Abstract
Molecular and cellular markers associated with malignant disease are frequently identified in healthy individuals. The relationship between these markers and clinical disease is not clear, except where a neoplastic cell population can be identified as in myeloma/monoclonal gammopathies of undetermined significance (MGUS). We have used the distinctive phenotype of chronic lymphocytic leukemia (CLL) cells to determine whether low levels of these cells can be identified in individuals with normal complete blood counts. CLL cells were identified by 4-color flow cytometric analysis of CD19/CD5/CD79b/CD20 expression in 910 outpatients over 40 years old. These outpatients were age- and sex-matched to the general population with normal hematologic parameters and no evident history of malignant disease. CLL phenotype cells were detectable in 3.5% of individuals at low level (median, 0.013; range, 0.002- 1.458 × 109 cells/L), and represented a minority of B lymphocytes (median, 11%; range, 3%-95%). Monoclonality was demonstrated by immunoglobulin light-chain restriction in all cases with CLL phenotype cells present and confirmed in a subset of cases by consensus-primer IgH-polymerase chain reaction. As in clinical disease, CLL phenotype cells were detected with a higher frequency in men (male-to-female ratio, 1.9:1) and elderly individuals (2.1% of 40- to 59-year-olds versus 5.0% of 60- to 89-year-olds, P = .01). The neoplastic cells were identical to good-prognosis CLL, being CD5+23+20wk79bwk11a−22wksIgwkCD38−, and where assessed had a high degree (4.8%-6.6%) of IgH somatic hypermutation. The monoclonal CLL phenotype cells present in otherwise healthy individuals may represent a very early stage of indolent CLL and should be useful in elucidating the mechanisms of leukemogenesis.
Introduction
The identification of neoplastic cells or markers of neoplasia in “normal” or subclinical states has been critical in developing methods for early disease identification, as well as for studying the mechanisms of oncogenesis and disease progression. Such markers can be broadly categorized into 3 groups: serum markers such as prostate-specific antigen (PSA; prostate cancer)1; molecular markers, usually balanced translocations such as t(14;18)2-4 or t(9;22)5,6 associated with follicular lymphoma and chronic myeloid leukemia (CML), respectively; or a neoplastic cell type, such as the abnormal plasma cells that are present in both myeloma and its premalignant counterpart, monoclonal gammopathy of undetermined significance (MGUS).7-9
Molecular markers are clearly central to the pathogenesis of disease, and their identification can lead directly to effective therapeutic strategies, such as signal transduction inhibitors (imatinib mesylate [STI571]) in CML.10 11 Although translocations may be detected in a high proportion of healthy individuals, the characteristics of the cell(s) carrying the mutation are not known. It is therefore difficult to determine the relationship between the presence of a molecular marker and malignant disease.
More useful information, both clinically and scientifically, can be obtained if it is possible to identify a neoplastic cell population, as in myeloma/MGUS. Longitudinal studies have demonstrated the pathogenetic relationship between MGUS and myeloma.12Comparison of neoplastic cells from patients with MGUS with those from healthy individuals can identify aberrations that occur in the early stages of oncogenesis, for example, translocations into the immunoglobulin switch region and dysregulation of interleukin 6 receptor expression.13-15 Comparison of plasma cells from patients with MGUS with those from patients with myeloma can identify the factors responsible for malignant transformation, for example, deletions of 13q and ras mutations.16 17
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world, affecting 2 to 6 individuals per 100 000 per year.18 Conventional classification requires more than 5 × 109/L circulating monoclonal B lymphocytes with a CD5+CD23+ phenotype and weak or no surface immunoglobulin expression.19 There is no known translocation or serum marker associated with CLL, and the only methods of identifying the neoplastic cells are through morphology, immunophenotype, or polymerase chain reaction (PCR) amplification of their unique immunoglobulin heavy-chain gene rearrangement. Until recently, all these techniques have required the presence of an excess of neoplastic cells for identification, and hence are unsuitable for detecting minimal CLL cell levels. However, we have developed a flow cytometry technique that uses a sequential gating strategy to accurately and sensitively identify total B lymphocytes at the level of 0.002%. CLL cells are then discriminated from normal B lymphocytes by their higher CD5 and lower CD20 and CD79 expression. CLL cells are uniquely identifiable by this technique in all patients with clinical disease even when they represent as few as 0.5% of total B lymphocytes.20 As such, this test is suitable for screening normal individuals to identify CLL phenotype cells. The aim of this study was therefore to identify the prevalence of CLL phenotype cells in a series of 910 hospital outpatients who had normal hematologic parameters and no history of malignant disease.
Patients, materials, and methods
Patients
Ethical approval was obtained from the local review board to collect anonymous samples from specimens sent for routine full blood count analysis. EDTA peripheral blood samples (910 total) from 425 males and 485 females were selected. Inclusion criteria were: (1) samples had a normal leukocyte count and differential, normal platelet count, and normal hemoglobin level; (2) samples were not sent to assess the presence of any malignancy; (3) patients had not previously been seen at a hematology, oncology, or transplantation clinic; and (4) samples were less than 24 hours old.
Samples were chosen from general practice, ophthalmology, gynecology, cardiology, dermatology, orthopedic preoperative patients, or patients presenting to the emergency department with chest pain, shortness of breath, or trauma. Once selected, only the sex, age, leukocyte count and differential, platelet count, and hemoglobin levels were recorded. Samples were balanced to represent the age and sex distribution of the normal United Kingdom population. In addition, diagnostic immunophenotypic analysis of 108 sequential patients with CLL has been included.
4-color flow cytometry
Leukocytes were prepared from EDTA peripheral blood by incubation with a 10-fold excess of NH4Cl (8.6 g/L in distilled water) for 5 minutes, centrifuged at 200g for 5 minutes, and washed twice in FACSFLOW/0.3% bovine serum albumin. Leukocytes (106) were incubated with 1 of 2 antibody cocktail mixtures: (1) CD20 fluorescein isothiocyanate (FITC), CD79b phycoerythrin (PE), CD19 Cyanine 5.18 (Cy5)/PE, and CD5 allophycocyanin (APC); (2) anti-κ FITC, anti-λ PE, CD19 Cy5/PE, and CD5 APC. A minimum of 200 000 total leukocytes was acquired and analyzed using CELLQuest software on a FACSort cytometer (BD Biosciences, Oxford, United Kingdom).
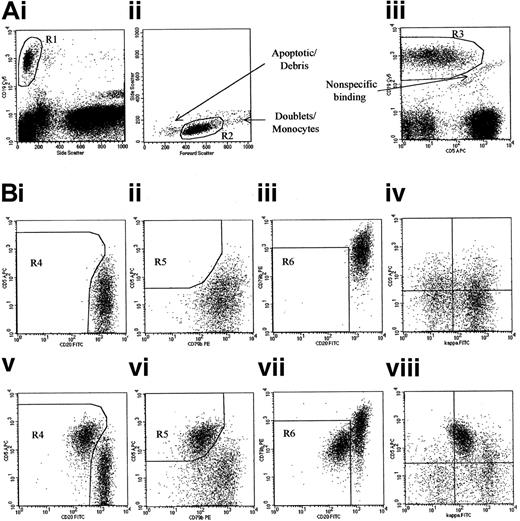
The sequential gating strategy is shown in Figure1. An initial region (R1) was set around cells with high CD19 expression and low side scatter (granularity); a second region (R2) was set on the physical characteristics of the CD19+ cells, excluding small apoptotic cells/debris as well as doublets/monocytes that can bind antibodies nonspecifically; finally a third region (R3) was set to exclude cells that were binding equivalent amounts of both CD19 and CD5—such cells fall in a diagonal line that indicates nonspecific binding. Further analysis was performed on cells that fell within all 3 of these regions. Total B lymphocytes were identified by their CD19 and light scatter characteristics. CLL phenotype cells were identified within the B-lymphocyte population by their high CD5 and low CD20 and CD79b expression. A population was classified as being CLL phenotype if there were over 50 events in all 3 “CLL” regions and their immunoglobulin light-chain expression was abnormal (κ/λ < 0.5 or > 4.0, or > 25% lacking surface immunoglobulin). A minimum of 50 events was required to define a population, because detection at this level was reproducible between operators.
Identification of monoclonal CLL phenotype cells in normal individuals.
(A) The gating strategy used to identify B lymphocytes and exclude contamination is shown. (B) The analysis of B-lymphocyte phenotype in 2 representative individuals, 1 with only normal B lymphocytes present (plots i-iv) and 1 with CLL phenotype cells present (plots v-viii). Cells were classified as having a CLL phenotype if a discrete population with stronger CD5, weaker CD20, and weaker CD79b than the other “normal” B lymphocytes was present in all 3 “CLL” regions (R4, R5, and R6, shown in plots i/v, ii/vi, and iii/vii, respectively). This phenotypic profile is unique to CLL, and as such this assay can detect CLL cells when they represent as few as 0.002% of total leukocytes, or 0.5% of Blymphocytes in all samples.20Plot viii demonstrates weak κ expression on the population of CD5+ cells. The remaining cells have a normal distribution of κ+ and κ− cells similar to that seen in plot iv, which shows the distribution in an individual with no CLL phenotype cells. Although the test is specific for CLL cells, monoclonal populations with a non-CLL phenotype may be detected if there are sufficient neoplastic cells to perturb the normal κ/λ ratio.
Identification of monoclonal CLL phenotype cells in normal individuals.
(A) The gating strategy used to identify B lymphocytes and exclude contamination is shown. (B) The analysis of B-lymphocyte phenotype in 2 representative individuals, 1 with only normal B lymphocytes present (plots i-iv) and 1 with CLL phenotype cells present (plots v-viii). Cells were classified as having a CLL phenotype if a discrete population with stronger CD5, weaker CD20, and weaker CD79b than the other “normal” B lymphocytes was present in all 3 “CLL” regions (R4, R5, and R6, shown in plots i/v, ii/vi, and iii/vii, respectively). This phenotypic profile is unique to CLL, and as such this assay can detect CLL cells when they represent as few as 0.002% of total leukocytes, or 0.5% of Blymphocytes in all samples.20Plot viii demonstrates weak κ expression on the population of CD5+ cells. The remaining cells have a normal distribution of κ+ and κ− cells similar to that seen in plot iv, which shows the distribution in an individual with no CLL phenotype cells. Although the test is specific for CLL cells, monoclonal populations with a non-CLL phenotype may be detected if there are sufficient neoplastic cells to perturb the normal κ/λ ratio.
Extended phenotyping was performed on cases containing CLL phenotype cells that could be discriminated from normal B lymphocytes using only CD19 and either CD5 or CD20. Cells were then incubated with CD19 Cy5/PE, CD5, or CD20 APC, and the following antibody pairs: CD3 FITC and CD3 PE (controls); anti-κ FITC and anti-λ PE; CD5 or CD20 FITC and CD79b PE; CD11a FITC and CD23 PE; IgM FITC and CD38 PE; CD10 FITC and CD22 PE; and IgG FITC and CD27 PE. Antibodies were prepared in-house except CD20 FITC and CD79b (Coulter, Oxford, United Kingdom) and CD20 APC, CD22 PE, IgM FITC, IgG FITC, and CD27 PE (BD Biosciences).
Immunoglobulin heavy-chain gene PCR amplification and sequencing
DNA was obtained from samples containing CLL phenotype cells and from age- and sex-matched normal controls (2 controls per individual with CLL phenotype cells) by standard phenol-chloroform extraction and ethanol precipitation. Amplification across the rearranged IgH genes was performed as reported previously21 using consensus family-specific VH primers and a consensus JHprimer, fluorescently labeled for fingerprint analysis or unlabeled for sequencing. For VH sequencing, PCR products were then enzymatically treated with shrimp alkaline phosphatase (2.0 U/μL) and exonuclease I (10.0 U/μL) and subjected to cycle sequencing with the ABI Prism BigDye Terminator Ready Reaction Kit (Foster City, CA). PCR products were electrophoresed on either a 6% or 4.25% polyacrylamide gel (for clonality assessment or sequence analysis, respectively) in an ABI Prism 377 DNA Sequencer. Analysis was performed using ABI Genescan or Sequencing Analysis 3.0 software. Sequences were aligned and compared to germ line IgH sequences on IgBlast website, which provides analysis of immunoglobulin sequences in Genbank (http://www.ncbi.nih.gov/igblast/).
Results
Identification of cells with a CLL phenotype in healthy individuals
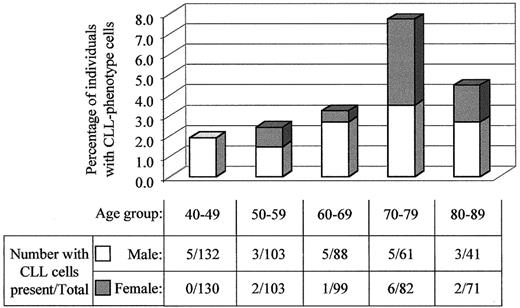
Cells with a CLL phenotype and evidence of light-chain restriction were detected in 21 of 425 men and 11 of 485 women, or 3.5% of total. The prevalence increased with age, from 2.1% in the individuals between 40 and 60 years old to 5.0% for individuals over 60 (χ2P = .01). The highest prevalence was found in 70- to 79-year-old individuals, with 8.2% of men and 7.3% of women having a detectable CLL phenotype population. The male predominance was consistent for all age groups, although least pronounced in the 70- to 79-year-old group. Figure2 shows the prevalence for each age and sex group. The absolute numbers of CLL phenotype cells were low, with a median of 0.013 × 109 cells/L, ranging from 0.003 to 1.458 × 109 cells/L. Furthermore, the CLL phenotype cells represented a minor proportion of total B lymphocytes in most cases, at a median of 11%, ranging from 3% to 95% of total B lymphocytes.
The prevalence of CLL phenotype cells increases with age and shows a high male-female ratio.
The percentage of men and women with CLL phenotype cells is shown graphically according to age group. Below, the table shows numbers of individuals with CLL phenotype cells as well as the total number of individuals analyzed from each age group.
The prevalence of CLL phenotype cells increases with age and shows a high male-female ratio.
The percentage of men and women with CLL phenotype cells is shown graphically according to age group. Below, the table shows numbers of individuals with CLL phenotype cells as well as the total number of individuals analyzed from each age group.
In addition to the individuals with CLL phenotype cells, monoclonal B lymphocytes with normal expression of CD5/20/79b (non-CLL phenotype) were detected due to a perturbation of κ/λ ratio in a further 9 (1.0%) of the 910 of individuals. These were mostly elderly individuals (median age, 78 years; range, 49-88 years). The cells were identified by light-chain restriction only, and we have previously shown that such an assay is approximately 2 orders of magnitude less sensitive in a polyclonal background than a disease-specific assay such as that used to detect CLL cells in this study.20 It is possible that the non-CLL monoclonal B lymphocytes represent early stages of follicular or marginal zone lymphoma and that, due to the relative insensitivity of the technique, more individuals have clonal non-CLL cells present.
Analysis of monoclonality by IgH-PCR
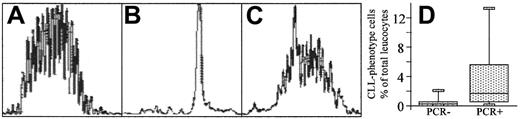
Monoclonality was confirmed by PCR amplification of the immunoglobulin heavy-chain gene using consensus primers to JH and Fr1, Fr2, or Fr3. Insufficient sample was available to analyze both extended phenotyping (see below) and IgH-PCR, so PCR analysis was performed on a proportion of the samples (n = 20) with detectable CLL phenotype cells. A monoclonal rearrangement was demonstrated in 8 of 12 amplifiable cases, which is similar to the reported detection rate in clinical CLL.22,23Unamplifiable or polyclonal samples had significantly lower numbers of CLL cells (Figure 3), below the limits of detection.24 Samples from 40 individuals with no CLL phenotype cells that were matched for age and sex all had polyclonal IgH rearrangements. All cases with a monoclonal excess detected by surface light-chain expression also had a monoclonal rearrangement detected by IgH-PCR. Thus all cases demonstrated surface light-chain restriction, and monoclonality was confirmed by IgH-PCR analysis in samples containing sufficient CLL phenotype cells for the sensitivity of the assay.
Confirmation of monoclonality by fluorescent IgH-PCR analysis.
The detection of monoclonality by IgH-PCR analysis of 3 representative individuals is shown. DNA from samples containing CLL phenotype cells and from age- and sex-matched normal controls (2 controls per individual with CLL phenotype cells) was amplified using a fluorescent JH primer and consensus primers to framework VH regions. PCR products show a range of sizes with a normal distribution separated by 3 base pairs in normal individuals (A), a single peak in individuals whose B lymphocytes are all CLL phenotype (B), or a large single peak on an otherwise normal distribution in individuals with both CLL phenotype and polyclonal B lymphocytes (C). PCR results for patients with CLL-phenotype cells detectable by flow cytometry are shown (D). The flow assay is more sensitive than PCR detection20 and as such, IgH-PCR analysis rarely identified a monoclonal population in samples with low levels (< 0.5%) of CLL phenotype cells (D).
Confirmation of monoclonality by fluorescent IgH-PCR analysis.
The detection of monoclonality by IgH-PCR analysis of 3 representative individuals is shown. DNA from samples containing CLL phenotype cells and from age- and sex-matched normal controls (2 controls per individual with CLL phenotype cells) was amplified using a fluorescent JH primer and consensus primers to framework VH regions. PCR products show a range of sizes with a normal distribution separated by 3 base pairs in normal individuals (A), a single peak in individuals whose B lymphocytes are all CLL phenotype (B), or a large single peak on an otherwise normal distribution in individuals with both CLL phenotype and polyclonal B lymphocytes (C). PCR results for patients with CLL-phenotype cells detectable by flow cytometry are shown (D). The flow assay is more sensitive than PCR detection20 and as such, IgH-PCR analysis rarely identified a monoclonal population in samples with low levels (< 0.5%) of CLL phenotype cells (D).
Extended phenotypic analysis of the monoclonal B lymphocytes in healthy individuals
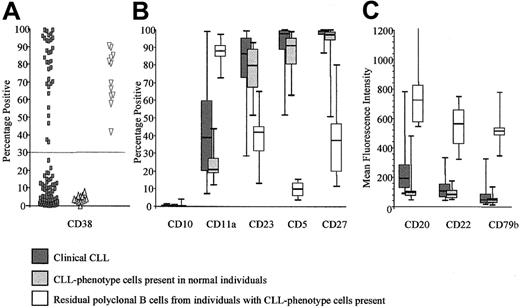
Extended phenotyping was performed on samples from 12 individuals in whom the monoclonal B lymphocytes could be distinguished from normal B lymphocytes on the basis of CD5 or CD20 expression alone. The antigens studied were CD10, CD11a, CD22, CD23, and CD27 in addition to CD5, CD20, CD79b, and surface κ and λ. The monoclonal cells present in these otherwise normal individuals were phenotypically identical to clinical CLL in all cases studied (Figure4).
CLL phenotype cells in normal individuals are phenotypically identical to clinical CLL cells.
The comparison of antigen expression is shown between clinical CLL samples (n = 108), CLL phenotype cells from normal individuals (n = 10), and the residual polyclonal B lymphocytes from individuals with CLL phenotype cells present (n = 10). Clinical CLL samples at presentation were analyzed as part of their routine diagnosis. Apart from CD38 expression, the clinical CLL cells and the CLL phenotype cells from normal individuals show a very similar phenotypic profile, which is distinct that from normal B lymphocytes. (A) The percentage of B cells expressing CD38, which has a bimodal distribution in presentation CLL. Cases showing more than 30% expression have a poor prognosis.33 Fewer than 10% of the CLL phenotype cells present in normal individuals express CD38, suggesting an indolent phenotype. The residual B lymphocytes express variable levels of CD38. (B) The percentage of B cells expressing CD10, CD11a, CD23, CD5, and CD27. CD10 is not detected on circulating normal or CLL phenotype B cells. CD11a is expressed weakly by clinical CLL cells and CLL phenotype cells from normal individuals, whereas normal B lymphocytes are virtually all positive. In contrast, the majority of clinical CLL cells and CLL phenotype cells from normal individuals express CD5, CD23, and CD27, whereas a variable proportion of normal B lymphocytes express these antigens. (C) The fluorescence intensity for CD20, CD22, and CD79b is shown. These antigens are expressed by all 3 B-cell types, but the level of expression differs. Thus, clinical CLL cells and CLL phenotype cells from normal individuals express very low levels of CD20, CD22, and CD79b, whereas all 3 antigens are expressed at a much higher level by normal B lymphocytes.
CLL phenotype cells in normal individuals are phenotypically identical to clinical CLL cells.
The comparison of antigen expression is shown between clinical CLL samples (n = 108), CLL phenotype cells from normal individuals (n = 10), and the residual polyclonal B lymphocytes from individuals with CLL phenotype cells present (n = 10). Clinical CLL samples at presentation were analyzed as part of their routine diagnosis. Apart from CD38 expression, the clinical CLL cells and the CLL phenotype cells from normal individuals show a very similar phenotypic profile, which is distinct that from normal B lymphocytes. (A) The percentage of B cells expressing CD38, which has a bimodal distribution in presentation CLL. Cases showing more than 30% expression have a poor prognosis.33 Fewer than 10% of the CLL phenotype cells present in normal individuals express CD38, suggesting an indolent phenotype. The residual B lymphocytes express variable levels of CD38. (B) The percentage of B cells expressing CD10, CD11a, CD23, CD5, and CD27. CD10 is not detected on circulating normal or CLL phenotype B cells. CD11a is expressed weakly by clinical CLL cells and CLL phenotype cells from normal individuals, whereas normal B lymphocytes are virtually all positive. In contrast, the majority of clinical CLL cells and CLL phenotype cells from normal individuals express CD5, CD23, and CD27, whereas a variable proportion of normal B lymphocytes express these antigens. (C) The fluorescence intensity for CD20, CD22, and CD79b is shown. These antigens are expressed by all 3 B-cell types, but the level of expression differs. Thus, clinical CLL cells and CLL phenotype cells from normal individuals express very low levels of CD20, CD22, and CD79b, whereas all 3 antigens are expressed at a much higher level by normal B lymphocytes.
Assessment of CLL-specific prognostic factors in the monoclonal CLL phenotype cells present in healthy individuals
The degree of hypermutation of the immunoglobulin VHgene and the level of CD38 expression are potent prognostic indicators in CLL. Approximately 30% to 50% of patients show more than 2% VH mutation, and less than 30% expression of CD38 by the CLL cells; these patients have an extremely good prognosis. In 3 of 12 amplifiable samples, the level of monoclonal CLL phenotype cells was sufficiently high and the polyclonal background sufficiently low that a single band was generated by fluorescent IgH-PCR. In these cases, it was possible to reamplify the sample using nonfluorescent Fr1 and Fr2 primers and sequence the products directly. Sequences were performed at least twice on independently amplified PCR product, and showed 100% identity. In all cases, the sequence showed significant levels of somatic hypermutation (4.8, 6.6, and 8.0% deviation from germ line). The VH families used were VH3-21 in 2 cases and VH3-74 in the other, which are rarely used in germ line or mutated clinical CLL.25 In a further 12 cases, it was possible to assess CD38 expression. In all cases, there was less than 5% expression in comparison to CD3 control, suggesting that the CLL phenotypes have similar characteristics to clinical disease with a good prognosis (Figure 4).
Discussion
In this study, we have identified a monoclonal population of B lymphocytes with the characteristics of CLL cells in 3.5% of individuals over the age of 40. Monoclonality was demonstrated by immunoglobulin light-chain restriction and IgH-PCR analysis. The assay used to detect the CLL phenotype cells has been validated on clinical samples for the detection of minimal levels of CLL cells in a polyclonal background.20 The prevalence increases with age, and there is an approximately 2:1 male-to-female predominance, both similar to clinical CLL.26 The extended phenotype of the neoplastic cells is identical to that of clinical CLL, and where assessed the neoplastic cells show the characteristics of the indolent disease variety. These results indicate that the CLL phenotype cells represent an abnormal clonal population rather than a monoclonal reactive response, and they are most likely to represent the earliest stages of indolent CLL.
Genetic disease markers, such as the t(14;18) translocation of follicular lymphoma or the Philadelphia chromosome t(9;22) of CML, are also found in normal individuals. However, it is not known whether the translocations occur in a relevant cell type; for example, it has been suggested that the t(14;18) translocation may occur in a variety of non–B-cell leukocytes.27 Similarly, individuals with raised serum factors associated with malignancy, such as PSA, do not have an identifiable clone of cells. Although clearly of scientific relevance, it is difficult or impossible to identify the relationship between such markers and clinical disease. However, the observation that CLL phenotype cells are found in “normal” individuals, and that these cells are indistinguishable from the neoplastic cells found in patients with the disease, including by clonality studies, demonstrates that cells of a relevant lineage are involved in the clonal process.
In addition to these individuals with CLL phenotype cells, monoclonal B lymphocytes with normal expression of CD5/20/79b (therefore not CLL phenotype) were identified in a further 1.0% of individuals. It is possible that such cells represent early stages of either follicular or marginal zone lymphoma, suggesting that all the common chronic lymphoproliferative disorders, for example, myeloma, CLL, and follicular and probably marginal zone lymphoma, have a relatively frequent premalignant counterpart. This supports the “multihit” hypothesis of oncogenesis and suggests that the CLL phenotype cells found in “normal” individuals should provide an extremely useful resource for delineating the genetic events responsible for disease progression.
This study was designed to determine the prevalence of CLL phenotype cells in individuals with normal complete blood counts who were granted anonymity to comply with ethical constraints. As such, it was beyond the scope of this study to perform the longitudinal studies required to identify whether any of the individuals with CLL phenotype cells progress to clinical disease. However, the relationship between MGUS and myeloma may provide a model for the kinetics of progression. Approximately 25% of patients with MGUS show disease progression to myeloma after a median of 10 years. The neoplastic cells are rarely a transient feature, and disease levels are either constant with time or show steady progression at presentation or at a later time point.28 If the CLL phenotype cells show a gradual increase in number, then it is likely that some individuals will develop clinical disease. However, this proportion will be at most small: the prevalence of CD38-CLL is approximately 0.03% (based on an estimated incidence of 3/100 000 with a median survival of 10 years). This is 100 times less than that of “subclinical” CLL, suggesting that fewer than 1% will show clinical disease progression. Longitudinal studies are ethically difficult, but further analysis of the “subclinical” disease model is vital to understanding disease pathogenesis, as epitomized by analysis of the neoplastic cells in MGUS and myeloma. Translocations into the immunoglobulin switch regions have been reported to be an early and potentially initiating event in both disorders, whereas the genetic aberrations responsible for accelerated disease progression are becoming apparent by comparison of MGUS, myeloma, and plasma cell leukemia.13,29-31 Despite being the most common lymphoproliferative disorder in the Western world, the oncogenic events responsible for CLL remain unclear, and molecular aberrations have only been identified by analysis of global cytogenetic changes.32
The degree of somatic hypermutation in the immunoglobulin heavy-chain gene and the level of CD38 expression by the neoplastic cells are powerful prognostic factors in CLL.25,33 34 In the cases assessed, the CLL phenotype cells have demonstrated a high degree of VH mutation. Furthermore, in all 10 samples assessed there was no detectable CD38 expression. If the degree of somatic hypermutation and absence of CD38 expression is a consistent feature of the CLL phenotype cells present in normal individuals, this suggests that these cells represent the early stages of the indolent variety of CLL. No CD38+ patients were identified, possibly because CD38+ CLL is more proliferative and hence patients are more likely to present with a lymphocytosis. The lack of a “subclinical” CD38+/unmutated CLL population supports the hypothesis that clinical CLL contains 2 distinct entities.
The identification of CLL phenotype cells in normal individuals has many implications for understanding the development of malignant disease. The cells are readily identifiable and may be isolated to examine gene/protein expression profiles in relation to normal B lymphocytes, to identify the early stages of oncogenesis, and to clinical CLL to identify the mechanisms of disease progression.
Supported by Yorkshire Cancer Research and the Leukaemia Research Fund of Great Britain.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andy C. Rawstron, Haematological Malignancy Diagnostic Service, Algernon Firth Building, Leeds General Infirmary, Leeds LS1 3EX, United Kingdom; e-mail:andy.rawstron@hmds.org.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal