The CHK2 gene codifies for a serine/threonine kinase that plays a central role in DNA damage response pathways. To determine the potential role of CHK2 alterations in the pathogenesis of lymphoid neoplasms we have examined the gene status, protein, and mRNA expression in a series of tumors and nonneoplastic lymphoid samples. A heterozygous Ile157Thr substitution, also present in the germ line of the patient, was detected in a blastoid mantle cell lymphoma (MCL). CHK2 protein and mRNA expression levels were similar in all types of lymphomas and reactive samples, and these levels were independent of the proliferative activity of the tumors. However, 5 tumors, one typical MCL, 2 blastoid MCLs, and 2 large cell lymphomas, showed marked loss of protein expression, including 2 samples with complete absence of CHK2 protein. These 2 lymphomas showed the highest number of chromosomal imbalances detected by comparative genomic hybridization in the whole series of cases. However, no mutations, deletions, or hypermethylation of the promoter region were identified in any of these tumors. mRNA levels were similar in cases with low and normal protein expression, suggesting a posttranscriptional regulation of the protein in these tumors. CHK2 gene and protein alterations were not related to p53 and ATMgene status. In conclusion, CHK2 alterations are uncommon in malignant lymphomas but occur in a subset of aggressive tumors independently of p53 or ATM alterations. The high number of chromosomal imbalances in tumors with complete absence of CHK2 protein suggests a role of this gene in chromosomal instability in human lymphomas.
Introduction
Lymphoid malignancies are a heterogeneous group of disease entities characterized by distinctive clinical, morphologic, immunophenotypical, and genetic features. Cytogenetic studies have identified a series of primary genetic aberrations usually associated with particular disease entities. Most of these alterations are chromosomal translocations involving specific oncogenes, the activation of which is considered to be an important initial event in the development of the tumors.1 De novo aggressive lymphomas and transformation of indolent neoplasms into more aggressive variants are usually associated with increasing number of chromosomal alterations and the development of complex karyotypes, suggesting that mechanisms involved in the maintenance of genomic integrity are probably impaired and may participate in the evolution of these tumors.2-4
Recent studies have elucidated the main regulatory pathways involved in DNA repair response, linking DNA damage sensors to different downstream effector elements involved in cell cycle control, cell death, and DNA repair. Alterations in these regulatory pathways may lead to genomic instability and development of chromosomal alterations promoting the malignant transformation of cells and tumor progression.5Some of these regulatory elements are tumor suppressor genes with an important role in the pathogenesis of human tumors. Particularly, p53 inactivation has been recognized as a frequent phenomenon in the development of aggressive variants of lymphoid neoplasms.6-8 On the other hand, mutations in theATM gene are responsible for the ataxia-telangiectasia syndrome, a rare hereditary disorder predisposing to the development of lymphoid malignancies.9 In addition, ATM is also inactivated in a variety of sporadic lymphoproliferative neoplasms.10-15 Interestingly, mutations in this gene have been recently associated with defective chromosome damage repair and increasing number of chromosomal imbalances in B-cell chronic lymphocytic leukemia (B-CLL) and mantle cell lymphoma (MCL), respectively, suggesting that inactivation of genes involved in DNA damage repair pathways may play an important role in the development of chromosomal instability in lymphoid neoplasms.16 17However, the possible role of other genes involved in the DNA damage signaling pathways in human lymphomas has not been well examined.
CHK2, the mammalian homologue of yeast Rad53 andCds1 genes, encodes for a serine/threonine kinase that becomes rapidly phosphorylated and activated in response to DNA damage by the upstream ATM kinase. Different studies have now identified a number of CHK2 substrates including p53, CDC25A, CDC25C, and BRCA1, mediating cell cycle arrest at G1/S, intra-S, and G2/M phases, and promoting DNA repair mechanisms.18 The consideration of CHK2 as a new tumor suppressor gene has been supported by the identification of germ line mutations in a subset of families with Li-Fraumeni syndrome (LFS) carrying a wild-type p53 and the finding of somatic mutations in occasional sporadic tumors.19-28 However, the number of CHK2 mutations identified in sporadic neoplasms is relatively scarce. This low number of CHK2 alterations has been suggested to be due in part to frequent defects in other elements of the same regulatory pathways in the tumors. In LFS, CHK2 andp53 inactivation seem to occur as alternative pathogenetic mechanisms. The possible relationship between these 2 genes and the upstream regulator ATM in sporadic tumors is not well known. On the other hand, the possible existence of other types of gene aberrations in addition to point mutations or alterations in gene expression patterns in the tumors has not been well examined.
To determine the potential role of CHK2 gene alterations in the pathogenesis of lymphoid neoplasms, we have analyzed the gene status and protein and mRNA expression in a series of tumors, including a subset of lymphomas in which p53 and ATMabnormalities had been previously examined. Our findings indicate that CHK2-decreased protein expression levels and gene alterations are uncommon in malignant lymphomas but occur in a subset of aggressive tumors independently of the p53 or ATM gene status.
Materials and methods
Case selection
Tumor specimens from 81 non-Hodgkin lymphomas (NHLs) were obtained from the Department of Pathology of the Hospital Clinic, University of Barcelona, on the basis of the availability of frozen samples for molecular studies (Table 1). These tumors included 9 chronic lymphocytic leukemias (CLL), 5 large B-cell lymphomas transformed from CLL (Richter syndrome [RS]), 14 typical mantle cell lymphomas (T-MCL), 10 blastoid variants of mantle cell lymphomas (B-MCL), and 43 primary large B-cell lymphomas (LCL). Of these samples, 66 were obtained at diagnosis in untreated patients whereas samples of 4 T-MCL, 5 LCL, and the 4 RS were obtained in patients previously treated with chemotherapy. In 2 cases, no information about previous treatment was available. Most of these tumors had been examined for alterations of the p53 andATM genes, and chromosomal imbalances had been analyzed by comparative genomic hybridization (CGH).2,16 29 A series of nonneoplastic lymphoid samples were also examined, including 6 tonsils, 2 spleens, 6 reactive lymph nodes, and 6 samples of normal peripheral blood lymphocytes.
CHK2 gene alterations and expression in a series of malignant non-Hodgkin lymphomas
| Diagnosis . | Gene alterations1-153 . | Loss of protein expression1-153 . | mRNA expression . | Proliferation, Ki-67* . | Chromosomal imbalances, CGH† . | |||
|---|---|---|---|---|---|---|---|---|
| n . | mean ± SD . | n . | mean ± SD . | n . | mean ± SD . | |||
| Chronic lymphocytic leukemia | 0/9 | 0/7 | 7 | 0.7 ± 0.3 | 5 | 14.0 ± 8.2 | 2 | 4 ± 2.8 |
| Richter syndrome | 0/4 | 0/4 | 3 | 1.3 ± 0.3 | 3 | 44.8 ± 11.9 | 4 | 7.2 ± 5.1 |
| Mantle cell lymphoma | ||||||||
| Typical | 0/14 | 1/7 | 7 | 1.3 ± 0.4 | 6 | 13.5 ± 8.1 | 13 | 5.5 ± 3.2 |
| Blastoid | 1/10‡ | 2/7 | 7 | 0.9 ± 0.3 | 7 | 51.1 ± 28.0 | 9 | 9.2 ± 4.7 |
| Large cell lymphoma | 0/23 | 2/34 | 10 | 1.2 ± 0.8 | 9 | 56.5 ± 30.1 | 10 | 3.1 ± 3.9 |
| Total | 1/60 | 5/59 | 34 | 30 | 38 | |||
| Diagnosis . | Gene alterations1-153 . | Loss of protein expression1-153 . | mRNA expression . | Proliferation, Ki-67* . | Chromosomal imbalances, CGH† . | |||
|---|---|---|---|---|---|---|---|---|
| n . | mean ± SD . | n . | mean ± SD . | n . | mean ± SD . | |||
| Chronic lymphocytic leukemia | 0/9 | 0/7 | 7 | 0.7 ± 0.3 | 5 | 14.0 ± 8.2 | 2 | 4 ± 2.8 |
| Richter syndrome | 0/4 | 0/4 | 3 | 1.3 ± 0.3 | 3 | 44.8 ± 11.9 | 4 | 7.2 ± 5.1 |
| Mantle cell lymphoma | ||||||||
| Typical | 0/14 | 1/7 | 7 | 1.3 ± 0.4 | 6 | 13.5 ± 8.1 | 13 | 5.5 ± 3.2 |
| Blastoid | 1/10‡ | 2/7 | 7 | 0.9 ± 0.3 | 7 | 51.1 ± 28.0 | 9 | 9.2 ± 4.7 |
| Large cell lymphoma | 0/23 | 2/34 | 10 | 1.2 ± 0.8 | 9 | 56.5 ± 30.1 | 10 | 3.1 ± 3.9 |
| Total | 1/60 | 5/59 | 34 | 30 | 38 | |||
Percent of positive cells.
Comparative genomic hybridization.
Heterozygous Ile157Thr germ line substitution.
Numbers indicated are number of alterations/number of patients examined.
RNA extraction and reverse transcriptase–polymerase chain reaction (RT-PCR)
Total RNA was obtained from frozen tissues using guanidine isothiocyanate extraction and cesium chloride gradient centrifugation. cDNA was synthesized from 1 μg total RNA using random hexamers (TaqMan reverse transcription reagents; Applied Biosystems, Warrington, United Kingdom) and following the manufacturer specifications. TheCHK2 complete coding region was amplified by PCR in 9 partly overlapping portions using the primers and PCR conditions previously described.24 All the amplifications were carried out in a 2400 Perkin-Elmer thermocycler (Norwalk, CT).
Single-stranded conformation polymorphism (SSCP) and sequencing analysis
SSCP analysis was used to screen for CHK2 mutations in the complete coding region using the 9 overlapping cDNA fragments obtained by RT-PCR. The PCR products were diluted in formamide-dye loading buffer, and electrophoresed on a 15% nondenaturing polyacrylamide gel in both the presence and absence of 5% glycerol at room temperature. The gels were developed using a silver staining procedure as previously described.30 RT-PCR products of cases showing an altered mobility were direct sequenced using cycle sequencing BigDye terminator chemistry (Applied Biosystems). Sequencing reactions were run on a Perkin-Elmer ABI-377 automated sequencer. All nucleotidic changes were confirmed by sequencing both strands. RNA extracted from the HCT15 colon cancer cell line carrying 2 biallelic missense mutations was used as a positive control for CHK2gene mutations.26
To determine the germ line status of CHK2 gene, DNA from normal cells was obtained from microdissected samples as described.31 Microdissection was carried out using the Laser Pressure Catapulting technique of the Robot-microbeam system (PALM GmbH, Benried, Germany).
To examine the possible presence of the Ile157Thr change in normal population a PCR-SSCP analysis was performed. DNA obtained from 104 healthy Spanish blood donors was subjected to PCR reaction using the following conditions: 35 cycles at 94°C for 40 seconds, 60°C for 40 seconds, and 72°C for 40 seconds. PCR amplifications were performed using CHK2 forward (5′-AGGAGAGCTGGTAATTTGGTCAT-3′) and reverse (5′-TCTGCTTAGTGACAGTGCA-3′) primers flanking the genomic region in which the Ile157Thr mutation was located. These PCR products were analyzed using an SSCP strategy as described above.
Protein extraction and Western blot analysis
Nuclear protein extracts were obtained from 11 CLLs, 4 RSs, 14 MCLs, and 34 LCLs in which additional frozen tissue was available. In addition, protein extracts obtained from 2 reactive spleens, 6 tonsils, 6 lymph nodes, and 6 samples of normal peripheral blood lymphocytes were also analyzed. Protein extract from the HCT15 cell line was used as a negative control for CHK2 protein expression. Cryostat frozen sections were lysed in ice-cold buffer containing 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.1% Nonidet P-40, 0.5 mM dithiothreitol, 2 μg/mL leupeptine, 5 μg/mL aprotinine, and 0.5 mM phenylmethylsulfonyl fluoride for 15 minutes. After centrifugation at 3500 rpm, the precipitated nuclei were lysed in a buffer containing 80 mM Tris-HCl (pH 6.8), 2% sodium dodecylsulphate, and 10% glycerol. A quantity of 50 μg of nuclear protein was run per lane on a 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel and electroblotted to a nitrocellulose membrane (Amersham, Buckinghamshire, United Kingdom). The membranes were blocked by overnight incubation in 5% dry milk and 0.1% Tween-20 at 4°C. The blocked membranes were then incubated with the monoclonal antibody anti-CHK2 (clone 2CHK01; Neomarkers, Fremont, CA), washed with phosphate-buffered saline containing 0.1% Tween-20, and incubated with a sheep anti–mouse secondary antibody conjugated to horseradish peroxidase (Amersham). A monoclonal antibody anti–α-tubulin (Santa Cruz Biotechnologies, Santa Cruz, CA) was used as a loading control. After washing, antibody binding was detected by chemoluminiscence detection procedures according to the manufacturer's recommendations (ECL; Amersham). Intensities of the CHK2 proteins were normalized to the α-tubulin band signal.
Analysis of CHK2 promoter hypermethylation by methylation-specific PCR
DNA methylation patterns in the CpG island of CHK2were determined by chemical modification of unmethylated, but not the methylated, cytosines to uracil, and subsequent PCR using primers specific for either methylated or the modified unmethylated DNA as previously described.32 A quantity of 1 μg DNA was denatured by NaOH and modified by sodium bisulfite. DNA samples were then purified using Wizard DNA purification resin (Promega, Madison, WI), again treated with NaOH, precipitated with ethanol, and resuspended in water. Primer sequences for the unmethylated reaction were 5′-TTGTTTTTTTTTTTTGTAGGTTAGATTTTGAT-3′ (forward primer) and 5′-AAACTACTTCAACCTTATAAACTAATACAAACAACA-3′ (reverse primer), and for the methylated reaction were 5′-GTTTTTTTTTTTCGTAGGTTAGATTTCGAC-3′(forward primer) and 5′-CTTCAACCTTATAAACTAATACGAACGACG-3′ (reverse primer). The annealing temperature was 55°C. Placental DNA treated in vitro with Sss I methyltransferase (New England Biolabs, Beverly, MA) was used as positive control for methylated alleles of CHK2, and DNA from normal lymphocytes was used as negative control for methylated alleles of CHK2. Controls without DNA were performed for each set of PCR. A quantity of 10 μL of each PCR reaction was directly loaded onto nondenaturing 6% polyacrylamide gels, stained with ethidium bromide, and visualized under UV illumination.
Real-time quantitative PCR analysis of CHK2gene
We subjected 100 ng of genomic DNA to real-time PCR analysis. Sequences of the CHK2 detection probe and primers were designed using the Primer Express software (Applied Biosystems) as follows: CHK2 forward: 5′-CGGATGTTGAGGCTCACGA-3; CHK2reverse: 5′-TATGCCCTGGGACTGTGAGG-3′. The CHK2 probe, 5′-AGCGTTACCCAGTCCCAAGGCTCC-3′, was labeled with 6-carboxy-fluorescein (FAM) as the reporter dye, 6-carboxy-tetramethyl-rhodamine (TAMRA) as the quencher fluorescent, and 3′-phosphorylated in order to prevent elongation during PCR (Applied Biosystems). To avoid interferences with any of the multiple homologous CHK2 sequences described within the genome, which comprises exons 10 to 14, the probe was specially designed in exon 1.33 The quantitative assay amplified 100 ng of DNA as template in 2 to 4 replicates, using the Taqman Universal Master Mix, 200 nM of each CHK2 primers and 300 nM of CHK2 probe. Amplification conditions were 2 minutes at 50°C for uracil N-glycosylase (UNG) activation, and 10 minutes at 95°C for TaqGold activation and predenaturation, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. All reactions were performed in an ABI PRISM 7700 Sequence Detector System (Applied Biosystems). Because lymphoid malignancies usually showed complex karyotypes we used 3 genes as endogenous controls:β-actin, β2-microglobulin, and albumin, as previously described.34 35CHK2 levels were related to the signal of the endogenous control genes in each case and amplification levels of DNA obtained from normal samples were used as controls.
Real-time quantitative RT-PCR
A quantity of 1 μg total RNA treated with DNAse was transcribed into cDNA using MMLV-reverse transcriptase (BRL, Gaithersburg, MD) and random hexamers, following manufacturer's directions. PCR reactions were performed using the same CHK2primers, CHK2 probe, and conditions described above. Theβ-glucoronidase gene (GUS; Applied Biosystems) was used as endogenous control as recommended by the manufacturer.GUS and CHK2 expression was related to a standard curve derived from serial dilutions of a cDNA from tonsil sample. The relative units (RU) of CHK2 expression were defined as the mRNA levels of these genes normalized to the GUS expression level in each case. An RNA mix obtained from 6 nonneoplastic lymphoid tissues (2 tonsils, 2 reactive lymph nodes, and 2 spleens) was used as reference control to normalize CHK2/GUSamplification ratios.
Immunohistochemistry
To determine the proliferation rate of tumor samples, an immunohistochemical analysis of Ki-67 antigen was assessed. Immunohistochemistry was performed on formaldehyde-fixed, paraffin-embedded material with the EnVision+System Peroxidase (DAB) method (DAKO, Carpinteria, CA). The antigen retrieval was done by heating samples on ethylenediaminetetraacetic acid (EDTA) buffer in a microwave oven. Primary antibody against Ki-67 (MIB-1; Immunotech, Marseille, France) was used at a 1/400 dilution. Quantitative immunohistochemical analysis was performed by counting at least 1000 cells in random high-power fields of each case. Cells were considered positive for Ki-67 when evident nuclear staining could be identified.
Statistical analysis
Because of the nonnormal distribution of the sample and the small size of some subsets of tumors, the statistical analysis evaluation was performed using nonparametric tests. Comparison between chromosomal imbalances and inactivation of CHK2,ATM, and p53 genes and between mRNA expression levels in different groups of NHLs was performed using the Kruskal-Wallis test. To determine whether CHK2 mRNA expression levels could correlate with proliferation, a regression analysis was assessed. Level of significance was set at .05 for all analyses and all calculations were performed with the SPSS software package (version 10.0; SPSS, Chicago, IL).
Results
CHK2 mutational analysis
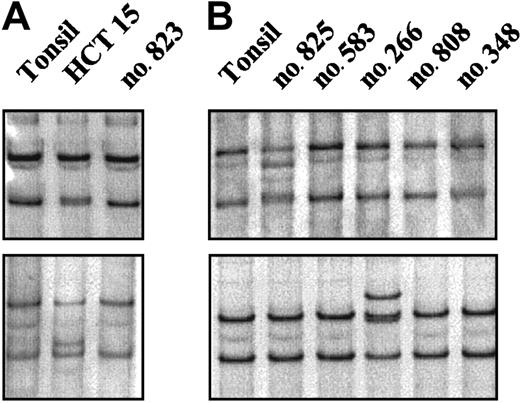
A mutational analysis of the entire coding region of theCHK2 gene was performed in 60 NHLs (Table 1). The 3′ terminal exons and introns of CHK2 are duplicated on multiple chromosomes with a high grade of sequence conservation complicating the mutational analysis of genomic DNA. Because these duplicated sequences are not expressed, we performed the mutational analysis on mRNA using an RT-PCR–SSCP method. As a positive control we included mRNA extracted from the HCT15 colon cancer cell line. This screening method allowed the detection of the biallelic mutations of the HCT15 cell line, including the Ala247Asp mutation that had been initially missed using denaturing high performance liquid chromatography (Figure1A).20,26 In the NHLs examined, an abnormally migrating band was identified in 4 cases. Of these tumors, 3 showed the same mobility shift. Sequencing analysis demonstrated the previously described silent polymorphism at codon 84 (GAA to GAG). These cases were one CLL, one blastoid MCL, and one LCL. The other anomalous migrating band was found in a blastoid MCL and corresponded to a previously described heterozygous mutation at codon 157 (ATT to ACT; Figure 1B). This change results in a substitution of isoleucine by threonine (Ile157Thr) within the highly conserved forkhead homology-associated (FHA) domain. Normal DNA from this patient could be obtained from normal tissue microdissected out of frozen tissue sections. Sequencing analysis showed that the Ile157Thr substitution was also present in heterozygosity in the germ line. CHK2 protein expression similar to other tumors was observed in this case (see “Protein expression analysis”). The Ile157Thr change has been identified in the germ line in some kindreds with LFS.20 26 To know if this patient could represent a member of an LFS family, the clinical history was reviewed. This patient was a 75-year-old man who one year before the diagnosis of the blastoid MCL had developed an infiltrating urinary bladder carcinoma. No evidence of cancer in previous family members was reported and 2 daughters (45 and 50 years old) were healthy with no evidence of cancer, making the consideration of LFS unlikely.
RT-PCR–SSCP analysis of the
CHK2 gene. (A) The abnormal mobilities observed in the HCT15 cell line corresponded to the previously described missense mutations Ala247Asp (top) and Arg145Trp (bottom). (B) Case no. 825 (top) showed a distinct mobility shift that corresponded to an aminoacid substitution at codon 157 (ATT>ACT) leading to an Ile→Thr change. Case no. 266 (bottom) showed an altered mobility that was also found in 2 more lymphomas corresponding to a silent polymorphism at codon 84 (GAA>GAG).
RT-PCR–SSCP analysis of the
CHK2 gene. (A) The abnormal mobilities observed in the HCT15 cell line corresponded to the previously described missense mutations Ala247Asp (top) and Arg145Trp (bottom). (B) Case no. 825 (top) showed a distinct mobility shift that corresponded to an aminoacid substitution at codon 157 (ATT>ACT) leading to an Ile→Thr change. Case no. 266 (bottom) showed an altered mobility that was also found in 2 more lymphomas corresponding to a silent polymorphism at codon 84 (GAA>GAG).
To test if the Ile157Thr mutation might be an uncommon polymorphism present in the healthy Spanish population we screened 208 alleles from 104 healthy blood donors using a PCR-SSCP method. The analysis revealed that this substitution was not present in the Spanish cancer-free population.
Protein expression analysis
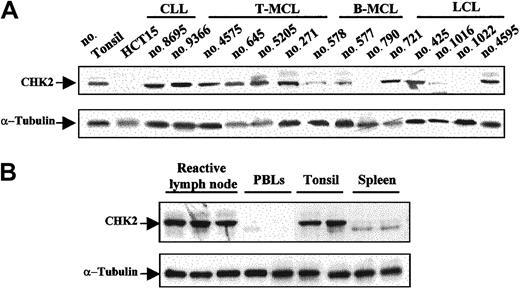
CHK2 protein expression was examined by Western blot in 59 malignant lymphomas, 6 reactive tonsils, 6 lymph nodes, 2 spleens, and 6 samples of normal peripheral blood lymphocytes. Protein extract from the colorectal HCT15 cell line was used as a negative control. Relative similar levels of CHK2 protein were observed in all reactive tissues and in almost all tumor samples (Figure 2A-B). Curiously, spleen samples showed a particular CHK2 protein pattern that may correspond to different CHK2 protein phosphorylation states (Figure 2B). Absent or very low levels of CHK2 protein were observed in the HCT15 cell line and in normal peripheral blood lymphocytes (Figure 2A-B). There were 2 aggressive lymphomas, one LCL and one blastoid variant of MCL, that showed a complete absence of CHK2 protein. There were 3 additional tumors, one typical MCL, one blastoid MCL, and one LCL, that had a marked decrease of protein expression (Figure 2A). The tissue samples of all these cases had been obtained at diagnosis in patients who had not been previously treated.
Western blot analysis of CHK2 protein in NHLs and normal lymphoid tissues.
(A) There were 2 aggressive lymphomas (no. 790 and no. 1022) that lacked CHK2 protein expression. There were 3 additional tumors (no. 578, no. 577, and no. 1016) that showed low levels of expression, whereas the remaining samples had protein levels similar to those observed in normal tonsil. The HCT15 colon cancer cell line was used as negative control. (B) Normal reactive lymphoid tissues showed similar CHK2 protein expression levels, while in normal peripheral blood lymphocytes (PBLs) very low or absent CHK2 expression was detected. Normal spleen samples showed low expression of CHK2 protein with a particular pattern, probably related to phosphorylation differences. Expression of α-tubulin in the same samples used as a loading control is shown in the bottom panels.
Western blot analysis of CHK2 protein in NHLs and normal lymphoid tissues.
(A) There were 2 aggressive lymphomas (no. 790 and no. 1022) that lacked CHK2 protein expression. There were 3 additional tumors (no. 578, no. 577, and no. 1016) that showed low levels of expression, whereas the remaining samples had protein levels similar to those observed in normal tonsil. The HCT15 colon cancer cell line was used as negative control. (B) Normal reactive lymphoid tissues showed similar CHK2 protein expression levels, while in normal peripheral blood lymphocytes (PBLs) very low or absent CHK2 expression was detected. Normal spleen samples showed low expression of CHK2 protein with a particular pattern, probably related to phosphorylation differences. Expression of α-tubulin in the same samples used as a loading control is shown in the bottom panels.
ATM gene inactivation had been previously examined in the MCLs included in this study. In addition, we examined theATM gene status in the LCL lacking CHK2 protein. Interestingly, the 2 cases with complete absence of CHK2 protein expression had ATM in the germ line (Table2). Contrary to the dissociation between ATM and CHK2 inactivation, p53 mutation was detected in the blastoid MCL with no CHK2 protein expression, whereas the LCL lacking CHK2 protein showed wild-type p53 (Table 2).
CHK2 protein expression and gene alterations in non-Hodgkin lymphomas
| Case no. . | Histology . | CHK2 . | Chromosomal imbalances (CGH) . | ATM . | p53 . | |
|---|---|---|---|---|---|---|
| Protein . | Gene . | |||||
| 1022 | LCL | − | GL | 13 | WT | WT |
| 790 | B-MCL | − | GL | 20 | WT | Mut |
| 1016 | LCL | −/+ | GL | 0 | NA | WT |
| 577 | B-MCL | −/+ | GL | 9 | Mut | Del |
| 578 | T-MCL | −/+ | GL | 3 | WT | WT |
| 266 | B-MCL | + | Ile157Thr/GL | 4 | WT | WT |
| Case no. . | Histology . | CHK2 . | Chromosomal imbalances (CGH) . | ATM . | p53 . | |
|---|---|---|---|---|---|---|
| Protein . | Gene . | |||||
| 1022 | LCL | − | GL | 13 | WT | WT |
| 790 | B-MCL | − | GL | 20 | WT | Mut |
| 1016 | LCL | −/+ | GL | 0 | NA | WT |
| 577 | B-MCL | −/+ | GL | 9 | Mut | Del |
| 578 | T-MCL | −/+ | GL | 3 | WT | WT |
| 266 | B-MCL | + | Ile157Thr/GL | 4 | WT | WT |
LCL indicates large cell lymphoma; B-MCL, blastoid mantle cell lymphoma; T-MCL, typical mantle cell lymphoma; Del, homozygous deletion; GL, germ line; WT, wild type; Mut, mutated; NA, not available; −, absence of protein expression; and −/+, protein down-regulation.
Genomic alterations had been examined by comparative genomic hybridization in 38 tumors (Table 1). The 2 lymphomas with complete absence of CHK2 protein expression had 20 and 13 chromosomal imbalances, which represent the highest number of chromosomal alterations in the whole series of tumors examined (17 ± 4.9). The case with 20 imbalances had an additional p53 mutation (Table 2). p53 and ATM inactivation had been examined in 21 and 19 additional cases, respectively. In 9 cases with inactivation of ATM, p53, or both genes, there was also a high number of chromosomal imbalances (9 ± 3.1). The number of genomic aberrations observed in lymphomas with wild-typeCHK2, p53, and/or ATM genes was significantly lower (5 ± 2.2) than those observed in tumors with inactivation of these genes (Kruskal-Wallis test,P = .011).
CHK2 gene deletion and hypermethylation analysis
To determine if mechanisms other than gene mutation could cause the decreased levels of CHK2 protein detected by Western blot, we first performed an analysis of possible gene deletions by real-time quantitative PCR in 11 cases, including the 5 lymphomas with decreased protein expression. To avoid amplification of duplicatedCHK2 gene sequences, we selected for the study a region from exon 1 only present in chromosome 22. Real-time PCR analysis showed no evidence of CHK2 gene deletion in any of the samples analyzed. In that sense, tumors with normal or decreased protein expression showed CHK2/β-actin, β2-microglobulin,and albumin amplification ratios similar to those observed in samples from normal peripheral blood lymphocytes and reactive tissues consistent with presence of both CHK2alleles.
To know if the decreased protein levels detected in these cases could be associated with hypermethylation of the gene, DNA from 12 lymphomas including the 5 cases with absent or low protein expression were subjected to a methylation-specific PCR. The results showed no evidence of hypermethylation in any of the samples analyzed. DNA obtained from other lymphomas expressing normal levels of CHK2 protein and normal peripheral blood lymphocytes were also analyzed and no hypermethylation could be detected in any sample (data not shown).
CHK2 mRNA expression
CHK2 mRNA levels were examined by real-time quantitative RT-PCR in 34 NHLs (7 CLLs, 3 RSs, 7 typical MCLs, 7 blastoid MCLs, and 10 LCLs), including all cases with low or no CHK2 protein expression. Nonneoplastic lymphoid tissues were also analyzed. Similar levels of CHK2 mRNA expression were observed in the different types of lymphomas and reactive lymphoid tissues (Kruskal-Wallis test; P > .05; Table 1, Figure3). On the other hand, concordantly to the protein levels detected by Western blot, extremely lowCHK2 mRNA expression was detected in normal peripheral blood lymphocytes. The lymphoma samples with lower or lacking CHK2 protein expression showed levels of mRNA expression relatively similar to other tumors with no protein alteration.
Real-time quantitative RT-PCR
CHK2 mRNA expression analysis (median and range) in NHLs. Similar CHK2 mRNA expression levels were observed in nonneoplastic reactive lymphoid tissues and different types of NHLs (Kruskal-Wallis test, P > .05). Results are depicted as the ratio of absolute CHK2 to GUS mRNA transcript numbers (RU). Bars indicate SD.
Real-time quantitative RT-PCR
CHK2 mRNA expression analysis (median and range) in NHLs. Similar CHK2 mRNA expression levels were observed in nonneoplastic reactive lymphoid tissues and different types of NHLs (Kruskal-Wallis test, P > .05). Results are depicted as the ratio of absolute CHK2 to GUS mRNA transcript numbers (RU). Bars indicate SD.
To study if the variable CHK2 mRNA expression levels found in NHLs could be related to the proliferative activity of the tumors, histologic sections were immunostained for the proliferation marker Ki-67. RS and blastoid MCLs had a significant higher proliferation rate than CLLs and typical MCLs, respectively. LCLs also showed a high proliferation rate relatively similar to RS and blastoid MCLs (Table1). A regression analysis of the proliferative rate of the tumors andCHK2 mRNA levels showed no correlation between these 2 parameters (r = −0.042, P > .05), suggesting that CHK2 mRNA expression is independent of the proliferative activity of the tumors.
Discussion
In this study, we have examined CHK2 gene alterations and both protein and mRNA expression in a series of human lymphoid neoplasms, reactive tissues, and normal lymphoid samples. Gene alterations or decreased protein expression were detected in 6 cases, one typical MCL, 3 blastoid variants of MCL, and 2 LCLs. One blastoid MCL had a CHK2 gene alteration, corresponding to the nonconservative change Ile157Thr (ATT to ACT), previously described in families with LFS.22 In addition, complete absence or marked decrease in levels of protein expression with no apparent alteration of the gene structure were detected in the other 5 tumors.
The central role of the CHK2 gene in DNA damage response pathways, downstream of ATM and upstream of p53, and the detection ofCHK2 gene mutations in a subset of families affected by LFS have suggested that this gene may be a potential tumor suppressor gene also involved in the pathogenesis of sporadic tumors. Mutational analysis of CHK2 gene in human neoplasms have been performed only in a limited number of tumor types. These studies have identified a low number of mutations including missense mutations and single nucleotide deletions in colorectal, breast, lung, and vulval carcinomas, a diffuse large B-cell lymphoma, a myelodisplastic syndrome, an acute myelocytic leukemia, and a mantle cell lymphoma.21-25,27,28 Mutational studies of theCHK2 gene in human neoplasms are hampered in part by the complicated analysis of genomic DNA due to the extensive duplication of nonexpressed CHK2 sequences in the genome.33
The unique gene alteration observed in our study was the nonconservative change Ile157Thr (ATT to ACT) in a blastoid MCL. This substitution occurs within the FHA domain, a highly conserved protein interaction domain essential for CHK2 activation after DNA damage, and it was first described as a heterozygous germ line mutation in an LFS variant lackingp53 mutation.20 The pathogenetic significance of this mutation has been controversial in part because of the finding of this change in approximately 6% of healthy controls in Finland.36 However, we have not detected this mutation in any of the 104 control Spanish individuals analyzed. These results are concordant with a recent study that has not detected the Ile157Thr mutation in 200 control individuals from the Boston area.26 Moreover, recent functional studies have demonstrated that CHK2 protein carrying the Ile157Thr change is defective in binding the downstream targets CDC25A and p53, suggesting that the regulatory function of this mutant protein may be compromised.37,38 The Ile157Thr CHK2substitution detected in our case was also present in the germ line of the patient. Although the patient developed a bladder carcinoma as a second neoplasia, the clinical history does not allow us to establish this case as a family cancer syndrome. Similarly to this case, Hangaishi et al28 have recently described a 15–base pair (bp) CHK2 gene deletion in an MCL that seemed to be also present in the patient's germ line. In that sense, our results and this finding raise the question of a possible role for theCHK2 gene in hereditary susceptibility to some of these tumors.
Several studies have analyzed the mutational status of CHK2 in human neoplasms. However, the information on mRNA and protein expression in different types of cancer is very limited. A previous study of protein expression in normal human tissues and cell lines has indicated that CHK2 protein is relatively stable and it is expressed at similar levels in all phases of the cell cycle, as well as in quiescent and terminally differentiated cells.39 In our study, we demonstrated that CHK2 protein levels were relatively similar in different types of malignant lymphomas, including both low and highly proliferative neoplasms such as CLL and blastoid MCL or LCL, indicating that CHK2 protein expression in these tumors is independent of the proliferative activity and the histologic type of the tumors. CHK2 protein expression levels were similar in malignant lymphomas and reactive lymphoid tissues, whereas quiescent peripheral blood lymphocytes had very low or absent protein expression.
Interestingly, we observed a marked decrease of protein expression in 5 tumors (9%), 2 of them showing a complete absence of protein and the other 3 with a marked reduction in protein expression. Several CHK2 mutations have been shown to facilitate destabilization and increased degradation of CHK2 protein in human tumors.26,39-41 However, no mutations, deletions, or hypermethylation of the promoter region were identified in any of these cases. On the other hand, mRNA levels were relatively similar in cases with low and normal protein expression, indicating that other mechanisms may regulate the stability of the protein in these lymphomas. A recent study in testicular germ-cell tumors has also demonstrated a reduction and lack of protein expression in a subset of invasive neoplasms in contrast with the high protein levels observed in other tumors, preinvasive lesions, and normal germ cells, suggesting that decreased CHK2 protein expression levels may be involved in the progression of these neoplasms.39
The CHK2 gene is an important regulatory element in DNA damage response pathways interacting with ATM andp53 genes, among others. Murine and human cells deficient inATM and p53 genes present a high number of chromosomal abnormalities and genetic instability.42 In that sense, we have recently shown that ATM inactivation is associated with increasing number of chromosomal imbalances in typical MCL.16 However, the possible relationship between CHK2 alterations and genomic instability in human tumors is not known. Recent studies using mutant constructs of RAD53, theSaccharomyces cerevisiae homologue of CHK2, have shown that inactivation of this gene resulted in an increased rate of gross chromosomal abnormalities including interstitial deletions, deletions of chromosomal arms, and nonreciprocal translocations.43 In the present study, we have observed that the 2 lymphomas with complete absence of CHK2 protein expression had the highest number of chromosomal imbalances in the whole series of tumors, suggesting that CHK2 inactivation may play a role in chromosomal instability in human lymphomas. One of the cases had also a concomitant p53 inactivation but in the second one ATM and p53 were in the germ line. The number of alterations in these 2 cases was relatively higher than those observed in tumors with mutations in ATM, p53, or both, and much higher than in tumors with no alterations in these genes. Although the number of cases is small, these observations may indicate that complete inactivation of CHK2 in malignant lymphomas may facilitate accumulation of genomic aberrations.
Recent studies have demonstrated inactivation of the ATMgene in a relatively high number of MCLs and CLLs. On the other hand, p53 aberrations are a frequent phenomenon in aggressive lymphomas. The fact that these genes participate in a common DNA damage response pathway with CHK2 raises the question of the possible alternative or cooperative alterations of these genes in the pathogenesis of human tumors. In fact, CHK2 and p53 mutations have been identified as mutually exclusive in LFS families. In the present study, we have included a subset of tumors in which we had previously examined the status of ATM and p53 genes. Interestingly, the 2 cases with complete absence of CHK2 protein had a wild-typeATM whereas p53 was mutated in the MCL but was wild type in the LCL. The other cases with a partial inactivation of CHK2 showed different patterns of inactivation of these genes. The presence of simultaneous alterations of CHK2 and p53 in human tumors seems to confirm experimental observations indicating that concomitant mutations in these genes may provide additional selective advantages to tumor cells.37
The mRNA analysis of this series of tumors demonstrated that theCHK2 expression levels in NHLs were not significantly variable and showed similar patterns in different types of lymphomas and reactive nonneoplastic tissues, whereas quiescent normal lymphocytes showed very low levels of CHK2 mRNA expression. These findings are concordant with the relatively constant protein levels detected in the different types of lymphomas and reactive normal lymphoid samples, and the low expression observed in normal peripheral blood lymphocytes. In spite of different studies on activation mechanisms of CHK2 protein, no information on regulatory mechanisms of mRNA expression is available. Finally, our study also demonstrates thatCHK2 mRNA levels are not related to the proliferative activity of the tumors.
In conclusion, our findings indicate that CHK2-decreased protein expression and CHK2 gene alterations are uncommon in malignant lymphomas but occur in a subset of aggressive tumors independently of p53 or ATM aberrations. The high number of chromosomal imbalances in tumors with a complete absence of protein expression suggest that inactivation of this gene may play a role in chromosomal instability in malignant lymphomas.
Prepublished online as Blood First Edition Paper, August 1, 2002; DOI 10.1182/blood-2002-04-1078.
Supported by the Comision Interministerial de Ciencia y Tecnologia (SAF 2002/03261), European Commission contract QLRT-1999-30687, FIS 01/3046, and Generalitat de Catalunya (2000SGR118). F.T., S.H., and S.B. were supported by the Spanish Ministery of Science and Technology and the Josep Carreras International Foundation against Leukemia. M.S. was supported in part by Dako Co.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Elias Campo, Laboratory of Pathology, Hospital Clinic, Villarroel 170, 08036-Barcelona, Spain; e-mail:campo@medicina.ub.es.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal