Abstract
One important question in stem cell biology of childhood acute lymphoblastic leukemia (ALL) is whether immature CD34+CD19− cells are part of the leukemic cell clone. CD34+CD19− cells from the bone marrow of 9 children with TEL/AML1-positive ALL were purified by flow sorting and subjected to reverse transcriptase–polymerase chain reaction (RT-PCR), fluorescence in situ hybridization, and methylcellulose cultures. In 3 of 8 patients analyzed by RT-PCR, noTEL/AML1-positive cells could be detected in the CD34+CD19− cell fraction. Altogether, the percentage of TEL/AML1-positive cells was low: 1.6% (n = 8; SD 2.2%) by nested real-time RT-PCR and 2.5% (n = 5; SD 2.6%) by fluorescence in situ hybridization. This correlated with the percentage of contaminating CD19+ leukemic cells in the CD34+CD19− cell fraction in 6 control sorts (mean 4.6%, SD 3.6%), indicating that the low levels of leukemic cells detected in the CD34+CD19− cell fraction could be attributed to sorter errors. Methylcellulose cultures in 3 patients provided further evidence that CD34+CD19− cells represent a candidate normal cell population. The clonogenicity of the CD34+CD19− cell fraction was similar to normal progenitors, including growth of primitive granulocyte, erythroid, macrophage, megakaryocyte colony-forming units. Each of 92 colonies from cultures with CD34+CD19− cells tested negative for TEL/AML1. In conclusion, our data support the hypothesis that the leukemia inTEL/AML1-positive childhood ALL originates in a CD19+ lymphoid progenitor. This has many therapeutic implications, eg, for purging of autologous stem cell products, flow cytometric monitoring of minimal residual disease, and targeting immunotherapy against the leukemic cell clone.
Introduction
For a long time it has been assumed that the leukemic transformation in childhood B-cell precursor acute lymphoblastic leukemia (ALL) occurs at the level of a lymphoid progenitor cell.1 Presence and stability of clonal immunoglobulin and T-cell receptor gene rearrangements in ALL have been regarded as strong evidence to support this hypothesis.
Contrasting this theory, molecular and functional analyses of purified immature progenitor cell populations provide increasing evidence for the involvement of a more primitive cell in the leukemic process. Cells with a CD34+CD38− stem cell–like immunophenotype were shown to harbor the leukemia-specific T-cell receptor Vδ2-Dδ3 rearrangements2 and were able to transfer Philadelphia chromosome–positive ALL onto immunodeficient nonobese diabetic/severe combined immunodeficiency mice.3In addition, CD34+CD19−CD33−CD38−cells were found to contain variable percentages of leukemic blasts by fluorescence in situ hybridization (FISH),4 indicating that there may be heterogeneity of stem cell involvement in ALL. This is in accordance with studies investigating clonal diversity at the level of immunoglobulin rearrangements. Several leukemic subclones with unrelated DJ rearrangements could be detected, particularly in patients with a poor prognosis.5 These data suggest that at least in some patients with ALL the leukemia originates from a primitive progenitor/stem cell that has not yet initiated immunoglobulin gene rearrangement.
However, these investigations may not reflect the situation in more common and prognostically favorable ALL subtypes, ie, in children with an overall event-free survival close to 80% at 5 years.6In the present study, we therefore focused on the childhood ALL subgroup characterized by the translocation t(12;21)(p13;q22). This chromosomal translocation leads to formation of the TEL/AML1 fusion oncogene and is the most common genetic aberration in childhood B-cell precursor ALL, occurring in approximately 20% to 25% of patients.7 Presence of t(12;21) is usually regarded as a prognostically favorable marker8 and correlates with in vitro sensitivity of the leukemic blasts to asparaginase,9vincristine, dexamethasone, and serum deprivation.10 Due to the high incidence of t(12;21) in relapse patients11and a recent study in children with primary ALL showing no predictive value of t(12;21) regarding outcome,12 the exact role of t(12;21) as a prognostic marker needs, however, further prospective evaluation.13
To address the question of involvement of the immature progenitor/stem cell compartment in childhood ALL, flow cytometric analyses had been initiated to identify and distinguish uncommitted and early B-lineage progenitors. Expression of CD19 was found to be a good marker to distinguish CD34+CD19+ leukemic from more immature CD34+CD19− progenitor/stem cells in the bone marrow of patients with newly diagnosed B-cell precursor ALL.14,15 CD19 is part of the CD19/CD21 complex that modulates B-cell–receptor signaling.16 Its expression is an early event in B-cell development17 directly following DJH, but preceding or simultaneously occurring with VDJH, rearrangement.18 Analysis of immature CD34+CD19− cells for expression of CD45, CD117, and CD133 in the bone marrow from children with B-cell precursor ALL showed that these cells had an immunophenotype similar to normal progenitor/stem cells.14 15
To investigate whether these immature cells belong to the leukemic clone, CD34+CD19− cells were purified out of diagnostic bone marrow samples from children with ALL/t(12;21). Flow-sorted cells were analyzed by semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) for TEL/AML1expression, by FISH for the presence of the translocation t(12;21), and by methylcellulose cultures for clonogenicity.
Patients, materials, and methods
Patients and cell lines
Patients.
Diagnostic bone marrow samples were obtained from 7 children withTEL/AML1-positive and 8 children withTEL/AML1-negative B-cell precursor ALL treated at the Department of Pediatric Hematology and Oncology, University of Muenster. Two additional TEL-AML1-positive patients (nos. 008/B, 009/B) were provided by the ALL-BFM (ALL–Berlin-Frankfurt-Muenster) study group (W.-D. Ludwig, Berlin, Germany; and M. Schrappe, Hannover, Germany). Only diagnostic bone marrow samples prior to starting chemotherapy were used for analysis. The percentage of blasts in the 7 TEL/AML1-positive patients in the bone marrow ranged from 30% to 96.5% (mean 84.4%, SD 24.1%). In addition, bone marrow samples obtained from a patient with a solid tumor (for exclusion of tumor cell metastasis to the bone marrow, n = 1) or from leukemia patients during stable remission (n = 2) were used as controls. The investigation was approved by the ethics committee of the Medical Faculty, University of Muenster.
Cell lines.
The B-lineage precursor cell lines REH (TEL/AML1-positive)19 and BLIN-1 (TEL/AML1-negative)20 were used as positive and negative controls. REH cells were cultured in Iscoves modified Dulbecco medium (IMDM) (Biochrom, Berlin, Germany) and BLIN-1 cells in RPMI (Biochrom) supplemented with 10% fetal calf serum (FCS), 2 mMl-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (LifeTechnologies, Karlsruhe, Germany) at 37°C and 5% CO2 in a humidified atmosphere.
Processing of bone marrow samples
Cell sorting.
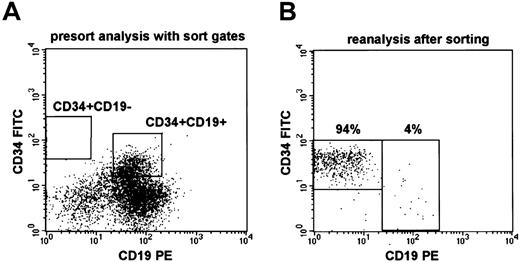
Frozen mononuclear cells (1 × 107 to 7 × 107 cells) from diagnostic bone marrow samples were thawed, washed with IMDM containing 10% FCS, and stained with saturating amounts of anti-CD19–phycoerythrin (J4.119; Beckman Coulter, Krefeld, Germany) and anti-CD34–fluorescein isothiocyanate (581; Beckman Coulter) antibody conjugates in a total volume of 100 to 200 μL IMDM plus 10% FCS for 20 minutes at 4°C. Cells were resuspended in IMDM with 10% FCS at a concentration of 107/1.5 mL. Analysis and cell sorting were performed on a FACS Vantage (Becton Dickinson, Heidelberg, Germany). Sorting gates were placed on CD34brightCD19− and CD34dim/brightCD19+ populations (Figure1A) using CellQuest software (Becton Dickinson).
Flow sorting of CD34+CD19−cells.
(A) Presort analysis and sort gates for CD34+CD19− and CD34+CD19+ cells in the leukemic bone marrow of patient no. 1523/00. (B) Reanalysis of flow-sorted CD34+CD19− cells from patient no. 1523/00 showing a purity of 94% and 4% contaminating CD19+ cells (sorter errors). The fluorescence intensity for CD34–fluorescein isothiocyanate was always lower on reanalysis, as expected from the initial sort gates.
Flow sorting of CD34+CD19−cells.
(A) Presort analysis and sort gates for CD34+CD19− and CD34+CD19+ cells in the leukemic bone marrow of patient no. 1523/00. (B) Reanalysis of flow-sorted CD34+CD19− cells from patient no. 1523/00 showing a purity of 94% and 4% contaminating CD19+ cells (sorter errors). The fluorescence intensity for CD34–fluorescein isothiocyanate was always lower on reanalysis, as expected from the initial sort gates.
Sorting of cells for RT-PCR analysis and isolation of RNA.
A total of 1000 cells from each population (CD34+CD19− and CD34+CD19+) and from cell lines (controls) were sorted directly into 100 μL RLT buffer (supplied with the RNeasy-Mini-Kit; Qiagen, Hilden, Germany); 250 μL RLT buffer and 3.5 μL β-mercaptoethanol were added, and the samples were passed through Qiashredder columns (Qiagen) according to the manufacturer's protocol. The lysate was frozen and stored at −80°C. Total RNA was extracted from the frozen lysates according to the RNeasy-Mini-Kit (Qiagen) protocol. RNA was finally eluted with 30 μL ribonuclease-free water and stored at −80°C.
Preparation of slides for FISH analysis.
From 600 to 2000 cells from each population were directly sorted into 20 μL drops of phosphate-buffered saline (PBS) that were placed on a grease-free glass slide. The slides were incubated for 10 minutes in a moist chamber to allow settling of the cells within the PBS drop and adherence to the glass slide. Subsequently, excessive PBS was carefully removed with a paper towel. The cells were fixed on the slide with ice-cold methanol/glacial acid (3:1, vol/vol). Air-dried slides were analyzed by FISH at the cytogenetic laboratory at the Department of Pediatric Hematology and Oncology in Gießen (S.R. and J.H.).
Purification of cells for colony assays.
For colony assays, 1500 to 3000 CD34+CD19−(depending on sample size) and 30 000 CD34+CD19+ cells were directly sorted into 300 μL IMDM.
Molecular and functional analysis of sorted cell populations
RT-PCR for TEL-AML1 and GAPDH.
For RT-PCR amplification of the TEL/AML1 fusion messenger RNA, a nested PCR was performed. RT-PCR amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) messenger RNA served as a quality control for the RNA preparation. GAPDHamplification and the first amplification round of TEL/AML1were performed by utilizing the OneStep RT-PCR-kit (Qiagen). A total of 5 μL (for TEL/AML1 RT-PCR) or 3 μL (for GAPDHRT-PCR) total RNA was transcribed into complementary DNA. The RT reaction and amplification were performed in a total volume of 50 μL containing 0.2 μM of each primer (TEL/AML1, sense: CTCTCATCGGGAAGACCTGG; antisense: AGCGGCAACGCCTCGCTCAT;GAPDH, sense: GACTGTGGATGGCCCCTCCGG; antisense: AGGTGGAGGAGTGGGTGTCGC), 400 μM of each deoxyribonucleoside triphosphate, 1 × OneStep RT-buffer, and OneStep RT-PCR enzyme mix. Reverse transcription and amplification conditions forTEL/AML1 were 30 minutes at 50°C (RT reaction), 15 minutes at 95°C, followed by 35 PCR cycles (30 seconds at 94°C, 30 seconds at 64°C, 60 seconds at 72°C). For GAPDH, amplification conditions were 30 minutes at 50°C (RT reaction), 15 minutes at 95°C, followed by 30 PCR cycles (30 seconds at 94°C, 45 seconds at 66°C, 60 seconds at 72°C). Both reactions were terminated by a final extension step of 10 minutes at 72°C. A total of 20 μL of the GAPDH amplification product was loaded onto a 0.8% agarose gel containing 0.5 μg/mL ethidium bromide.
A second PCR for TEL/AML1 amplification was performed in a total volume of 50 μL containing 5 μL of the first PCR product, 0.2 μM of each nested TEL/AML1 primer (sense: GAACCACATCATGGTCTCTG; antisense: ATCTTGCCTGGGCTCAGCGC), 200 μM of each deoxyribonucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.1 mg/mL bovine serum albumin, 0.05% Triton X-100, and 1 unit Taq. A hot-start (initial step 4 minutes at 94°C, and temperature then was allowed to cool down to 80°C, at which time point the Taq enzyme was added), directly followed by 35 PCR cycles (30 seconds at 94°C, 30 seconds at 64°C, 60 seconds at 72°C) and 10 minutes at 72°C for final extension, was performed. A total of 20 μL of amplification product was loaded onto a 0.8% agarose gel.
To quantify the relative amount of TEL/AML-1 transcripts and thus the percentage of leukemic cells in the different populations, the second PCR reaction was alternatively performed on the LightCycler instrument using the LightCycler-DNA-master-SYBR-green-I-kit (Roche, Mannheim, Germany). For this reaction, 2 μL amplification product from the first PCR was subjected to a second PCR according to the manufacturer's protocol using the hot-start method (Taq-Start antibody; Clontech, Heidelberg, Germany), using 0.2 μM of the second nested primer pair (see above), 3 mM MgCl2, and 1 × LightCycler-DNA-master-SYBR-green buffer. Amplification conditions were set to 2 minutes at 95°C, followed by 40 PCR cycles (1 second at 95°C, 5 seconds at 60°C, 15 seconds at 72°C). Quantification ofTEL/AML1 transcripts was done by using 1000 sorted cells from standard dilutions of REH cells in TEL/AML1-negative cells (100%, 10%, 1%, 0.1%, 0% REH cells) as controls and applying the LightCycler quantification software (Roche) that compares the crossing points of the different PCR reactions.
Dual-color FISH for t(12;21).
FISH analyses were performed using the commercially available Vysis LSI TEL/AML1 extra signal dual-color probe (Vysis, Downers Grove, IL). The TEL probe directly labeled with SpectrumGreen covers approximately 350-kilobase telomeric sequences on chromosome 12 starting between exons 3 to 5 of TEL, whereas the 500-kilobase AML1 probe labeled with SpectrumOrange fluorophore spans the entire AML1 gene.
The slides were rehydrated in an alcohol series of 100%, 70%, 50%, 30% ethanol for 1 minute each, passed through 0.1 × SSC, and incubated in 2 × SSC for 30 minutes at 70°C. After a second 0.1 × SSC step, chromosomal DNA was denatured in 0.07 N NaOH for 1 minute at room temperature and chilled in 0.1 × SSC and 2 × SSC for 1 minute each. The slides were dehydrated in 30%, 50%, 70%, and 100% ethanol and air-dried. Probes were denatured and hybridized according to manufacturer's instructions. Hybridization was executed in a moist chamber at 37°C overnight. Posthybridization washes were performed in 2 × SSC for 10 minutes at room temperature, 1 × SSC at 72°C for 5 minutes, and 2 × SSC/Triton X-100 for 5 minutes at room temperature. The slides were passed through PBS, dehydrated in an ascending alcohol series, and air-dried. Cells were counterstained with 4,6-diamidino-2-phenylindole and mounted with Vectashield (Vector Laboratories, Burlingame, CA).
Analysis of FISH preparations was performed with a ZeissAxiophot epifluorescence microscope (Zeiss, Oberkochem, Germany) equipped with a 100 W mercury lamp and an appropriate filter combination.
For evaluation of the FISH results, 150 to 200 nuclei were analyzed. A nucleus without the TEL/AML1 gene fusion is expected to yield 2 green (TEL) and 2 red (AML1) signals separated from each other. With the occurrence of the translocation t(12;21) the fusion of TEL and AML1 genes is detected as a yellow fusion signal on the derivative chromosome 21, whereas derivative chromosome 12 is marked with a small red signal (residual AML1). Furthermore, the nucleus usually contains one green signal (native TEL) and one large red signal (native AML1). In the case of loss of heterozygosity of TEL, which has been described for the cell line REH, the normal green signal is absent.
Colony assays.
A total of 600 to 30 000 cells sorted into 300 μL IMDM was added to 2.7 mL MethoCult H4434 (StemCell Technologies/CellSystems, St Katharinen, Germany), which supports colony formation of myeloid progenitor cells. Cells were plated in duplicate or triplicate, depending on the amount of cells that were recovered from the patient samples. Colonies were counted and photographed 10 to 14 days after plating. Expression analysis of TEL/AML1 fusion transcript within colonies by RT-PCR was done by transferring individual colonies into 350 μL RLT buffer plus 3.5 μL β-mercaptoethanol (Qiagen, RNeasy-Mini-Kit). Preparation of RNA and RT-PCR analysis were performed as already described.
Results
RT-PCR for TEL/AML1 in immature cell populations
Extensive flow cytometric studies of immature cell populations in childhood ALL had shown that expression of the B-cell antigen CD19 is a suitable marker to distinguish immature progenitor/stem cells from the bulk leukemic cell population.14 15 Based on these analyses, immature CD34+CD19− and leukemic CD34+CD19+ cells (Figure 1A) were purified by flow sorting in 9 patients with TEL/AML1-positive ALL.
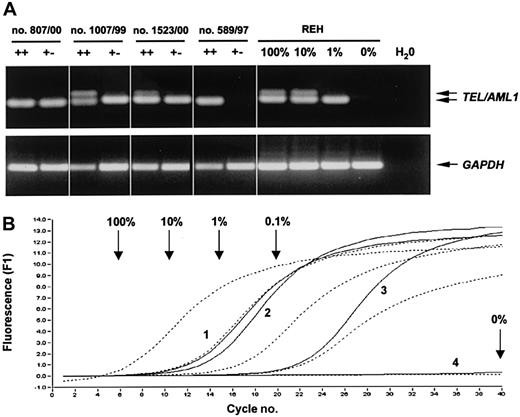
In 8 patients, we were able to sort 1000 CD34+CD19− and CD34+CD19+ cells from diagnostic bone marrow samples for RT-PCR analysis. As expected, a TEL/AML1 fusion product could be amplified from the CD34+CD19+cell population in all patients, confirming the leukemic character of this population (Figure 2A and data not shown). In contrast, in 3 of the 8 patients (nos. 589/97, 658/97, 008/B) no TEL/AML1 transcripts could be amplified from the CD34+CD19− population (Figure 2A and Table1).
RT-PCR analysis of flow-sorted CD34+CD19−cells.
(A) Representative agarose gel electrophoresis of the nested PCR forTEL/AML1 and GAPDH (single-step PCR) from 1000 sorted CD34+CD19− (+−), CD34+CD19+ (++) cells, and REH control cells. When high numbers of TEL/AML1 transcripts were present in the samples (eg, ++ cells in patient nos. 1007/99 and 1523/00; 100% and 10% REH controls), the slightly longer product from the first PCR reaction was coamplified (upper band). (B) Semiquantitative real-time analysis of the second PCR reaction for TEL/AML1. Arrows indicate the crossing points of the respective controls (broken lines), and numbers identify the different patients (solid lines; 1, no. 807/00; 2, no. 1007/99; 3, no. 1523/00; 4, no. 589/97).
RT-PCR analysis of flow-sorted CD34+CD19−cells.
(A) Representative agarose gel electrophoresis of the nested PCR forTEL/AML1 and GAPDH (single-step PCR) from 1000 sorted CD34+CD19− (+−), CD34+CD19+ (++) cells, and REH control cells. When high numbers of TEL/AML1 transcripts were present in the samples (eg, ++ cells in patient nos. 1007/99 and 1523/00; 100% and 10% REH controls), the slightly longer product from the first PCR reaction was coamplified (upper band). (B) Semiquantitative real-time analysis of the second PCR reaction for TEL/AML1. Arrows indicate the crossing points of the respective controls (broken lines), and numbers identify the different patients (solid lines; 1, no. 807/00; 2, no. 1007/99; 3, no. 1523/00; 4, no. 589/97).
RT-PCR and FISH analysis of sorted CD34+CD19− and CD34+CD19+ cell populations
| Patient no. . | Level of TEL/AML1-positive cells by RT-PCR . | Level of t(12;21)-positive cells by FISH . | |||
|---|---|---|---|---|---|
| Sorted cell populations . | Control cell lines . | ||||
| CD34+CD19−, % n = 8 . | CD34+CD19−, % n = 5 . | CD34+CD19+, % n = 6 . | BLIN-1 negative control, % n = 4 . | REH positive control, % n = 6 . | |
| 807/00 | 6.0 | 8.8 | 7.5* | 2.0 | 98.5 |
| 1523/00 | 0.1 | 12.0 | 96.0 | — | 98.0 |
| 1007/99 | 3.1 | — | 87.4 | 3.2 | 99.0 |
| 917/98 | 3.3 | — | — | — | — |
| 1318/97 | — | 1.0 | 94.5 | 3.3 | 98.6 |
| 658/97 | 0.0 | 5.4 | 96.0 | 4.0 | 92.0 |
| 589/97 | 0.0 | 8.9 | 99.2 | — | 98.0 |
| 008/B | 0.0 | — | — | — | — |
| 009/B | 0.6 | — | — | — | — |
| Median | 0.4 | 8.8 | 95.3 | 3.3 | 98.3 |
| Mean | 1.6 | 7.2 | 80.1 | 3.1 | 97.4 |
| SD | ± 2.2 | ± 4.2 | ± 35.8 | ± 0.8 | ± 2.6 |
| Patient no. . | Level of TEL/AML1-positive cells by RT-PCR . | Level of t(12;21)-positive cells by FISH . | |||
|---|---|---|---|---|---|
| Sorted cell populations . | Control cell lines . | ||||
| CD34+CD19−, % n = 8 . | CD34+CD19−, % n = 5 . | CD34+CD19+, % n = 6 . | BLIN-1 negative control, % n = 4 . | REH positive control, % n = 6 . | |
| 807/00 | 6.0 | 8.8 | 7.5* | 2.0 | 98.5 |
| 1523/00 | 0.1 | 12.0 | 96.0 | — | 98.0 |
| 1007/99 | 3.1 | — | 87.4 | 3.2 | 99.0 |
| 917/98 | 3.3 | — | — | — | — |
| 1318/97 | — | 1.0 | 94.5 | 3.3 | 98.6 |
| 658/97 | 0.0 | 5.4 | 96.0 | 4.0 | 92.0 |
| 589/97 | 0.0 | 8.9 | 99.2 | — | 98.0 |
| 008/B | 0.0 | — | — | — | — |
| 009/B | 0.6 | — | — | — | — |
| Median | 0.4 | 8.8 | 95.3 | 3.3 | 98.3 |
| Mean | 1.6 | 7.2 | 80.1 | 3.1 | 97.4 |
| SD | ± 2.2 | ± 4.2 | ± 35.8 | ± 0.8 | ± 2.6 |
The leukemic cells from patient no. 807/00 wereTEL/AML1-positive by RT-PCR.
To quantify the amount of TEL/AML1-positive cells in the CD34+CD19− fraction in all 8 patients, the second reaction of the nested RT-PCR was transferred to a LightCycler instrument and the individual PCR reactions were quantified in real-time by the SYBR green method (Figure 2B). As shown in Table 1, only low levels (n = 8, range 0%-6.0%, mean 1.6%) ofTEL/AML1-expressing leukemic cells could be detected within the CD34+CD19− fraction.
However, there are several possibilities by which this RT-PCR–based quantification may underestimate the level ofTEL/AML1-positive leukemic cells within the CD34+CD19− cell fraction: (1) The quality of RNA in the clinical samples may have been poor and partially degraded although GAPDH was successfully amplified from all samples. (2) Putative immature CD34+CD19− leukemic stem cells, unlike blasts from the bulk leukemic population, may not expressTEL/AML1 or only at low levels. (3) Primary leukemic cells in general may express lower copy numbers of the TEL/AML1 fusion transcripts compared with the REH cell line that was used as positive control. (4) Finally, exact quantification by the described method may have been hampered by the fact that the first PCR reaction was not quantified by real-time PCR. Linearity of the amplification during this first PCR reaction is likely—because of the results with the different REH controls (0%, 0.1%, 1%, 10%, and 100%)—but cannot be proven.
FISH analysis for t(12;21) of sorted cell populations
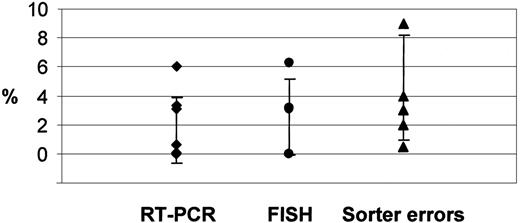
Therefore, to quantify the level of leukemic cells by a DNA-based method, FISH analysis for the presence of t(12;21) was performed on the different subpopulations sorted directly onto glass slides. Consistent with the RT-PCR results, only low levels of cells with a TEL/AML1 gene fusion could be detected within the CD34+CD19−population of 5 patients, while the CD34+CD19+cells were predominantly leukemic (Table 1). The lower cutoff level for FISH analysis in accordance with diagnostic standards was defined by taking the mean background of sorted TEL/AML1-negative BLIN-1 cells and adding 3 SDs. By this definition, the cutoff level was 5.7%. Taking this cutoff level into account, on average 2.5% cells carrying the translocation t(12;21) (range, 0%-6.3%) were detected within the immature CD34+CD19− population (Figure 3). If only the background of 3.1% false positive signals was subtracted from the individual values, only a slightly higher mean percentage of 4.5% cells with a TEL/AML1 fusion signal (range, 0%-8.9%) was measured. In 1 patient (no. 1318/97) the percentage of leukemic cells in the CD34+CD19− cell fraction was below the lowest background value detected in the negative cell line BLIN-1 and in another patient (no. 658/97) below the cutoff level of 5.7%. Thus, FISH analysis confirms the RT-PCR results detecting only low levels of cytogenetically abnormal cells within the immature prelymphatic cell compartment.
Quantification of leukemic cells by RT-PCR and FISH analysis within the sorted CD34+CD19−population compared with contaminating CD19+ cells due to sorter errors.
The symbols (♦, RT-PCR; ●, FISH analysis; ▴, sorter errors) indicate individual values. The error bars show the SD.
Quantification of leukemic cells by RT-PCR and FISH analysis within the sorted CD34+CD19−population compared with contaminating CD19+ cells due to sorter errors.
The symbols (♦, RT-PCR; ●, FISH analysis; ▴, sorter errors) indicate individual values. The error bars show the SD.
Colony assays with sorted cell populations
To investigate if the CD34+CD19− cells not only lack the cytogenetic aberration of the leukemic cell clone but also display the differentiation pattern of normal progenitor cells, CD34+CD19− cells were isolated and seeded into methylcellulose cultures. These conditions supported the growth of myeloid progenitors of all lineages.
The clonogenicity of thawed and subsequently flow-sorted CD34+CD19− cells in control bone marrow was 6.5% (n = 3, SD 4.8%) (Table 2). Similarly, sorted CD34+CD19− cells from 7TEL/AML1-negative ALL patients had a median clonogenicity of 4.0% (SD 2.5%) (Table 2). This included the whole spectrum of myeloid colonies, including granulocyte colony-forming units (CFU-Gs), granulocyte macrophage CFUs (CFU-GMs), erythroid burst-forming units (BFU-Es), and granulocyte, erythroid, macrophage, megakaryocyte CFUs (CFU-GEMMs). Normal and leukemic CD19+ B-lineage cells did not grow under these conditions (clonogenicity < 0.03% and < 0.01%, respectively).
Clonogenicity of sorted CD34+CD19−and CD34+CD19+ cells in methylcellulose cultures
| Sample source . | Percentage of clonogenic cells . | |
|---|---|---|
| CD34+CD19− . | CD34+CD19+ . | |
| Control bone marrow (n = 3) | 6.5 (± 0.8) | < 0.03 |
| TEL/AML1-negative ALL (n = 7) | 4.0 (± 2.5) | < 0.01 |
| TEL/AML1-positive patients | ||
| Patient no. 1523/00 | 6.1 | 0 |
| Patient no. 807/00 | 15.3 | < 0.01 |
| Patient no. 589/97 | 5.5 | 0 |
| Sample source . | Percentage of clonogenic cells . | |
|---|---|---|
| CD34+CD19− . | CD34+CD19+ . | |
| Control bone marrow (n = 3) | 6.5 (± 0.8) | < 0.03 |
| TEL/AML1-negative ALL (n = 7) | 4.0 (± 2.5) | < 0.01 |
| TEL/AML1-positive patients | ||
| Patient no. 1523/00 | 6.1 | 0 |
| Patient no. 807/00 | 15.3 | < 0.01 |
| Patient no. 589/97 | 5.5 | 0 |
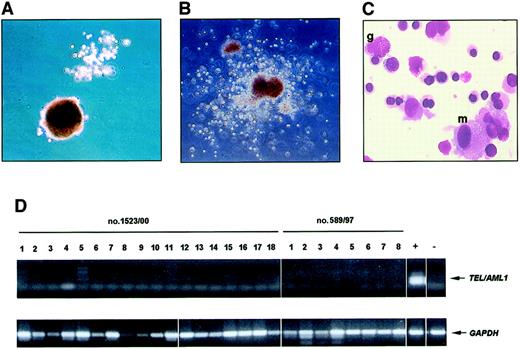
In 3 TEL/AML1-positive patients (nos. 589/97, 807/99, 1523/00), enough cells were available for colony assays. From 600 to 3000 CD34+CD19− cells and 30 000 CD34+CD19+ cells from each patient were sorted and seeded into methylcellulose cultures. The clonogenicity of the CD34+CD19− cells ranged from 5.5% to 15.3% (Table 2), while CD19+ leukemic cells showed no colony formation. These cultures included growth of CFU-Gs, CFU-GMs, BFU-Es, and immature multilineage CFU-GEMMs (Figure4A-C).
Analysis of multilineage colonies from flow-sorted CD34+CD19− cells.
The upper left photos show a CFU-G together with a BFU-E (A) and a multilineage CFU-GEMM (B) in methylcellulose cultures initiated with purified CD34+CD19− cells from patient nos. 589/97 (A) and 1523/00 (B). A Pappenheim-stained smear of the CFU-GEMM shows the presence of granulocytic (g), macrophage (m), and erythroid cells (C). Original magnification A-B, × 50; C, × 1000. A representative RT-PCR analysis of 18 colonies from patient no. 1523/00 and 8 colonies from patient no. 589/97 shows that CFU derived from sorted CD34+CD19− cells lack expression of TEL/AML1 (D).
Analysis of multilineage colonies from flow-sorted CD34+CD19− cells.
The upper left photos show a CFU-G together with a BFU-E (A) and a multilineage CFU-GEMM (B) in methylcellulose cultures initiated with purified CD34+CD19− cells from patient nos. 589/97 (A) and 1523/00 (B). A Pappenheim-stained smear of the CFU-GEMM shows the presence of granulocytic (g), macrophage (m), and erythroid cells (C). Original magnification A-B, × 50; C, × 1000. A representative RT-PCR analysis of 18 colonies from patient no. 1523/00 and 8 colonies from patient no. 589/97 shows that CFU derived from sorted CD34+CD19− cells lack expression of TEL/AML1 (D).
To demonstrate that these progenitors not only exhibited a normal proliferation and differentiation pattern but also lack expression ofTEL/AML1, single colonies were isolated and subjected to RT-PCR. None of 92 colonies isolated from methylcellulose cultures with purified CD34+CD19− cells (patient nos. 589/97 31 and 1523/00 61 colonies) showed expression of TEL/AML1(Figure 4D). The molecular analysis of these colonies provides further evidence that CD34+CD19− cells represent a genetically and functionally normal residual progenitor cell population.
Purity of flow sorting
Diagnostic bone marrow samples were chosen for analysis, though the probability of sorter errors due to the high percentage of leukemic cells was expected to be high.2 However, our own flow cytometric analyses of immature cells in good-prognosis childhood ALL had suggested that normal progenitor/stem cells may be predominant within this compartment.14,15 Moreover, immature leukemic (stem) cells—similar to acute myeloid leukemias21 22—may be rare. Consequently, remission bone marrow samples in children with a rapid response to chemotherapy and a low relapse rate were expected to contain few if any putative CD19− immature leukemic (stem) cells.
Direct quantification of the expected sorter errors except in patient no. 1523/00 was impossible because the number of sorted CD34+CD19− cells from theTEL/AML1-positive patient samples was too low for reanalysis. Therefore, control sorts with diagnosticTEL/AML1-negative ALL bone marrow samples were performed applying the same gating strategy, to estimate the purity of CD34+CD19− cells after flow sorting and to quantify the percentage of contaminating CD19+ leukemic cells in these sorts. All 5 control patients had a CD19+B-cell precursor ALL with a similar bone marrow blast cell count (mean 85.6%, SD 8.4%) compared with the TEL/AML1-positive patients (mean 84.4%, SD 24.1%).
Considering the rarity of the CD34+CD19− cell population in leukemic bone marrow samples at diagnosis, the purity in these control sorts was high: 94% in patient no. 1523/00 and 89% to 99% (n = 5, mean 93.4%, SD 4.4%) in the 5 control patients. Accordingly, the level of contaminating CD19+ cells in these sorts, including patient no. 1523/00 (4% CD19+ cells after flow sorting, compare Figure 1B), varied between 0.5% and 9% (n = 6, mean 4.6%, SD 3.6%). This correlates with the number ofTEL/AML1-positive cells detected in the CD34+CD19− cell population by RT-PCR and FISH, suggesting that the low levels of leukemic cells in this population can be attributed to contamination through sorter errors (Figure 3).
Discussion
Previous flow cytometric studies of immature CD34+CD19− cells in the bone marrow of children with B-cell precursor ALL had demonstrated that this population is characterized by an immunophenotype identical to normal progenitor/stem cells. Unlike the bulk leukemic population, the immature cells showed expression of CD4514 in CD45-negative ALL,23,24 of CD117,15 which is generally absent on B-lineage leukemic blasts,25,26 and of CD133.15 Based on these studies we hypothesized that immature CD34+CD19− cells in the bone marrow of children with ALL may represent a candidate normal progenitor/stem cell population. As has been assumed for a long time,1this also implies that the leukemia in typical childhood ALL originates in a CD19+ lymphoid progenitor.
Childhood ALL, however, is a heterogeneous disease. On one hand, there are patients with Philadelphia chromosome–positive ALL/t(9;22) with a dismal prognosis, particularly if unresponsive to steroids.27,28 On the other hand, there is the vast majority of children with a favorable outcome and cure rates close to 80%.6 Accordingly, there may also be heterogeneity in stem cell involvement in ALL, as has recently been suggested by several studies,2-5 necessitating analyses of the progenitor/stem cell compartment in well-defined ALL subgroups.
Here, we analyzed the CD34+CD19−progenitor/stem cell compartment in patients with a molecularly defined subtype that is common in childhood ALL. In 9 children withTEL/AML1-positive ALL, CD34+CD19− cells were purified and characterized by RT-PCR, FISH analysis, and methylcellulose cultures. The percentage of leukemic cells in this population was low. In 3 of 8 patients analyzed by nested RT-PCR, the percentage ofTEL/AML1-positive leukemic cells was below the limit of detection. By semiquantitative real-time RT-PCR, the mean percentage of leukemic cells in the flow-sorted CD34+CD19−cell fraction in all 8 patients was estimated to be 1.6%, ranging from zero to a maximum of 6.0% (Table 1). Due to the limitations of quantifying leukemic cells in clinical samples by RNA-based PCR methods (see “Results”) these data, however, needed cytogenetic confirmation.
When rare leukemic cells carrying clonal chromosomal translocations are quantified by FISH analysis of interphase nuclei, one has to take into account that by chance 2 fluorescent probes lie closely together, forming a false positive fusion signal. Therefore, in accordance with diagnostic standards, the lower cutoff for this method was defined by the mean percentage of false-positive signals in control tissue that is negative for the respective translocation (here, sorted BLIN-1 cells) plus 3 SDs. Applying this definition, an average percentage of 2.5% leukemic cells carrying the translocation was detected in the sorted CD34+CD19− cell fraction from 5 patients with ALL/t(12;21). Even a more conservative estimation, ie, simply subtracting the mean background of 3.1% from the individual measurements as shown in Table 1, led to an only marginally higher percentage of 4.5% leukemic cells in this population. Thus, these data confirm the RT-PCR–based estimations that only low levels of leukemic cells could be detected in the CD34+CD19− cell fraction in these 9 children with TEL/AML1-positive ALL.
Functional analysis of the CD34+CD19− cells demonstrated that this population represents a residual normal progenitor/stem cell population. Flow-sorted CD34+CD19− cells showed a similar clonogenicity in methylcellulose cultures as compared with cells with an identical immunophenotype from control bone marrow (Table 2). The whole range of myeloid colonies, including CFU-Gs, CFU-GMs, BFU-Es, and primitive multilineage CFU-GEMMs (Figure 4A-C), were formed in methylcellulose cultures with purified CD34+CD19− cells from leukemic bone marrow. Moreover, none of 92 colonies isolated from methylcellulose cultures with sorted CD34+CD19− cells (from patient nos. 589/97 and no. 1523/00) and analyzed by RT-PCR expressedTEL/AML1 (Figure 4D). Although the culture conditions did not support the growth of lymphoid cells, aTEL/AML1-positive pluripotent cell should have been able to differentiate into myeloid colony-forming progenitors. These experiments therefore provide further evidence that the colonies originated from residual normal progenitor cells and not from a putative TEL/AML1-positive CD34+CD19− (pre-)leukemic stem cell that still had the potential for pluripotent differentiation.
The purity of flow-sorted cell populations largely depends on the frequency of the target population in the processed sample. Thus, isolation of rare CD34+CD19− cells from leukemic bone marrow (< 1% of all cells) was expected to be associated with a high frequency of sorter errors. Enrichment of the CD34+CD19− cells preceding the cell sort, eg, by B-cell depletion with magnetic beads, was impossible due to the expected cell loss and the small sample size in pediatric patients. It was therefore imperative to quantify the frequency of the sorter errors. Diagnostic bone marrow samples from patient no. 1523/00 and 5 patients with TEL/AML1-negative B-cell precusor ALL were chosen as controls. Using the same gating as applied for isolation of CD34+CD19− cells for RT-PCR, FISH, and colony assays, a mean contamination of the CD34+CD19−sorted cell fraction with CD19+ leukemic cells of 4.6% (range, 0.5% to 9%) was observed. This was in the same range as the percentage of leukemic cells detected by RT-PCR and FISH analysis inTEL/AML1-positive patients (Figure 3).
An additional technical limitation of flow sorting of rare events is a consequence of the log-normal distribution of antigen expression leading to overlap of different populations.29 In particular, when rare events (in our analysis, CD34+CD19− cells) are amplified against the background of a dominant population (in our analysis, leukemic CD34+CD19+ cells), events belonging to the main population but with an antigen expression (here, CD19) of 2 SDs below its mean will be amplified.14 We therefore conclude that the low levels of leukemic cells detected in the flow-sorted CD34+CD19− population can be explained by sorter errors and overlap of the populations rather than involvement of the immature progenitor/stem cell compartment in the leukemic process.
These results are consistent with the hypothesis that the leukemia in childhood TEL/AML1-positive B-cell precursor ALL originates from a CD19+ lymphoid progenitor cell. This does not contradict recent studies showing involvement of a more primitive progenitor/stem cell compartment, particularly in high-risk patients.2-5 In large clinical trials, response to prednisone has shown to be the single most important prognostic marker predicting response to therapy and outcome.6,30 It can thus be speculated that in good-prognosis ALL, such asTEL/AML1-positive ALL, the leukemia originates in a lymphoid, steroid receptor–positive progenitor cell that is prone to undergo apoptosis. On the other hand, in patients with prognostically unfavorable subtypes, the leukemia—like in myeloid leukemias21,22 31—may arise in a pluripotent progenitor/stem cell resistant to apoptosis. This implies that the biology of the leukemia is not only determined by the molecular events (eg, chromosomal translocations) leading to the malignant transformation but also by the cellular microenvironment (ie, the pattern of gene expression) of the progenitor cell in which the transformation occurs.
It will be important to determine if the involvement/noninvolvement of the more primitive progenitor/stem cell compartment is specific for certain chromosomal translocations (eg, frequent involvement in ALL/t(9;22) versus noninvolvement in ALL/t(12;21)) or independent of the chromosomal translocations leading to heterogeneity within molecularly defined ALL subtypes. As has recently been shown, even inTEL/AML1-positive ALL there appears to be heterogeneity in the stage of immunoglobulin heavy chain rearrangement and thus the level of differentiation of the lymphoid progenitor in which the translocation occurs.32 Additional studies in larger numbers of patients and different ALL subtypes within prospective clinical trials are therefore necessary to define the frequency and clinical relevance of involvement of the immature prelymphatic progenitor/stem cell compartment in childhood ALL.
In addition to gaining a better understanding of the stem cell biology of ALL, this has many therapeutic implications. In some children with prognostically unfavorable ALL subtypes or with relapse, autologous stem cell transplantation is regarded as one possible treatment option if no allogeneic donor is available. Transplantation protocols often include purging of the autologous stem cell products to minimize the potential risk of reinfusing leukemic cells.33 34 However, purging strategies based on the expression of lymphoid markers will only be effective if all leukemic cells, including putative immature ALL stem cells, express the respective marker.
Detection of minimal residual disease in childhood ALL has been correlated with a high risk of relapse.35 36 Immunologic methods based on leukemia-associated immunophenotypes may, however, lack sensitivity if the most primitive leukemic progenitors have a phenotype distinct from the bulk leukemic population.
Most importantly, exact definition of the leukemic phenotype is a prerequisite for the development of effective immunotherapy. CD19 is considered a promising candidate target antigen because its expression is highly B-lineage specific. CD19 is under the control of the B-cell transcription factors Pax537 and human early B-cell factor–like protein,38 and CD19 knock-out mice show no abnormalities except for the defects in B-cell function.39However, if the leukemia originates in a CD19−progenitor/stem cell, the most primitive leukemic cells will be spared and initiate regrowth of the leukemia from this immature progenitor/stem cell compartment.
In conclusion, this investigation provides molecular and functional evidence that in TEL/AML1-positive childhood ALL, immature CD34+ cells that lack expression of B-cell markers are not part of the leukemic cell clone. Our data support the hypothesis that the leukemia in typical childhood ALL originates in a CD19+lymphoid progenitor.1 This has many implications for understanding the biology of TEL/AML1-positive B-cell precursor ALL and for targeting therapy against the leukemic cell clone.
We gratefully thank Dr B. Bürger, Prof Dr J. Kienast, Dr T. Lapidot, and Dr C. Rössig for critically reviewing the manuscript and Prof Dr W.-D. Ludwig and Prof Dr M. Schrappe from the ALL-BFM study group for providing 2 additional patient samples. We acknowledge Christina Böth-Sauerwein, Sabine Gräf-Höchst, Thomas Jung, and Tanja Möllers for technical help.
Supported by DFG grant Vo 476/3-3 (J.V.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Josef Vormoor, Klinik und Poliklinik für Kinderheilkunde, Pädiatrische Hämatologie/Onkologie, Universitätsklinikum Münster, Albert-Schweitzer-Str 33, 48129 Münster, Germany; e-mail:vormoor@uni-muenster.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal