When the primary was not labeled, another incubation with a secondary PE-labeled antibody was carried out (PE–mouse anti–rat immunoglobulin κ light chain; Pharmingen, Erembodegem, Belgium). After washing, cells (n = 10 000) were subjected to fluorescence-activated cell sorting (FACS) analysis with a FACSVantage (Becton Dickinson, Franklin Lakes, NJ). In some experiments, GFP+ cells were enriched by FACS. The primary antibodies specific for CD4 (rat IgG2b, κ), CD19 (rat IgG2a, κ), CD31 (rat IgG2a, κ), CD11b (rat IgG2b, κ), major histocompatibility complex class II (MHC class II) (rat IgG2b, κ), CD11c (hamster IgG1, λ), CD62E (rat IgG2a, κ), CD54 (hamster IgG1, κ), CD45R/B220 (rat IgG2a, κ), CD80 (hamster IgG2, κ), CD8a (rat IgG2a, κ), CD86 (rat IgG2a, κ), and CD28 (hamster IgG2, λ; all purchased from Pharmingen), and F4/80 (rat IgG2b) (Serotec, Oxford, United Kingdom) were PE-conjugated. The antibodies specific for CD105 (rat IgG2a, κ) and CD106 (rat IgG2a, κ) were nonconjugated. Controls correspond to gated GFP+ cells that were stained with a PE-conjugated antibody specific for an irrelevant antigen (eg, tri-nitrophenol [TNP]). These control antibodies were isotype-matched with each of the PE-conjugated antibodies that are specific for the various membrane markers (listed above). For the secondary immunostaining (CD105 and CD106), controls correspond to an isotype-matched, unconjugated primary antibody in conjunction with a secondary PE-conjugated antibody (PE–mouse anti–rat immunoglobulin, κ light chain). The actual percentage of positive cells was determined with the use of appropriate FACS software (Cellquest, Becton Dickinson) by subtracting the background profiles from the experimental profiles. Consequently, only the fluorescent cells that fall outside the background area are counted and considered positive.
Results
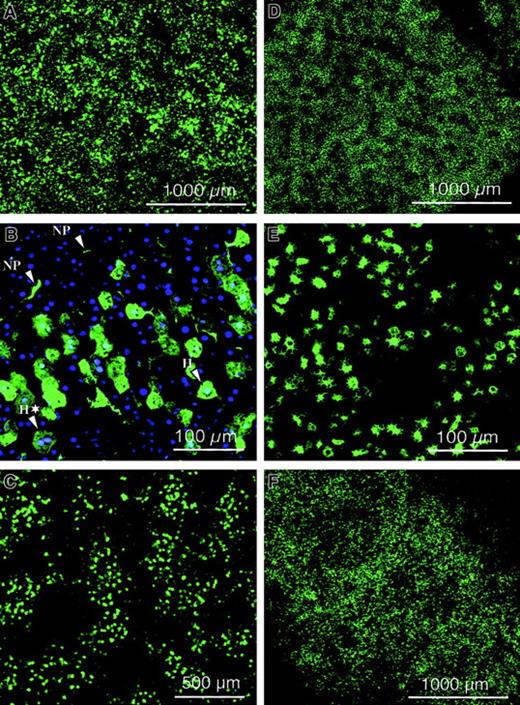
Improved lentiviral vectors containing the central DNA flap and WPRE were produced by transient transfection of 293T cells with the HIV-GFP vector plasmid and the gag-pol, rev, and VSV-G expression constructs (Figure 1). Consequently, none of the HIV-1 accessory proteins (Vif, Vpr, Vpu, and Nef) were present in the vector particles. Typically, vector titers were between 106 and 5 × 106 TU/mL, and upon concentration by ultrafiltration, titers of 2 × 109TU/mL could be achieved. Lentiviral transduction was analyzed in quiescent mouse livers by injecting a total vector dose of 109 TU HIV-GFP into the tail vein of adult SCID mice. Confocal microscopic analysis was performed on dissected organs after 1 week (Figure 2A,D,E) or 3 months (Figure2C,F) and revealed intense and widespread GFP fluorescence in the liver (Figure 2A-C) and spleen (Figure 2D-F), whereas other organs were negative for GFP. The appearance of the transduced splenocytes was predominantly macrophagelike/dendriticlike, with characteristic pseudopodia (Figures 2 and 3). Hence, long-term GFP expression could be achieved in liver and spleen following intravenous administration of lentiviral vectors in SCID mice. The results indicate that the VSV-G–pseudotyped lentiviral vector preferentially transduced liver cells, including hepatocytes and nonparenchymal cells (Figures 2 and 3). Transduced cells were found at the highest frequency in the periportal areas of the lobuli, where the hepatic arterial and portal vessels branch out into the liver parenchyma (Figure 2C). It therefore appeared that lentiviral vectors preferentially transduced hepatocytes and nonparenchymal cells that are in the proximity of the vasculature. Negligible fluorescence was detected in the organs of negative control animals injected with PBS.
Biodistribution of lentiviral vector.
Confocal microscopic analysis was performed on dissected organs of adult SCID mice injected intravenously with 109 TU HIV-GFP vector, after 1 week (A,D,E) or 3 months (C,F) and revealed intense and widespread GFP fluorescence in the liver (A-C) and spleen (D-F). NP and H refer to nonparenchymal cells and hepatocytes, respectively. Green fluorescence corresponds to GFP-expressing cells. Scale bar is depicted. Panel B corresponds to cryosectioned liver 1 week after injection of 109 TU HIV-GFP vector into adult SCID mice after nuclear staining with DAPI. A typical binuclear hepatocyte was indicated by H*. Scale bar is depicted.
Biodistribution of lentiviral vector.
Confocal microscopic analysis was performed on dissected organs of adult SCID mice injected intravenously with 109 TU HIV-GFP vector, after 1 week (A,D,E) or 3 months (C,F) and revealed intense and widespread GFP fluorescence in the liver (A-C) and spleen (D-F). NP and H refer to nonparenchymal cells and hepatocytes, respectively. Green fluorescence corresponds to GFP-expressing cells. Scale bar is depicted. Panel B corresponds to cryosectioned liver 1 week after injection of 109 TU HIV-GFP vector into adult SCID mice after nuclear staining with DAPI. A typical binuclear hepatocyte was indicated by H*. Scale bar is depicted.
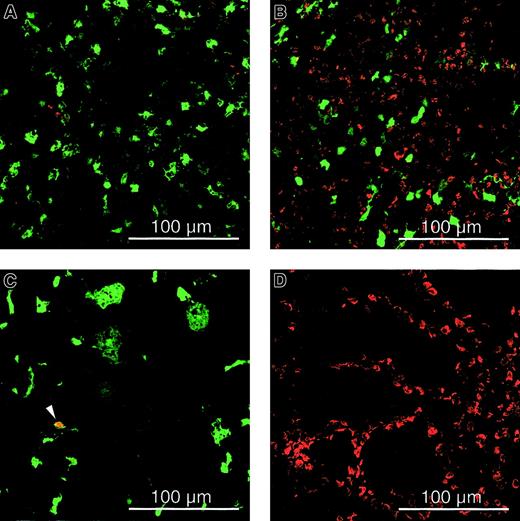
Cell cycle analysis of transduced cells.
Immunohistochemistry on spleen (A,B), liver (C), and gut (D) sections for assessing BrdU incorporation from adult SCID (A,C,D) and Balb/c (B) mice injected intravenously with 109 TU HIV-GFP and implanted with BrdU-osmotic pumps. The analysis was performed 1 week after transduction, and green fluorescence corresponds to GFP-expressing cells. The red cells are BrdU+ cells that have proliferated, and the arrow defines lentivirally transduced nonparenchymal cells that had divided and stained BrdU+(cells that are not expressing GFP but have divided are red; cells that are expressing GFP but are not dividing are green; and cells that are expressing GFP and have divided are yellow). Scale bar is depicted.
Cell cycle analysis of transduced cells.
Immunohistochemistry on spleen (A,B), liver (C), and gut (D) sections for assessing BrdU incorporation from adult SCID (A,C,D) and Balb/c (B) mice injected intravenously with 109 TU HIV-GFP and implanted with BrdU-osmotic pumps. The analysis was performed 1 week after transduction, and green fluorescence corresponds to GFP-expressing cells. The red cells are BrdU+ cells that have proliferated, and the arrow defines lentivirally transduced nonparenchymal cells that had divided and stained BrdU+(cells that are not expressing GFP but have divided are red; cells that are expressing GFP but are not dividing are green; and cells that are expressing GFP and have divided are yellow). Scale bar is depicted.
The distinction between hepatocytes and nonparenchymal cells following DAPI staining was based on morphological criteria. It is well known that hepatocytes (designated H in Figure 2B) have a polyhedral appearance and are larger than the nonparenchymal cells (designated NP in Figure 2B), which have a typical irregular shape.39 We confirmed this independently using hematoxylin and eosin staining, consistent with previous reports.39 Additional evidence that these distinctive morphological features are characteristic of hepatocytes is based on the observations that these cells are occasionally binucleate (designated H* in Figure 2B). Hepatocytes, in contrast to nonparenchymal cells, are known to occasionally undergo nuclear duplication and division without cell division.39,40 In previously published studies, the same morphologic criteria were used to determine the number of transduced hepatocytes relative to the number of nuclei following nuclear staining, which provides a reasonably accurate estimate of the total number of cells per section.40 The transduced hepatocytes expressed GFP and consequently became fluorescent green as shown in Figure 2B. The number of GFP+ hepatocytes relative to the total number of (DAPI-stained) nuclei corresponded to 8.0% ± 6.0% (mean ± SD).
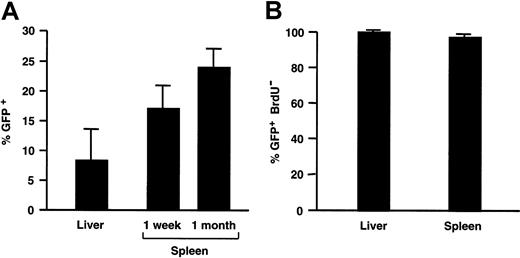
The number of transduced GFP+ splenocytes was determined by FACS analysis, since single-cell suspensions could easily be obtained from the spleen. The frequency of GFP+ cells in the spleen of SCID mice injected with 109 TU HIV-GFP corresponded to 17% ± 2% (n = 2) of the total splenocyte population 1 week after injection and 24% ± 3% (n = 3) 1 month after injection (Figures4A and 7B).
Transduction efficiency analysis.
(A) The percentage of GFP+ cells in transduced liver and spleen from recipient adult SCID mice injected with 109 TU HIV-GFP. (B) Fraction of transduced GFP+ cells in liver and spleen that are nondividing as measured by lack of BrdU incorporation.
Transduction efficiency analysis.
(A) The percentage of GFP+ cells in transduced liver and spleen from recipient adult SCID mice injected with 109 TU HIV-GFP. (B) Fraction of transduced GFP+ cells in liver and spleen that are nondividing as measured by lack of BrdU incorporation.
To evaluate the cell-cycle status of the transduced cells, recipient mice had been implanted 1 day prior to injection of the lentiviral vector with a minipump that continuously produced BrdU at a rate of 1 mg/d for a week, after which mice were killed. Immunohistochemistry for BrdU incorporation revealed that the vast majority of transduced liver cells (98% ± 2%), hepatocytes (97% ± 9%), and splenocytes (97% ± 3%) were nondividing (Figures 3 and4B). Vector injection did not increase the percentage of dividing cells (data not shown). In the controls, there was widespread and intensely positive BrdU-specific staining in cycling duodenal epithelia (Figure 3D). Although only a few proliferating splenocytes can be detected in the spleen of immunodeficient SCID mice (Figure 3A), there are many proliferating cells in the spleen of normal Balb/c mice (Figure 3B). This confirms that the paucity of replicating (BrdU+) cells in the spleen of immunodeficient mice is inherent to their immunodeficient status and is not due to a limitation of the sensitivity of the BrdU-staining method used.
To detect integrated vector DNA in the liver and spleen of treated mice, a nested PCR was performed with the use of primers specific for the lentiviral vector and for ubiquitous repeats found in the mouse genome that belong to the B2 family repeats.38Consequently, only lentiviral sequences that are integrated in the target cell chromosomes yield a PCR fragment. This was confirmed by the results shown in Figure 5. Adult mice that were injected with 109 TU/mL HIV-GFP lentiviral vectors were killed 1 to 3 months later and showed a 121-bp and a 166-bp fragment specific for the 5′ and 3′ end of the integrated lentiviral genomes in liver and spleen (Figure 5).
Genomic integration analysis.
To detect integrated vector DNA in the liver and spleen of treated mice, a nested PCR was performed with primers specific for the lentiviral vector and for mouse B2 family repeats. Adult SCID mice were injected with 109 TU/mL HIV-GFP lentiviral vectors, which were killed after 1 month (lanes 2,4,6) or 3 months (lanes 8,9). Mice injected with PBS served as control (lanes 1,3,5,7). The integrated lentiviral genomes in liver and spleen yielded a 121-bp and a 166-bp fragment at their 5′ end (left gel) and 3′ end (right gel), respectively. The top panel corresponds to the PCR products from the B2-vector PCR reaction. The middle panel corresponds to the nested PCR products with the use of the lentiviral vector-specific primers. The lower panel is from control nested PCR performed with 10 ng nonamplified genomic DNA from liver and spleen of treated and control mice. No DNA was added in the PCR reactions in lane C.
Genomic integration analysis.
To detect integrated vector DNA in the liver and spleen of treated mice, a nested PCR was performed with primers specific for the lentiviral vector and for mouse B2 family repeats. Adult SCID mice were injected with 109 TU/mL HIV-GFP lentiviral vectors, which were killed after 1 month (lanes 2,4,6) or 3 months (lanes 8,9). Mice injected with PBS served as control (lanes 1,3,5,7). The integrated lentiviral genomes in liver and spleen yielded a 121-bp and a 166-bp fragment at their 5′ end (left gel) and 3′ end (right gel), respectively. The top panel corresponds to the PCR products from the B2-vector PCR reaction. The middle panel corresponds to the nested PCR products with the use of the lentiviral vector-specific primers. The lower panel is from control nested PCR performed with 10 ng nonamplified genomic DNA from liver and spleen of treated and control mice. No DNA was added in the PCR reactions in lane C.
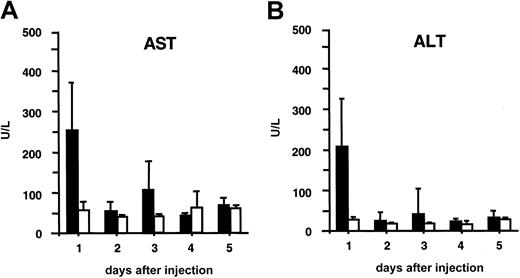
Liver toxicity in recipient mice was analyzed by measuring serum AST and ALT levels (Figure 5). A short-term, transient increase to moderately elevated levels was detected in serum AST and ALT concentration, but only 24 hours after injection (Figure6), after which AST/ALT levels were normal again. None of the recipient mice died from exposure to this lentiviral vector dose.
Liver toxicity analysis.
Hepatocellular cytotoxicity was analyzed by measuring serum AST (A) and ALT (B) levels. The black bars correspond to AST (A) or ALT (B) levels in plasma from adult SCID mice injected intravenously with 109 TU HIV-GFP. The white bars correspond to normal AST (A) or ALT (B) levels in plasma from control mice injected with PBS.
Liver toxicity analysis.
Hepatocellular cytotoxicity was analyzed by measuring serum AST (A) and ALT (B) levels. The black bars correspond to AST (A) or ALT (B) levels in plasma from adult SCID mice injected intravenously with 109 TU HIV-GFP. The white bars correspond to normal AST (A) or ALT (B) levels in plasma from control mice injected with PBS.
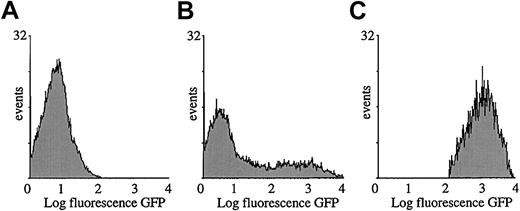
Morphologic microscopic analysis suggested that the transduced splenocytes consisted predominantly of APCs (Figure 2E). To precisely determine their phenotype, transduced splenocytes were subjected to cytofluorimetric analysis with the use of antibodies specific for antigens that are expressed predominantly on APCs. As mentioned above, 20% to 30% of the splenocytes were GFP+ as determined by FACS analysis, 1 week or 1 month after injection of the lentiviral vector (109 TU) in the SCID mice (Figure 4A and7B). Fluorescence was not significantly different from background levels in control mice injected with PBS (Figure 7A). The GFP expression profile of the gated cell fraction is shown in Figure 7C. This GFP+ cell fraction was then stained for different cell differentiation antigens.
Cytofluorimetric analysis of transduced splenocytes.
SCID mice were injected with 109 TU HIV-GFP. GFP expression profile of total splenocyte population injected with PBS (A) or vector (B). Gated GFP expression profile of GFP+ cells by means of FACSvantage (C). Analysis was performed 1 week after transduction.
Cytofluorimetric analysis of transduced splenocytes.
SCID mice were injected with 109 TU HIV-GFP. GFP expression profile of total splenocyte population injected with PBS (A) or vector (B). Gated GFP expression profile of GFP+ cells by means of FACSvantage (C). Analysis was performed 1 week after transduction.
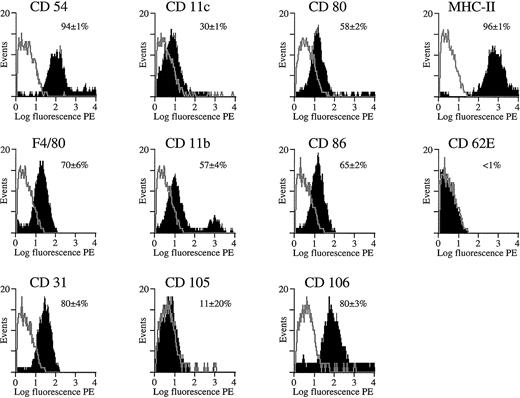
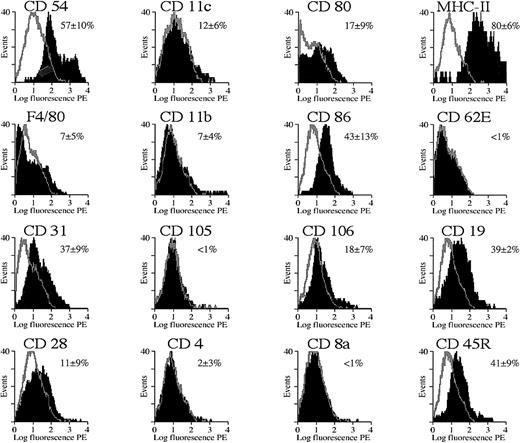
The FACS profiles in Figures 8 and9 show the expression of various membrane markers on the gated splenocyte subpopulation that expresses GFP. The black area in the overlaid FACS profiles corresponds to the expression of specific membrane markers on the basis of immunostaining with the use of PE-conjugated antibodies specific for membrane markers or with the use of indirect staining (for CD105 and CD106 only). Controls included gated GFP+ cells that were stained with an isotype-matched antibody specific for an irrelevant antigen (TNP) either directly conjugated with PE or used in conjunction with a secondary PE-conjugated antibody (as controls for CD105 and CD106). These controls yielded a low fluorescent background signal (gray overlays), as expected. The percentage of positive cells was determined with the use of appropriate FACS software by subtracting the background profiles from the experimental profiles. Consequently, only the fluorescent cells that fall outside the background area (ie, gray contour; Figures 8 and 9) were counted and considered positive. The mean percentage of fluorescent cells ± SD was mentioned in Figures 8 and 9, and one representative profile for each of the membrane markers was depicted.
Cytofluorimetric analysis of transduced splenocytes in SCID mice.
Expression of cell differentiation antigens by cytofluorimetric analysis on gated GFP+ splenocyte population from SCID mice shown in Figure 7C. The black area in the overlaid FACS profiles corresponds to the expression of specific membrane markers on the basis of immunostaining by means of PE-conjugated antibodies specific for each of these different membrane markers (CD54, CD11c, CD80, MHC class II, F4/80, CD11b, CD86, CD62E, CD31) or via indirect staining (CD105 and CD106). Controls included gated GFP+ cells that were stained with an isotype-matched antibody specific for an irrelevant antigen (TNP) either directly conjugated with PE or used in conjunction with a secondary PE-conjugated antibody (as control for CD105 and CD106) (gray contours). The percentage of positive cells was determined with the use of appropriate FACS software by subtracting the background profiles from the experimental profiles. Consequently, only the fluorescent cells that fall outside the background area (gray contour) were counted and considered positive. The mean percentage of fluorescent cells ± SD was mentioned, and one representative profile for each of the markers was depicted. Analysis was performed 1 week after transduction.
Cytofluorimetric analysis of transduced splenocytes in SCID mice.
Expression of cell differentiation antigens by cytofluorimetric analysis on gated GFP+ splenocyte population from SCID mice shown in Figure 7C. The black area in the overlaid FACS profiles corresponds to the expression of specific membrane markers on the basis of immunostaining by means of PE-conjugated antibodies specific for each of these different membrane markers (CD54, CD11c, CD80, MHC class II, F4/80, CD11b, CD86, CD62E, CD31) or via indirect staining (CD105 and CD106). Controls included gated GFP+ cells that were stained with an isotype-matched antibody specific for an irrelevant antigen (TNP) either directly conjugated with PE or used in conjunction with a secondary PE-conjugated antibody (as control for CD105 and CD106) (gray contours). The percentage of positive cells was determined with the use of appropriate FACS software by subtracting the background profiles from the experimental profiles. Consequently, only the fluorescent cells that fall outside the background area (gray contour) were counted and considered positive. The mean percentage of fluorescent cells ± SD was mentioned, and one representative profile for each of the markers was depicted. Analysis was performed 1 week after transduction.
Cytofluorimetric analysis of transduced splenocytes in Balb/c mice.
Expression of cell differentiation antigens by cytofluorimetric analysis on gated GFP+ splenocyte population from Balb/c mice. See Figure 8 legend for details. In addition, T-and B-cell markers were included with the use of PE-conjugated antibodies specific for each of these different membrane markers (CD19, CD28, CD4, CD8a, CD45R/B220). Analysis was performed 4 days after transduction.
Cytofluorimetric analysis of transduced splenocytes in Balb/c mice.
Expression of cell differentiation antigens by cytofluorimetric analysis on gated GFP+ splenocyte population from Balb/c mice. See Figure 8 legend for details. In addition, T-and B-cell markers were included with the use of PE-conjugated antibodies specific for each of these different membrane markers (CD19, CD28, CD4, CD8a, CD45R/B220). Analysis was performed 4 days after transduction.
Cytofluorimetric analysis of the gated GFP+ cell fraction in the SCID mice (n = 3) injected with 109 HIV-GFP confirmed that most were APCs (Figure 8). The majority of GFP+ cells (more than 90%) expressed high levels of MHC class II antigens (Figure 8). MHC class II is known to be expressed predominantly on APCs, particularly macrophages, dendritic cells, and B cells but also on some activated T cells. In addition, the costimulatory molecules CD80 (B7-1) and CD86 (B7-2) (Figure 8), which are characteristic of APCs, were also expressed on the GFP+cells. The adhesion molecules CD11b and CD11c, and the F4/80 antigen, which are often used as markers for APCs, were expressed on the GFP+ cell fraction (Figure 8). The cell-adhesion molecules CD54 (intercellular adhesion molecule–1 [ICAM-1]), CD31 (platelet/endothelial cell adhesion molecule [PECAM-1]), and CD106 (vascular cell adhesion molecule–1 [VCAM-1]) were expressed on the majority of the transduced GFP+ splenocytes. These markers are known to be expressed on APCs but also on endothelial cells. However, the endothelial cell–specific markers CD62E (E-selectin) and CD105 (endoglin) were not expressed on the GFP+populations, suggesting that splenic endothelial cells were not transduced.
Immunodeficient mice have only a limited number of T and B cells compared with immunocompetent animals (SCID mice [n = 3]: CD4+, 9% ± 6%; CD8a+, 10% ± 9%; CD28+, less than 1%; CD19+, 3% ± 4%. Balb/c mice [n = 3]: CD4+, 40% ± 2%; CD8a+, 20% ± 3%; CD28+, 24% ± 4%; CD19+, 63% ± 3%). FACS analysis was subsequently carried out in immunocompetent Balb/c mice with normal cell numbers in the different lineages. The transduced GFP+splenocytes from normal immunocompetent Balb/c mice (n = 5) (Figure9) expressed a variety of membrane markers, consistent with the phenotype of APCs (predominantly MHC class II, CD54, CD31, and CD86). This resembles the transduction pattern in immunodeficient mice. In addition, there was significant transduction of B cells, as confirmed with 2 independent B-cell markers: CD19 and CD45R/B220+. In contrast, CD4+ and CD8+ T cells were not transduced, which was confirmed by the lack of GFP fluorescence in the CD28+ T-cell fraction (Figure 9). There seemed to be fewer cells expressing APC markers in the Balb/c mice that were injected with the HIV-GFP vector than in noninjected Balb/c controls (Balb/c HIV-GFP: CD11b, 7% ± 4%; CD31, 37% ± 9%; CD54, 57% ± 10%; CD80, 17% ± 9%. Balb/c controls: CD11b, 24% ± 2%; CD31, 84% ± 3%; CD54, 84% ± 1%; CD80, 27% ± 2%). In addition, there seemed to be fewer transduced cells expressing APC markers in Balb/c compared with SCID mice (Figures 8 and 9). This suggests that the lentivirally transduced APCs might have been eliminated by an immune response in immuncompetent animals.
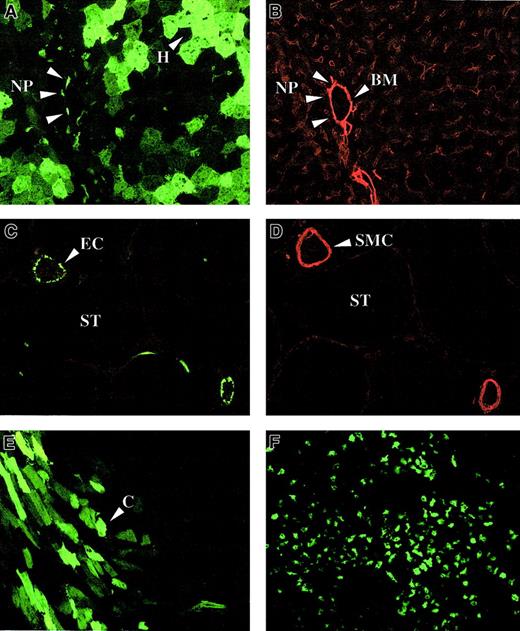
In neonatal FVIII-deficient mice, lentiviral transduction was efficient in liver (Figure 10A) and spleen (Figure 10F). The apparent clustering of transduced GFP+hepatocytes in the liver of neonatal recipients would be consistent with clonal expansion of transduced cells (Figure 10A). Both hepatocytes and nonhepatocytes were transduced (Figure 10A). The nonhepatocytes were near the vasculature, the basement membrane of which was visualized with laminin-specific antibodies (Figure 10B). These data suggest that the transduced nonhepatocytes could be perisinusoidal cells, Kupffer cells, or endothelial cells. In contrast to adult recipients, significant gene transfer could be detected in the heart (Figure 10E) and involved predominantly cardiomyocytes (Figure10E).
Lentiviral biodistribution in neonatal mice.
Immunohistochemistry on liver (A,B), testis (C,D), heart (E), and spleen (F) of newborn FVIII knockout mice injected intravenously with 4 × 107 TU HIV-GFP and killed 1.5 months (A-E) or 9 months (F) after injection. Vasculature was stained with antibodies specific for either laminin (B) or smooth muscle cell actin (D). Transduced hepatocytes (H), nonparenchymal (NP) cells, cardiomyocytes (C), and endothelial cells (EC) are shown. Smooth muscle cells (SMC), basement membrane (BM), and the seminiferous tubules (ST) are indicated. Panels A and B and panels C and D are adjacent sections.
Lentiviral biodistribution in neonatal mice.
Immunohistochemistry on liver (A,B), testis (C,D), heart (E), and spleen (F) of newborn FVIII knockout mice injected intravenously with 4 × 107 TU HIV-GFP and killed 1.5 months (A-E) or 9 months (F) after injection. Vasculature was stained with antibodies specific for either laminin (B) or smooth muscle cell actin (D). Transduced hepatocytes (H), nonparenchymal (NP) cells, cardiomyocytes (C), and endothelial cells (EC) are shown. Smooth muscle cells (SMC), basement membrane (BM), and the seminiferous tubules (ST) are indicated. Panels A and B and panels C and D are adjacent sections.
GFP expression in liver, spleen, and heart was detectable for at least 9 months after injection. In 1 neonatal recipient of 5, which was killed 1.5 months after injection, there was gene transfer in the gonads (Figure 10C). Using smooth muscle cell actin–specific antibodies (Figure 10D) to visualize the testicular vasculature, we determined that transduced GFP+ cells corresponded to endothelial cells in the testicular vasculature. No GFP+cells could be detected inside the seminiferous tubules where spermatogenesis occurs.
Discussion
In the present study, nondividing hepatocytes in adult recipient mice were transduced efficiently with an improved HIV-based self-inactivating lentiviral vector that contained the central polypurine/termination tract (cPPT/CTS) (or central DNA flap) and WPRE sequences. The central DNA flap facilitates intranuclear transcription of lentiviral genomes,20 and the WPRE increases transgene expression by a posttranscriptional mechanism.30 High and widespread hepatic GFP expression could be detected in nondividing hepatocytes and nonhepatocytes, obviating the need to artificially induce hepatocyte proliferation by partial hepatectomy19or by administration of hepatic growth factors.41 42
These results are in contrast to previous reports by Park et al18,19 indicating that efficient lentiviral transduction of hepatocytes requires cell cycling in vivo. This difference is probably due to our incorporation of the central DNA flap in the lentiviral backbone. This element enhances lentiviral transduction in dividing and nondividing cells, possibly by improving the nuclear translocation of the lentiviral genome.20,27-29,43,44 Furthermore, the use of the WPRE may have further increased GFP expression in hepatocytes in vivo. Finally, differences in vector production and/or the route of administration may at least partially account for the limited hepatotoxicity and absence of mortality, in contrast to the previous study.18
The present study is consistent with the work of Kafri et al,13 who observed local transduction of hepatocytes at the injection site of first-generation lentiviral vectors in the liver parenchyme. However, specific HIV-1 auxiliary proteins (Vpr, Vif) were required to achieve efficient hepatic gene transfer.13 In addition, previous studies with HIV variants containing mutations invpr and with simian immunodeficiency virus (SIV) variants containing mutations in the functionally equivalent vpx gene indicated a selective replication defect in nondividing cells.22,45,46 The present data suggest that incorporation of the central DNA flap obviates the need for these auxiliary proteins to achieve efficient hepatic lentiviral transduction. This offers an additional safety advantage since at least some of these HIV-1 auxiliary proteins may be cytotoxic.18,45 The present results are consistent with 2 recent studies indicating that the presence of central DNA flap facilitates lentiviral transduction into nondividing hepatocytes.40,47 The DNA flap may promote nuclear import by a mechanism independent of nucleophilic accessory proteins but still depends on the viral integrase.48 How the DNA flap promotes nuclear uptake of HIV DNA is unclear.20 The DNA flap may provide a docking site between the preintegration complex and the cellular nuclear import receptors that direct the preintegration complex to the nuclear pore complex. Alternatively, the flap could mediate interaction of the preintegration complex with nuclear pore proteins.
In previous studies, local physical injury resulting from direct injection in the liver parenchyma, and vector-associated toxic effects related to the auxiliary proteins, could have increased the number of hepatocytes progressing into the cell cycle.13,18 These cycling hepatocytes could become permissive for lentiviral-vector mediated gene transfer. However, it is unlikely that this is the case in the present study since injection of the vector did not significantly increase the number of dividing hepatocytes and since most transduced hepatocytes (more than 99%) were nondividing. Nevertheless, the increase in AST/ALT concentrations suggested that some hepatocellular injury may have been inflicted following the intravenous injection of the lentiviral particles However, only transient and relatively small increases in AST/ALT concentrations were apparent, and none of the recipient mice had died, which is consistent with the previously reported decreased hepatotoxicity of Vpr/Vif lentiviral vectors.18
In the extrahepatic tissues of adult mice, gene transfer was most pronounced in the spleen, predominantly in putative APCs. Indeed, splenocytes transduced with lentiviral vectors expressed high levels of MHC class II; the costimulatory molecule B7-2 (CD86); and the cell-adhesion molecules ICAM-1 (CD54), VCAM-1 (CD106), and PECAM-1 (CD31), which are all characteristic for APCs. In addition, most transduced splenocytes exhibited morphological features of phagocytes, such as pseudopodia formation. These results are consistent with the observation that wild-type HIV virus and other lentiviruses can readily transduce nonproliferating monocytes49 and that macrophages constitute a main component of the cellular reservoir for HIV and SIV. In addition to macrophagelike cells, transduced cells may belong to the dendritic cell population, which has many common membrane antigens with macrophages. The adhesion molecules such as CD106 (VCAM-1), CD31 (PECAM-1), and CD54 (ICAM-1) are highly expressed on APCs but also on endothelial cells, suggesting that the transduced GFP+ splenocyte population may include at least some endothelial cells originating from the splenic vasculature. However, the lack of any endothelial cell–specific markers, such as E-selectin (CD62E) and endoglin (CD105), on the GFP+ splenocyte population would suggest that splenic endothelial cells were probably not transduced with the lentiviral vectors. The lack of lentiviral gene transfer in mouse T lymphocytes is consistent with other reports showing that nonstimulated human T cells are refractory to lentiviral transduction and to infection with HIV-1.14 17
The majority of transduced splenocytes (more than 95%) were nondividing as confirmed by the lack of BrdU incorporation. This is consistent with earlier reports indicating that nondividing human monocyte–derived terminally differentiated macrophages can efficiently be transduced in vitro by first-generation lentiviral vectors.50 The present data suggest that lentiviral transduction of splenic APCs in vivo does not require the HIV-1 auxiliary proteins such as Vif or Vpr, supporting the hypothesis that the DNA flap may promote nuclear import by a mechanism independent of nucleophilic accessory virion proteins and depending on the viral integrase only.48
Although cell division and/or activation are not absolutely required for gene transfer, the present study shows that this requirement may be organ dependent. In particular, the heart was not permissive for lentiviral transduction in adult recipients, but significant gene transfer could be consistently detected in cardiomyocytes of neonatal recipients. This suggests that the proliferation and/or activation status of the cardiomyocytes may influence lentiviral transduction efficiency. Alternatively, the difference in cardiac transduction between neonates and adults could have been due to different accessibility of the vector to the cells since the blood vessels are leakier in the neonatal period. This may allow for a better extravasation of the vector particles from the vasculature. These results may pave the way toward possible gene therapy applications for inborn cardiac disorders or perhaps even other cardiac disorders, such as myocardial infarction, that are characterized by activated and/or proliferating cardiomyocytes.51 Recently, it has been shown that cardiomyocytes regenerate after a heart attack and that large-scale replication of heart muscle cells is apparent in the zones of the heart bordering the infarct area.51 It is possible that the increased gene transfer efficiencies and the differential biodistribution in neonates were at least partly due to the higher multiplicities of infection that could be achieved given the higher vector concentration in the plasma. Sporadic gene transfer in the testis could be detected in some of the neonatal recipients and involved primarily endothelial cells from the blood vessels located between the seminiferous tubules. No evidence of lentiviral gene transfer was apparent inside the seminiferous tubules, suggesting that the lentiviral vectors may be unable to cross the basement membranes surrounding the tissue where spermatogenesis occurs. This might imply that inadvertent gene transfer in the germ cells is unlikely following in vivo lentiviral vector administration.
The demonstration that lentiviral vectors can efficiently transduce liver cells has implications for the possible treatment of various genetic diseases that result in serum protein deficiencies, including hemophilia and hepatic disorders. In addition, in situ transduction of APCs with lentiviral vectors could be useful for genetic vaccination strategies against cancer and infectious and autoimmune diseases3-6 as well as for strategies to treat genetic or acquired APC-related disorders.8,3,7 In addition, these improved lentiviral vectors may lead to a better understanding of the pathologic mechanisms related to APC dysfunction. Nevertheless, the relatively efficient lentiviral transduction of APCs in vivo should be taken into account in the designing of vectors for gene therapy. Inadvertent transgene expression in transduced APCs may be undesirable in circumstances where an immune response against the transgene product should be avoided,52 eg, in hemophilia gene therapy. The current results suggest that the lentivirally transduced APCs might have been eliminated by an immune response in immuncompetent animals. The present study implies that the use of liver-specific promoters may be warranted to restrict transgene expression in hepatocytes.
We are grateful to E. Vangoidsenhoven, V. Vanslembrouck, M. L. Vanderhaegen, D. Van Bockstaele, M. Lenjou, and Z. Zhang for technical assistance. We thank Dr Vermylen and Dr Conway for reviewing the manuscript.
Supported by the Flemish Fund for Scientific Research (F.W.O) and the Flemish government. L.T. is supported by an Vlaams Institute voor de Bevordering van het Wetenschappelÿk Technologisch Onderzoek in de Industrie (I.W.T.) fellowship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Thierry VandenDriessche, Center for Transgene Technology and Gene Therapy, Flanders Interuniversity Institute for Biotechnology, University of Leuven, 49 Herestraat B-3000 Leuven, Belgium; e-mail: thierry.vandendriessche@med.kuleuven.ac.be.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal