Previous reports have suggested that the adenosine triphosphate–binding cassette protein ABCG2 (breast cancer resistance protein [BCRP], mitoxantrone resistance [MXR]) is associated with drug resistance in acute myeloid leukemia (AML). The aims of this study were to determine the level of ABCG2 mRNA expression necessary to produce drug resistance and to define the ABCG2 levels in normal bone marrow (BM), peripheral blood (PB), cord blood (CB), and adult AML blast cell populations. First, using transduced clonal cell lines expressing varying levels of ABCG2, we found that ABCG2 expression conferred resistance to mitoxantrone and topotecan, but not to idarubicin. Next, we developed a real-time reverse transcription–polymerase chain reaction assay for measuring ABCG2 mRNA expression levels in clinical samples. Normal BM and PB contained low levels of ABCG2 mRNA, while higher levels were measured in CB mononuclear cells, CD34+, and Ac133+populations, consistent with the known stem cell enrichment in these populations. Next, we studied the ABCG2 mRNA levels in 40 specimens from newly diagnosed adult AML patients. Only 7% of these samples contained ABCG2 mRNA levels within the range of our drug-resistant clone, although another 78% were higher than normal blood and bone marrow. Flow cytometry revealed very small subpopulations of ABCG2-expressing cells in the cases we examined. Our data suggest that high levels of ABCG2 mRNA expression in adult AML blast specimens are relatively uncommon and that ABCG2 expression may be limited to a small cell subpopulation in some cases.
Introduction
Drug resistance is a frequently encountered phenomenon contributing to treatment failure in acute leukemia. Several members of the adenosine triphosphate–binding cassette (ABC) family of transmembrane transporters have been associated with drug resistance in acute myeloid leukemia (AML), including P-glycoprotein,1,2 encoded by the multidrug resistance gene, MDR1, and the multidrug resistance protein, MRP1.3 Several studies have suggested that other transporters may be responsible for the multidrug resistance phenotype in some AML cases. Leith et al showed that 18% of adult AML specimens had drug efflux activity, which was not associated with MDR1 or MRP1 expression, suggesting the presence of a previously unidentified transporter.4
The transporter ABCG2, also designated BCRP and MXR, was recently described as a cause of the multidrug resistance phenotype.5 ABCG2 mRNA expression was identified in approximately one third of cases of adult AML by semiquantitative reverse transcription–polymerase chain reaction (RT-PCR).6 Another study detected ABCG2 protein expression in 27% of cases by flow cytometry.7 However, it is unclear if the ABCG2 mRNA expression measured in these samples was physiologically significant or due to contamination from normal hematopoietic cells in low-expressing cases. Because ABCG2 expression has subsequently been demonstrated in several types of normal hematopoietic cells, including erythroid progenitors, natural killer cells, and primitive stem cells,8 9 expression in AML blast samples could be due to contamination from normal hematopoietic cells.
The substrate profile of ABCG2 includes the antineoplastic drugs mitoxantrone and topotecan.10 Mitoxantrone is commonly used in AML treatment, suggesting that ABCG2 could play an important role in drug resistance in this disease. Idarubicin is another anthracycline drug that is sometimes used in the treatment of AML, and it has not been known whether ABCG2 expression can confer resistance to idarubicin.
ABCG2 is known to efflux the fluorescent dye, Hoescht 33342, but not rhodamine 123.8,11 This dye efflux activity can be used to identify hematopoietic stem cells using the “side population” (SP) assay,12,13 and previous studies have identified ABCG2 expression as a molecular determinant of the SP phenotype.8,9 This finding may be important in AML for several reasons. First, several investigators have found the frequent presence of SP cells in bone marrow from AML patients.14,15 While in both studies the SP cells were usually leukemic, Feuring-Buske et al demonstrated that specimens from most AML patients contained both leukemic and nonleukemic SP cells.15 Interestingly, these SP cells frequently did not express MDR1 and were capable of multilineage NOD/SCID engraftment. These studies provide further evidence that a transporter such as ABCG2 may be expressed in stem cells of both normal and leukemic origin.
To assess ABCG2 expression in AML patient samples, we developed clonal human AML cell lines by transduction with a retroviral vector containing the ABCG2 cDNA. Using these clonal cell lines, we identified a low-expressing clone, referred to as clone 3.3, that has readily detectable levels of ABCG2 mRNA by real-time RT-PCR analysis but very low levels of protein expression and no drug resistance. Another clone, 6.2 with ABCG2 mRNA expression much higher than clone 3.3, demonstrated relatively high levels of protein expression by monoclonal antibody labeling and mitoxantrone efflux assays. Resistance to several relevant cytotoxic drugs was then measured in these cell lines to develop a correlation between ABCG2 mRNA levels and functional drug resistance. The mRNA levels in these reference cell lines were also compared to those seen in normal hematopoietic cell populations and in 40 samples from adult AML patients. In several cases, blast cell populations were studied for ABCG2 protein expression using a monoclonal antibody we have recently generated. These expression and functional studies further clarify the role of ABCG2 expression in AML.
Materials and methods
Preparation of ABCG2-transduced clonal AML cell lines
OCI/AML3 cells16 were grown in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). Derivation of the human ABCG2 expression vector, HaABCG2, and the subsequent transfection of an ecotropic producer cell line has been previously described.8 The amphotropic packaging line, PA317, was transduced with supernatant from the producer cells. Transduced PA317 cells were sorted to 100% purity by flow cytometry for Hoescht 33342 efflux. Next, 8 × 106 irradiated PA317/HaABCG2 cells were cultured in 9 mL DMEM with 10% FBS, with the addition of 10 μL polybrene (Sigma, St Louis, MO). Added to the adherent producer cells for coculture were 5.0 × 105OCI-AML3 cells. Nonadherent AML cells were then transferred to fresh plates of producer cells at daily intervals for 4 days. Transduced OCI-AML3/HaABCG2 cells were sorted based on Hoechst 33342 efflux, using a sterile single-cell sorter (Becton Dickinson, Franklin Lakes, NJ), then grown in 96-well plates. Culture wells were visually inspected with inverted phase contrast microscopy to verify the presence of single cells. At 10 to 14 days, wells containing an expanded number of cells were transferred to 24-well plates. Culture medium was exchanged at weekly intervals until cell density was sufficient for transfer to 75 cm2 culture flasks.
Drug efflux assays
OCI-AML3 cell clones were used to study mitoxantrone efflux, as previously described.16 Briefly, 5 × 105cells were harvested from suspension culture and washed in 3 mL phosphate-buffered saline (PBS). Cells were then centrifuged for 5 minutes at 1500 revolutions per minute (rpm) and resuspended in 500 μL DMEM with 10% FBS containing 2 μg/mL mitoxantrone (Immunex, Seattle, WA) or 200 ng/mL idarubicin (Sigma) and incubated for 30 minutes at 37°C. Cells were washed twice in PBS at 4°C then resuspended in DMEM + 10% FBS and kept at 4°C until analysis by flow cytometry. Mitoxantrone was excited at a wavelength of 630 nm and detected at 670 nm. Idarubicin was excited at a wavelength of 485 nm and detected at 542 nm. Results were expressed as mean fluorescent intensity and as percentage of cells within the dim gate.
SP analyses and antibody-mediated inhibition of efflux
Thirty micrograms of 5D3 anti-ABCG2 antibody8(prepared by our laboratory) or isotype control (mouse IgG2b, Caltag, Burlingame, CA) were added per 1 million cells in a total volume of 100 μL serum-free DMEM and then incubated on ice for 30 minutes. After this incubation step, Hoechst 33342 (Sigma) was diluted in DMEM with 2% fetal calf serum (FCS) plus 1 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid) and added to cells for a final concentration of 5 μg/mL and a final volume of 1.0 mL per million cells. After incubation at 37°C for 90 minutes, the cells were centrifuged at 1200 rpm for 5 minutes and resuspended in cold Hanks balanced salt solution with 2% FCS + 1 mM HEPES. Cells were kept on ice until flow analysis for SP cells was performed as previously described.13
Normal hematopoietic cell assays
Under approved institutional protocols, 4 samples each of bone marrow, peripheral blood, and umbilical cord blood were obtained from healthy volunteer donors. Peripheral blood stem cells (PBSCs) were obtained from healthy volunteer donors after mobilization using filgrastim (G-CSF; Amgen, Thousand Oaks, CA) at a dose of 480 μg/day for 4 days. PBSC collection was performed by double-volume apheresis, using the Cobe Spectra LRS-Turbo7 (Cobe Laboratories, Lakewood, CO).
Mononuclear cells were obtained using ficoll separation (Histopaque-1077, Sigma) according to the manufacturer's recommendations. Following recovery of mononuclear cells, further purification was performed by resuspending in red cell lysis buffer (8.3 g/L ammonium chloride with potassium bicarbonate 1.0 g/L) for 15 minutes, centrifuging, then resuspending in PBS. RNA was extracted from the resulting cell pellet using RNA STAT-60 (Tel-Test, Friendswood, TX) according to the manufacturer's recommendations.
CD34 and Ac133 cell enrichments were performed on mobilized peripheral blood leukapheresis products using the CliniMACS instrument (CS2 CE/UL, Miltenyi Biotec, Gladbach, Germany) and CD34 and Ac133 CliniMACS reagents and using the manufacturer's protocols. RNA was extracted using RNA STAT-60.
AML blast samples
Bone marrow or peripheral blood leukapheresis samples were obtained from 40 patients with newly diagnosed AML at the M. D. Anderson Cancer Center in Houston, TX. Leukemic cells were obtained from bone marrow in 32 cases, and from peripheral blood leukapheresis in the other 8 cases. The samples contained between 30% and 98% blasts. 5 × 106 to 2 × 107 cells were frozen in medium containing 90% FCS and 10% DMEM. Cells were warmed and immediately centrifuged at 1500 rpm for 3 minutes. RNA was extracted from the resulting cell pellet using the RNA STAT-60.
Quantitative real-time RT-PCR
Levels of ABCG2 and β-actin expression were determined for each sample using quantitative real-time RT-PCR, in separate reactions. Briefly, to each well of a 96-well plate were added 100 ng RNA, 12.5 μL Universal 2 × PCR Master Mix (Applied Biosystems, Foster City, CA), 0.625 μL 40 × RT Buffer (Applied Biosystems), primers (300 nM each), probe (100 nM), and diethyl pyrocarbonate–treated water, to a total volume of 25 μL per well. For ABCG2, the sequence of the forward primer was 5′-TGCAACATGTACTGGCGAAGA-3′; the sequence of the reverse primer was 5′-TCTTCCACAAGCCCCAGG-3′; and the sequence of the probe was 5′-fam-TATTTGGTAAAGCAGGGCATCGATCTCTCA-tamra-3′. For β-actin, the sequence of the forward primer was 5′-AGTACTCCGTGTGGATCGGC-3′; the sequence of the reverse primer was 5′-GCTGATCCACATCTGCTGGA-3′; and the sequence of the probe was 5′-vic-CCATCCTGGCCTCGCTGTCCA-tamra-3′. Thermal cycler conditions included an initial reverse transcription step at 48°C for 30 minutes, followed by incubation at 95°C for 10 minutes, then 40 cycles alternating between 95°C for 15 seconds and 60°C for 1 minute. Results were collected and analyzed using an ABI Prism 7700 Sequence Detection System (Applied Biosystems).
Reaction data were expressed as cycle thresholds (Ct), which is the PCR cycle number at which the fluorescent signal in each reaction reaches a threshold above background. Samples were normalized for RNA loading by subtracting the Ct of ABCG2 from the Ct of β-actin. The resulting ΔCt was subtracted from the ΔCt of clone 3.3 to produce the ΔΔCt. Finally, the ΔΔCt was expressed as 2-(ΔΔCt) to allow comparisons as a percentage of clone 3.3. Results are reported as the mean of 3 separate assays. To determine whether the overall ABCG2 mRNA measurements were affected significantly by interpatient differences in proportions of blast cells, mRNA results also were normalized for the blast cell content. The corrected value = (ABCG2 mRNA level in sample/proportion blasts)−[ABCG2 mRNA level in normal bone marrow × (1−proportion blasts)]/proportion blasts.
Northern analysis
Northern analysis was performed as previously described.17 A full-length ABCG2 cDNA probe was prepared by restriction digest of the pHaABCG2 plasmid with the enzymesNcoI and SacII (Promega, Madison, WI). The probe was radiolabeled with dCTP32 using the NEBlot Kit according to the manufacturer's instructions (New England BioLabs, Beverly, MA) and hybridized at 42°C for 12 hours.
ABCG2 expression was normalized to β-actin expression using a β-actin probe. Northern blots were analyzed using the Storm phosphor imager system (Molecular Dynamics, Sunnyvale, CA) with ImageQuaNT software (Molecular Dynamics) for signal quantification and background subtraction.
Flow cytometry analysis
1 × 106 cells were harvested from suspension culture (for cell lines) or thawed bone marrow or leukapheresis sample (for patient specimens) and washed twice with 5 mL PBSA (1% bovine serum albumin in PBS). Cells were labeled with 1 μg of anti-ABCG2 monoclonal phycoerythrin-conjugated antibody 5D3 (E-Bioscience, San Diego, CA) at room temperature for 30 minutes. Cells were washed twice with 5 mL PBSA, then resuspended in 500 μL PBSA and analyzed by flow cytometry. Propidium iodide (PI) was added, and all analyses were performed on PI-dull (live) cells. Results were compared to labeling with a PE-conjugated mouse IgG2b isotype control antibody (Caltag). Results were expressed as the ratio of mean fluorescent intensity for 5D3 to isotype for cell lines. For patient samples, results are expressed as percent positive cells by 5D3 labeling, and also as the ratio of mean fluorescent intensity of 5D3 compared to isotype.
Drug sensitivity assays
The parental OCI-AML3 cell line and ABCG2-transduced clones designated 3.3 and 6.2 were harvested and placed in 96-well plates at a density of 1 × 105 cells in 50 μL medium per well. For drug sensitivity testing, mitoxantrone (Novantrone, Immunex), topotecan (Hycamtin, GlaxoSmithKline, Research Triangle Park, NC), and idarubicin (Sigma) were each diluted in DMEM medium with 10% FCS to 2 × working concentrations. To achieve the indicated concentrations, 50 μL of each dilution was added to the plated cells. Cells were incubated with drug at 37°C in 5% CO2 for 96 hours.
To assess the number of viable cells, a modified 3-(4,5-dimethyltriazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed. Briefly, 20 μL CellTiter 96 Aqueous solution (Promega) was added to each well and incubated at 37°C for 1 hour. Ultraviolet absorption was measured using a plate reader with a 492-nm filter, and absorbance was measured at 650 nm. Results were calculated by subtracting the background absorbance obtained with plain medium from the absorbance of each experimental sample. Percentage viability was determined by dividing the absorbance of each sample by the absorbance of untreated wells. Results are expressed as the means of 6 replicate determinations, relative to the amount of viability detected in the untreated wells.
Statistical methods
Statistics were calculated using GraphPad Prism, version 3.02 for Windows (GraphPad Software, San Diego, CA). Normality of results distribution was analyzed using the KS test. Normal hematopoietic cell populations were compared using the 2-tailed unpaired Studentt test. Nonparametric data sets from leukemic patient samples were compared with the Mann-Whitney U test.
Results
Characterization of ABCG2-expressing AML cell lines
To develop cell line standards for ABCG2 mRNA expression, we prepared clonal AML cell lines based on the parental AML cell line, OCI-AML3.16 These cells were transduced with the HaABCG2 retroviral vector,8 and single transduced cells were isolated using a single-cell sorter. Numerous clones were expanded and screened for ABCG2-mediated mitoxantrone efflux activity. Based on their respective low and high levels of efflux activity, 2 individual clones, designated 3.3 and 6.2, were chosen for subsequent studies.
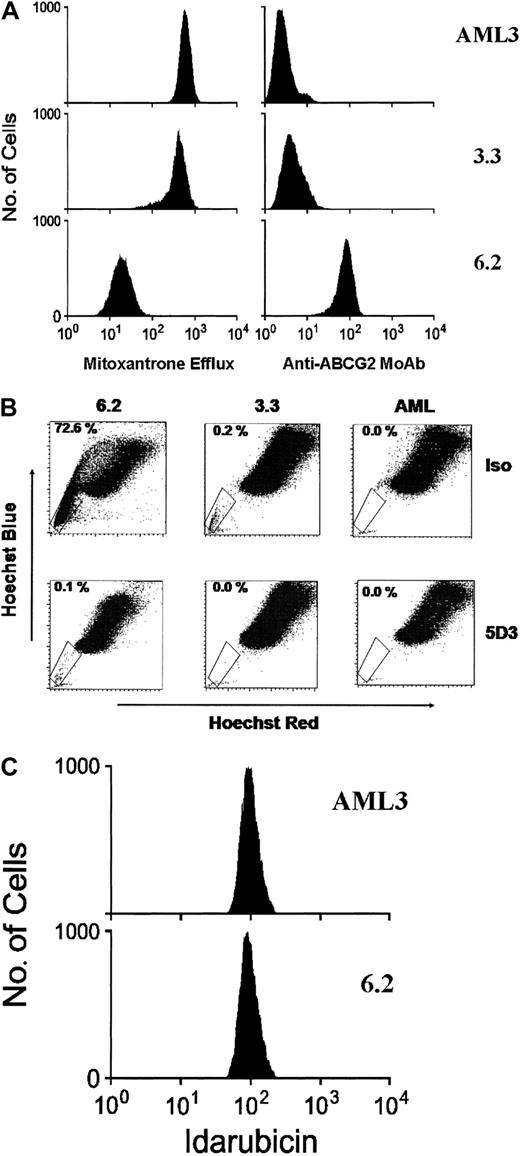
First, the clones were analyzed by flow cytometry for their capacity to efflux mitoxantrone. The parental OCI-AML3 cells showed no intrinsic efflux capacity, clone 3.3 had a trace amount of efflux, and clone 6.2 displayed strong efflux activity (Figure1A). The mean fluorescence intensities for the cell lines were 2298 for OCI-AML3, 2300 for clone 3.3, and 252 for clone 6.2. The percentage of cells appearing as mitoxantrone dim was 0.3% for parental OCI-AML3 cells, 21.7% for clone 3.3 cells, and 98.8% for clone 6.2 cells.
ABCG2 protein expression and function in cell lines.
ABCG2 protein expression and function measured in cell lines using (A) mitoxantrone efflux and anti-ABCG2 monoclonal antibody 5D3, (B) SP analysis and antibody inhibition, and (C) idarubicin efflux. Panel A shows results of flow cytometry for mitoxantrone, in which higher fluorescence levels indicate higher intracellular retention of the fluorescent antitumor agent mitoxantrone. Results also are shown for anti-ABCG2 monoclonal antibody 5D3–labeling of the cell lines. Panel B shows an SP analysis of the cell lines after incubation with either the 5D3 antibody (bottom row), which inhibits ABCG2 function, or an isotype control antibody (top row). The percentage of cells within the SP gate is shown in the upper left corner of each analysis. Panel C represents the results of flow cytometry for idarubicin in the parental cell line AML3 and clone 6.2.
ABCG2 protein expression and function in cell lines.
ABCG2 protein expression and function measured in cell lines using (A) mitoxantrone efflux and anti-ABCG2 monoclonal antibody 5D3, (B) SP analysis and antibody inhibition, and (C) idarubicin efflux. Panel A shows results of flow cytometry for mitoxantrone, in which higher fluorescence levels indicate higher intracellular retention of the fluorescent antitumor agent mitoxantrone. Results also are shown for anti-ABCG2 monoclonal antibody 5D3–labeling of the cell lines. Panel B shows an SP analysis of the cell lines after incubation with either the 5D3 antibody (bottom row), which inhibits ABCG2 function, or an isotype control antibody (top row). The percentage of cells within the SP gate is shown in the upper left corner of each analysis. Panel C represents the results of flow cytometry for idarubicin in the parental cell line AML3 and clone 6.2.
We analyzed the presence of ABCG2 surface protein by flow cytometry using an anti-ABCG2 monoclonal antibody, 5D3, which recognizes an external epitope on the plasma membrane of living cells.8OCI-AML3 showed no significant labeling with the 5D3 antibody, clone 3.3 showed trace labeling, while clone 6.2 was strongly positive (Figure 1A). These results show that clone 6.2 expressed relatively high levels of ABCG2 protein, while clone 3.3 expressed much lower levels. The mean fluorescence intensity ratios of 5D3 to isotype were 1:1 for OCI-AML3, 1.6:1 for clone 3.3, and 32.7:1 for clone 6.2. The percentage of cells within the positive gate were 0.0% for parental AML3 cells, 1.4% for clone 3.3, and 99.5% for clone 6.2. To ensure that clone 3.3 did not contain aberrantly localized ABCG2 protein, immunofluorescence studies were performed on the AML3, 3.3, and 6.2 cell lines using the fluorescein isothiocyanate–conjugated BXP-21 anti-ABCG2 antibody.18 This revealed no detectable labeling in parental AML3, strong membrane labeling in clone 6.2, and a low level of membrane labeling in clone 3.3 with no evidence of aberrant subcellular localization (data not shown).
To further test for ABCG2 function in these cell lines, we performed SP analyses by staining cells with Hoechst 33342 and using flow cytometry to quantify SP cell numbers.13 We and others have previously shown that enforced ABCG2 expression confers the SP phenotype to transduced cells.8 9 As expected, clone 6.2 showed a large increase in SP cells relative to the untransduced AML3 parental line, and clone 3.3 showed a much smaller increase in SP cells over the parental line (Figure 1B, top row). The 5D3 monoclonal antibody inhibits ABCG2 function, presumably by binding to an external epitope and preventing conformational changes in the transporter, and therefore provides a specific inhibitor assay for ABCG2 function. When cells were incubated with 5D3 prior to the SP analyses, both the 6.2 and 3.3 lines showed a marked decrease in SP cells (Figure 1B, bottom row). This result verifies the relative degrees of specific ABCG2 function in both the 6.2 and 3.3 cell lines.
We also analyzed the capacity of clone 6.2 to efflux the anthracycline, idarubicin, a commonly used drug in AML induction chemotherapy. In contrast to the results seen with mitoxantrone, no significant efflux of idarubicin was detected (Figure 1C).
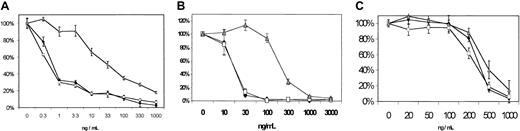
To measure drug resistance of the 2 clones, we used a modified MTT assay, which measured cell viability following 96 hours of continuous drug incubation with mitoxantrone, topotecan, or idarubicin. We found that clone 6.2 was highly resistant to mitoxantrone and topotecan, but not to idarubicin (Figure 2A-C). In contrast, clone 3.3 showed no significant resistance to any of the agents. These results demonstrate that (1) the level of ABCG2 expression in clone 6.2 confers significant resistance to mitoxantrone and topotecan, (2) the low level of ABCG2 expression in clone 3.3 does not confer drug resistance in these assays, and (3) idarubicin is not a substrate for ABCG2 and can kill the ABCG2-expressing 6.2 cells. We have not yet determined if this idarubicin sensitivity can be generalized to other ABCG2-expressing cell lines.
ABCG2 mRNA expression in AML clones
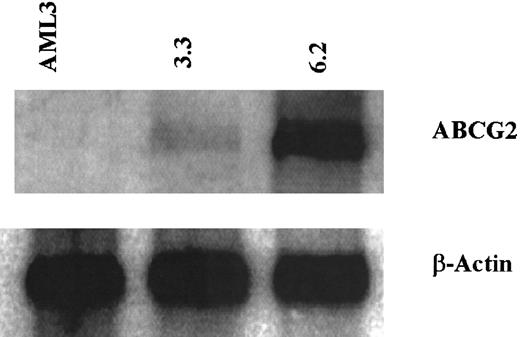
To measure ABCG2 mRNA expression levels in the AML clones, Northern analysis of cellular RNA was performed. Clone 6.2 had relatively high levels of ABCG2 transcript, while clone 3.3 had much lower levels of ABCG2 mRNA (Figure 3). The parental OCI-AML3 cells had no detectable ABCG2 mRNA expression. Using β-actin as an internal control to normalize for loading, the level of ABCG2 mRNA in clone 6.2 was 1900% that of clone 3.3, and parental OCI-AML 3 cells had 2.8% the level of clone 3.3.
Antineoplastic drug sensitivity in cell lines.
(A) Mitoxantrone, (B) topotecan, and (C) idarubicin. ♦ indicates OCI-AML3; ■, clone 3.3; , clone 6.2. Clones were incubated with continuous drug exposure at the indicated concentrations for 96 hours, at which time cell viability was assessed using a modified MTT reagent. Results are expressed as percentage of cells viable relative to the initial number of cells. Error bars represent the SD from 6 replicates of each dose.
, clone 6.2. Clones were incubated with continuous drug exposure at the indicated concentrations for 96 hours, at which time cell viability was assessed using a modified MTT reagent. Results are expressed as percentage of cells viable relative to the initial number of cells. Error bars represent the SD from 6 replicates of each dose.
Antineoplastic drug sensitivity in cell lines.
(A) Mitoxantrone, (B) topotecan, and (C) idarubicin. ♦ indicates OCI-AML3; ■, clone 3.3; , clone 6.2. Clones were incubated with continuous drug exposure at the indicated concentrations for 96 hours, at which time cell viability was assessed using a modified MTT reagent. Results are expressed as percentage of cells viable relative to the initial number of cells. Error bars represent the SD from 6 replicates of each dose.
, clone 6.2. Clones were incubated with continuous drug exposure at the indicated concentrations for 96 hours, at which time cell viability was assessed using a modified MTT reagent. Results are expressed as percentage of cells viable relative to the initial number of cells. Error bars represent the SD from 6 replicates of each dose.
ABCG2 and β-actin mRNA expression in cell lines by Northern analysis.
Total RNA was isolated from parental AML3 cells, clone 3.3, and clone 6.2. RNA was separated on a formaldehyde/agarose gel and transferred to a nylon membrane, then probed for ABCG2 mRNA (top row). The membrane was then stripped and probed for β-actin (bottom row).
ABCG2 and β-actin mRNA expression in cell lines by Northern analysis.
Total RNA was isolated from parental AML3 cells, clone 3.3, and clone 6.2. RNA was separated on a formaldehyde/agarose gel and transferred to a nylon membrane, then probed for ABCG2 mRNA (top row). The membrane was then stripped and probed for β-actin (bottom row).
To enable more sensitive measurements of ABCG2 mRNA expression levels in clinical samples, we developed a quantitative real-time RT-PCR assay using these cell line controls. The amplification range of the real-time RT-PCR assay was demonstrated to be linear for both the ABCG2 and β-actin mRNA signals over a range of 6 logarithmic dilutions of RNA from clone 6.2 (data not shown). This assay was also able to reliably detect the increased ABCG2 mRNA signal resulting from spiking parental OCI-AML3 cells with clone 6.2 cells at a frequency of one cell in 10 000 (data not shown).
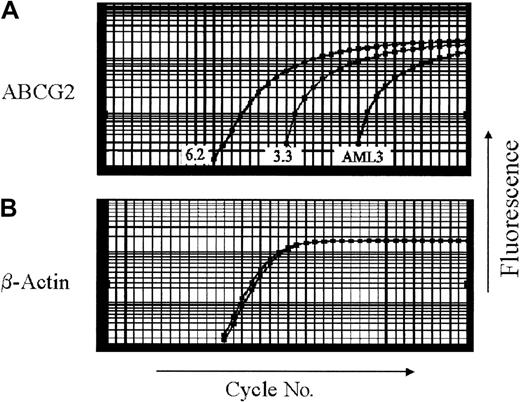
Expression in the 6.2 and 3.3 clonal cell lines was measured by real-time RT-PCR (Figure 4) and compared to results from Northern analysis. By real-time RT-PCR, untransduced OCI-AML3 cells contained 0.2% of the ABCG2 level found in clone 3.3, versus 2.8% by Northern analysis. By real-time RT-PCR, clone 6.2 contained 2500% of the ABCG2 mRNA level found in clone 3.3, versus 1900% by Northern analysis. Thus, the PCR assay correlated fairly well with the Northern blot analysis. The ABCG2 mRNA level seen in clone 3.3 defined a level of expression insufficient to confer drug resistance but readily detectable by the real-time RT-PCR assay.
ABCG2 mRNA expression by quantitative real-time RT-PCR in cell lines.
(A) ABCG2, (B) β-actin. RNA from parental AML3, clone 3.3, and clone 6.2 cells was analyzed by real-time RT-PCR. Amplification plots of real-time RT-PCR depict the fluorescent signal (y-axis) compared to reaction cycle number (x-axis). Ct values are defined as the reaction cycle number at which fluorescent signal crosses the threshold over background. To normalize for RNA loading, the Ct value for ABCG2 is subtracted from the Ct for β-actin, to obtain the ΔCt. The resulting ABCG2 level of clone 3.3 was used as the reference point for all subsequent real-time RT-PCR experiments.
ABCG2 mRNA expression by quantitative real-time RT-PCR in cell lines.
(A) ABCG2, (B) β-actin. RNA from parental AML3, clone 3.3, and clone 6.2 cells was analyzed by real-time RT-PCR. Amplification plots of real-time RT-PCR depict the fluorescent signal (y-axis) compared to reaction cycle number (x-axis). Ct values are defined as the reaction cycle number at which fluorescent signal crosses the threshold over background. To normalize for RNA loading, the Ct value for ABCG2 is subtracted from the Ct for β-actin, to obtain the ΔCt. The resulting ABCG2 level of clone 3.3 was used as the reference point for all subsequent real-time RT-PCR experiments.
ABCG2 mRNA expression in normal hematopoietic cell populations
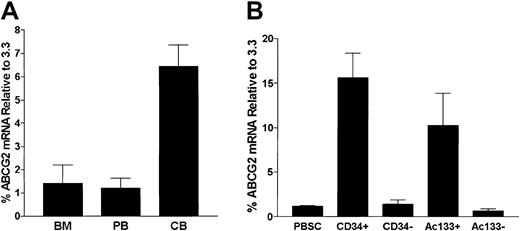
Expression of the murine ortholog of ABCG2 has been described previously in primitive stem cell populations from bone marrow, skeletal muscle, and embryonic stem cells of mice.8 Normal human hematopoietic populations also have been shown to express ABCG2 mRNA.9 Because leukemic bone marrow and peripheral blood specimens are likely to contain at least some residual normal cells with ABCG2 expression, we quantified this background level in order to interpret ABCG2 levels in leukemic samples. To define the ABCG2 mRNA expression in normal hematopoietic cells, we analyzed RNA from mononuclear cell preparations of human bone marrow (BM), peripheral blood (PB), and umbilical cord blood (CB) specimens by real-time RT-PCR. ABCG2 mRNA levels were relatively low in bone marrow (1.4% ± 0.8% that of clone 3.3), peripheral blood (1.2% ± 0.4%), and umbilical cord blood (6.4% ± 0.9%) populations (Figure 5A). These very low levels are consistent with our recent results showing that expression in murine hematopoietic tissues was relatively restricted to rare repopulating stem cells.8 ABCG2 mRNA expression in CB was significantly higher than levels in BM (P = .015) and PB (P = .002), consistent with the known higher stem cell frequency in cord blood.19
ABCG2 mRNA expression analysis by real-time PCR.
ABCG2 mRNA expression analysis by real-time PCR in (A) normal hematopoietic tissues and (B) normal stem cell populations. (A) Results are shown for bone marrow (BM), normal peripheral blood (PB), and human umbilical cord blood (CB), and expressed relative to the ABCG2/β-actin level measured in clone 3.3. (B) G-CSF–mobilized PBSCs were assayed and compared to immunopurified CD34+ and AC133+ cell populations. The corresponding immunodepleted fractions are also shown (CD34− and AC133−). Error bars indicate one standard deviation from the mean.
ABCG2 mRNA expression analysis by real-time PCR.
ABCG2 mRNA expression analysis by real-time PCR in (A) normal hematopoietic tissues and (B) normal stem cell populations. (A) Results are shown for bone marrow (BM), normal peripheral blood (PB), and human umbilical cord blood (CB), and expressed relative to the ABCG2/β-actin level measured in clone 3.3. (B) G-CSF–mobilized PBSCs were assayed and compared to immunopurified CD34+ and AC133+ cell populations. The corresponding immunodepleted fractions are also shown (CD34− and AC133−). Error bars indicate one standard deviation from the mean.
To determine the ABCG2 mRNA levels in human hematopoietic cell populations enriched for hematopoietic stem cells, we studied RNA samples from granulocyte colony-stimulating factor–mobilized peripheral blood stem cell (PBSC) populations magnetically immunopurified for either CD34 or Ac133 antigen expression (> 95% purity). The mean ABCG2 mRNA level of mobilized PB by real-time RT-PCR was 1.2% that of clone 3.3 (± 0.1%). Compared to unsorted PBSCs, CD34+ and Ac133+ populations contained significantly higher mean ABCG2 mRNA levels (Figure 5B), which were 15.6% ± 2.7% (P = .002), and 10.3% ± 3.6% (P = .015), respectively. These results are consistent with enrichment of primitive hematopoietic stem cells previously shown to be present in these populations.19 20The low ABCG2 levels relative to clone 3.3 are consistent with the specificity of ABCG2 expression in hematopoietic stem cells and the overall low proportion of stem cells in these immunopurified populations.
ABCG2 mRNA levels in adult AML specimens
To determine the range of ABCG2 mRNA expression in blast cells from adults with AML, RNA samples from 40 AML bone marrow or leukapheresis specimens from previously untreated adult AML patients were analyzed by real-time RT-PCR. Antileukemic induction therapy consisted of idarubicin and cytarabine, with or without the addition of fludarabine. Among patients who received antileukemic therapy and for whom survival information was available, 17 of 34 (50%) achieved a complete remission (CR) with initial induction therapy; 10 (29%) patients required additional induction therapy to achieve CR, while 7 (21%) patients failed to achieve remission. Of 27 patients who achieved CR, 11 (41%) experienced subsequent leukemic relapse. Thus, our patient cohort experienced clinical outcomes representative of the modern experience with adult AML.
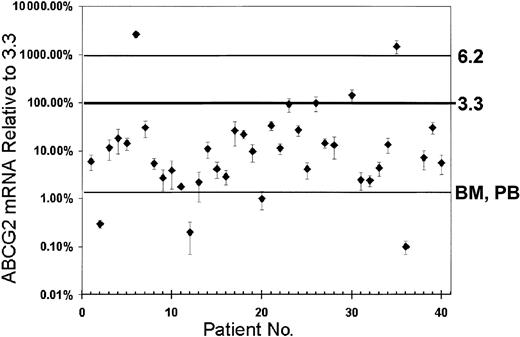
The results of ABCG2 mRNA quantification can be divided into 3 categories: low (lower than normal PB and BM), intermediate (greater than PB and BM but lower or equal to clone 3.3), and high (greater than clone 3.3). Of the patients, 18% had low ABCG2 mRNA levels, ranging from 0.01% to 2.0% of the levels in clone 3.3 (Figure6); 75% had intermediate ABCG2 mRNA levels (2.1 to 100% of clone 3.3). Three patients (7%) had high ABCG2 mRNA levels, ranging from 140% to 2600% of that seen in clone 3.3. Two of these had ABCG2 mRNA levels exceeding the level in clone 6.2. These high-expressing samples showed no PCR amplification when reverse transcriptase was not included in the reaction, indicating that the high ABCG2 levels were not the result of contamination by DNA. Patient characteristics and results of real-time RT-PCR analysis of patient samples for ABCG2 mRNA expression are summarized in Table 1. There were no obvious correlations between ABCG2 expression and clinical parameters or outcomes. While the percentage of blast cells in patient samples ranged from 31% to 98%, mathematical correction of the ABCG2 mRNA levels for the proportion on normal cells did not significantly change the category groupings. Altogether, these results showed that most cases of AML had ABCG2 mRNA levels that were significantly greater than those in normal bone marrow or peripheral blood, but that rarely approached the levels associated with our drug-resistant 6.2 clone.
ABCG2 mRNA expression levels in adult AML blast cell populations.
The ABCG2 mRNA levels of 40 AML samples were measured by real-time RT-PCR and normalized for beta-actin expression. As reference points, the levels associated with clone 6.2, clone 3.3, and normal human BM and PB also are shown. Error bars represent the SD for triplicate measurements. Percentage levels for mRNA measurements (y-axis) represent the percentage of ABCG2 mRNA signal, normalized to beta-actin mRNA, relative to the measurement obtained for clone 3.3.
ABCG2 mRNA expression levels in adult AML blast cell populations.
The ABCG2 mRNA levels of 40 AML samples were measured by real-time RT-PCR and normalized for beta-actin expression. As reference points, the levels associated with clone 6.2, clone 3.3, and normal human BM and PB also are shown. Error bars represent the SD for triplicate measurements. Percentage levels for mRNA measurements (y-axis) represent the percentage of ABCG2 mRNA signal, normalized to beta-actin mRNA, relative to the measurement obtained for clone 3.3.
Patient characteristics and results of real-time RT-PCR analysis of ABCG2 mRNA levels
| UPN . | Age . | Sex . | FAB . | Cytogenetics . | Source . | Blasts, % . | ABCG2, % of 3.3 . | Response . | Relapse . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 73 | M | M2 | 46,xy | BM | 63 | 6.0 | Resistant | No |
| 2 | 27 | M | M1 | 46,xy | PB | 93 | 0.3 | CR | Yes |
| 3 | 78 | F | M0 | 46,xx | BM | 32 | 11.6 | CR | No |
| 4 | 21 | F | M2 | 46,xx | BM | 50 | 18.4 | CR | No |
| 5 | 53 | F | M1 | t(8;16)(q24;q24) | BM | 30 | 14.3 | CR | No |
| 6 | 85 | M | M1 | 46,xy | BM | 40 | 2620.0 | No treatment | N/A |
| 7 | 69 | M | M1 | 46,xy | BM | 94 | 30.4 | Fail | N/A |
| 8 | 73 | M | M1 | 46,xy | BM | 91 | 5.5 | Resistant | No |
| 9 | 81 | F | M4 | 46,xx | BM | 76 | 2.7 | CR | No |
| 10 | 31 | M | N/A | del(7)(p13p15) | BM | 51 | 3.9 | Resistant | Yes |
| 11 | 66 | F | M4 | 46,xx | BM | 31 | 1.8 | CR | No |
| 12 | 75 | M | M4 | +8, inv(16)(p13q22) | BM | 74 | 0.2 | CR | Yes |
| 13 | 48 | M | M0 | 46,xy | BM | 66 | 2.2 | Fail | N/A |
| 14 | 66 | F | M4 | 46,xx | BM | 35 | 11.1 | CR | Yes |
| 15 | 55 | F | M1 | del(9)(q13;q22), complex | BM | 90 | 4.2 | Resistant | No |
| 16 | 68 | M | M1 | −5,−7, complex | PB | 98 | 2.9 | Fail | N/A |
| 17 | 85 | M | M2 | 46,xy | BM | 31 | 26.2 | No treatment | N/A |
| 18 | 36 | F | M4 | inv(16) | BM | 43 | 22.3 | CR | Yes |
| 19 | 68 | F | M1 | del(5)(q14), complex | BM | 35 | 9.7 | Resistant | No |
| 20 | 54 | F | M4 | +i(13)(Q8) | BM | 37 | 1.0 | Resistant | No |
| 21 | 69 | M | M4 | −5, −7, complex | BM | 52 | 33.1 | No treatment | N/A |
| 22 | 63 | M | M2 | del(7)(q34q36), complex | BM | 35 | 11.2 | Resistant | No |
| 23 | 56 | M | M4 | 46,xy | BM | 44 | 94.1 | CR | Yes |
| 24 | 51 | F | M0 | −5, complex | BM | 53 | 26.9 | N/A | N/A |
| 25 | 45 | F | M4 | 46,xx | BM | 37 | 4.1 | Fail | N/A |
| 26 | 69 | M | M1 | −17, −21 | BM | 94 | 99.2 | Fail | N/A |
| 27 | 20 | M | M1 | iso(17)(q10) | BM | 87 | 14.4 | CR | Yes |
| 28 | 20 | M | M2 | 46,xy | PB | 83 | 13.0 | CR | Yes |
| 29 | 68 | F | M2 | del(11)(p15) | BM | 32 | 0.0 | CR | Yes |
| 30 | 77 | F | M1 | del(5)(q31) | BM | 41 | 142.6 | Resistant | No |
| 31 | 54 | F | M4 | 46,xx | PB | 94 | 2.5 | Fail | N/A |
| 32 | 42 | F | M1 | −7 | BM | 68 | 2.4 | CR | Yes |
| 33 | 61 | F | M4 | iso(17) | BM | 44 | 4.4 | Resistant | No |
| 34 | 71 | M | M4 | −7 | BM | 50 | 13.6 | No treatment | N/A |
| 35 | 67 | M | M5 | t(9;11)(p21;q23) | BM | 50 | 1483.6 | Resistant | No |
| 36 | 22 | M | M4 | 22 | PB | 91 | 0.1 | CR | Yes |
| 37 | 74 | M | N/A | 46,xy | BM | 84 | 0.0 | No treatment | N/A |
| 38 | 50 | M | M4 | inv(16) | PB | 60 | 7.1 | CR | No |
| 39 | 73 | M | M1 | −7 | PB | 80 | 31.0 | CR | No |
| 40 | 54 | M | M4 | dic(7;17)(p12;p12) | PB | 39 | 5.6 | Fail | N/A |
| UPN . | Age . | Sex . | FAB . | Cytogenetics . | Source . | Blasts, % . | ABCG2, % of 3.3 . | Response . | Relapse . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 73 | M | M2 | 46,xy | BM | 63 | 6.0 | Resistant | No |
| 2 | 27 | M | M1 | 46,xy | PB | 93 | 0.3 | CR | Yes |
| 3 | 78 | F | M0 | 46,xx | BM | 32 | 11.6 | CR | No |
| 4 | 21 | F | M2 | 46,xx | BM | 50 | 18.4 | CR | No |
| 5 | 53 | F | M1 | t(8;16)(q24;q24) | BM | 30 | 14.3 | CR | No |
| 6 | 85 | M | M1 | 46,xy | BM | 40 | 2620.0 | No treatment | N/A |
| 7 | 69 | M | M1 | 46,xy | BM | 94 | 30.4 | Fail | N/A |
| 8 | 73 | M | M1 | 46,xy | BM | 91 | 5.5 | Resistant | No |
| 9 | 81 | F | M4 | 46,xx | BM | 76 | 2.7 | CR | No |
| 10 | 31 | M | N/A | del(7)(p13p15) | BM | 51 | 3.9 | Resistant | Yes |
| 11 | 66 | F | M4 | 46,xx | BM | 31 | 1.8 | CR | No |
| 12 | 75 | M | M4 | +8, inv(16)(p13q22) | BM | 74 | 0.2 | CR | Yes |
| 13 | 48 | M | M0 | 46,xy | BM | 66 | 2.2 | Fail | N/A |
| 14 | 66 | F | M4 | 46,xx | BM | 35 | 11.1 | CR | Yes |
| 15 | 55 | F | M1 | del(9)(q13;q22), complex | BM | 90 | 4.2 | Resistant | No |
| 16 | 68 | M | M1 | −5,−7, complex | PB | 98 | 2.9 | Fail | N/A |
| 17 | 85 | M | M2 | 46,xy | BM | 31 | 26.2 | No treatment | N/A |
| 18 | 36 | F | M4 | inv(16) | BM | 43 | 22.3 | CR | Yes |
| 19 | 68 | F | M1 | del(5)(q14), complex | BM | 35 | 9.7 | Resistant | No |
| 20 | 54 | F | M4 | +i(13)(Q8) | BM | 37 | 1.0 | Resistant | No |
| 21 | 69 | M | M4 | −5, −7, complex | BM | 52 | 33.1 | No treatment | N/A |
| 22 | 63 | M | M2 | del(7)(q34q36), complex | BM | 35 | 11.2 | Resistant | No |
| 23 | 56 | M | M4 | 46,xy | BM | 44 | 94.1 | CR | Yes |
| 24 | 51 | F | M0 | −5, complex | BM | 53 | 26.9 | N/A | N/A |
| 25 | 45 | F | M4 | 46,xx | BM | 37 | 4.1 | Fail | N/A |
| 26 | 69 | M | M1 | −17, −21 | BM | 94 | 99.2 | Fail | N/A |
| 27 | 20 | M | M1 | iso(17)(q10) | BM | 87 | 14.4 | CR | Yes |
| 28 | 20 | M | M2 | 46,xy | PB | 83 | 13.0 | CR | Yes |
| 29 | 68 | F | M2 | del(11)(p15) | BM | 32 | 0.0 | CR | Yes |
| 30 | 77 | F | M1 | del(5)(q31) | BM | 41 | 142.6 | Resistant | No |
| 31 | 54 | F | M4 | 46,xx | PB | 94 | 2.5 | Fail | N/A |
| 32 | 42 | F | M1 | −7 | BM | 68 | 2.4 | CR | Yes |
| 33 | 61 | F | M4 | iso(17) | BM | 44 | 4.4 | Resistant | No |
| 34 | 71 | M | M4 | −7 | BM | 50 | 13.6 | No treatment | N/A |
| 35 | 67 | M | M5 | t(9;11)(p21;q23) | BM | 50 | 1483.6 | Resistant | No |
| 36 | 22 | M | M4 | 22 | PB | 91 | 0.1 | CR | Yes |
| 37 | 74 | M | N/A | 46,xy | BM | 84 | 0.0 | No treatment | N/A |
| 38 | 50 | M | M4 | inv(16) | PB | 60 | 7.1 | CR | No |
| 39 | 73 | M | M1 | −7 | PB | 80 | 31.0 | CR | No |
| 40 | 54 | M | M4 | dic(7;17)(p12;p12) | PB | 39 | 5.6 | Fail | N/A |
Patient characteristics include patient number (UPN), age at diagnosis, sex, French-American-British (FAB) subtype, and cytogenetics data. Specimen descriptions include source of specimen and blast cell content of specimen. ABCG2 mRNA expression was measured by real-time RT-PCR, and results are expressed relative to the ABCG2/β-actin level of clone 3.3. Patient outcome data include response to initial induction chemotherapy, which are indicated as complete remission (CR), requiring additional induction therapy to achieve remission (Resistant), and induction failure (Fail). Among patients who achieved remission, the occurrence of subsequent relapse is indicated in the final column. N/A indicates not applicable.
ABCG2 protein expression in AML samples by flow cytometry
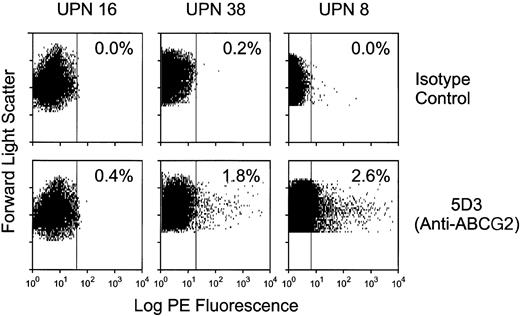
In order to determine the cellular distribution of ABCG2 expression within AML samples, we analyzed several available AML specimens by flow cytometry after labeling with the 5D3 anti-ABCG2 monoclonal antibody. In many cases, the viability was poor after thawing of these frozen samples, as indicated by PI staining. In 3 cases with a sufficient number of viable cells, very small subpopulations of ABCG2-positive cells were identified, which represented 2.6%, 0.4%, and 1.8% of total mononuclear cells for patients 8, 16, and 38, respectively (Figure7). The ratio of 5D3 to isotype mean fluorescent intensity was 3.7:1, 1.1:1, and 4.1:1 for patients 8, 16, and 38, respectively. These results suggest that the ABCG2 mRNA expression levels detected by real-time RT-PCR were due to minor subpopulations of ABCG2-expressing cells, rather than a homogenous distribution of leukemic cells with very low levels of ABCG2 expression.
ABCG2 surface protein expression in adult specimens.
Selected patient samples were analyzed by flow cytometry. Frozen samples were thawed and labeled with either anti-ABCG2 5D3 or isotype control antibody. The dot plots represent PE fluorescence on the x-axis and forward scatter on the y-axis. Each sample was gated to analyze only PI-negative (live) cells. The top row represents results of labeling with isotype control antibody; the bottom row represents results from labeling with anti-ABCG2 monoclonal antibody 5D3. The percent positive cells was based on a gate derived from the isotype control. The percentage of cells within the positive gate is shown for each sample.
ABCG2 surface protein expression in adult specimens.
Selected patient samples were analyzed by flow cytometry. Frozen samples were thawed and labeled with either anti-ABCG2 5D3 or isotype control antibody. The dot plots represent PE fluorescence on the x-axis and forward scatter on the y-axis. Each sample was gated to analyze only PI-negative (live) cells. The top row represents results of labeling with isotype control antibody; the bottom row represents results from labeling with anti-ABCG2 monoclonal antibody 5D3. The percent positive cells was based on a gate derived from the isotype control. The percentage of cells within the positive gate is shown for each sample.
Discussion
While ABCG2 expression in AML recently has been reported in several studies, our study addresses several important gaps in knowledge regarding this area. ABCG2 mRNA expression has been documented with RT-PCR in a number of cases,6 but it was not clear whether these levels of expression were significant with regard to drug resistance or if expression could have been due to contaminating normal hematopoietic cells. Another study measured ABCG2 protein expression in blast cell samples but did not indicate what proportion of AML cells was expressing the ABCG2 transporter.21 Another question is whether the ABCG2 protein can confer resistance to idarubicin, a commonly used induction-phase drug for treating AML.
Based on our analysis, a majority of AML patient samples showed an intermediate level of ABCG2 mRNA expression. This level was well below that shown to correlate with drug resistance and detectable surface protein but was significantly higher than the background levels in normal peripheral blood and bone marrow. Therefore, these intermediate levels cannot be explained by the presence of normal blood or bone marrow cells within the leukemic patient samples. There are 2 main scenarios that could explain these findings. First, ABCG2 could have been expressed homogenously among leukemic cells at low levels. Based on the measured mRNA levels in these intermediate samples, a homogenous pattern of ABCG2 expression would predict cellular expression levels below that seen in clone 3.3 and would therefore be insufficient to confer drug resistance or mitoxantrone efflux. Under this scenario, we would not expect significant drug resistance in these intermediate AML cases. Another possible explanation for the frequent intermediate expression levels is that ABCG2 expression could be heterogenous and limited to a subpopulation of leukemic cells. Our antibody staining results show that this latter scenario was true in all cases we examined by flow cytometry. In these cases, a small subpopulation of ABCG2-expressing cells were the main source for the ABCG2 mRNA signal detected in the blast cell samples. This suggests that the expression per cell in this small subpopulation was relatively high and could be above the threshold required for drug resistance. This could be clinically significant with regard to drug resistance if these rare cells were clonogenic leukemic stem cells.
There are several other lines of evidence to suggest that leukemic stem cells could express high levels of ABCG2. Several studies have shown both normal and leukemic cells within the SP fraction of AML bone marrow.14,15 Furthermore, the side population is often expanded in leukemic marrow.14 Because ABCG2 expression is a critical molecular determinant of the SP phenotype in normal stem cells,8 it is possible that leukemic stem cells also share this molecular phenotype. This idea is consistent with the recent report showing that ABCG2 protein expression was positively correlated with a primitive immunophenotype within AML blasts samples.21 Such a subpopulation might represent a reservoir of primitive leukemic stem cells with intrinsic drug efflux capacity and could potentially contribute to subsequent relapse in certain cases. This would be consistent with the recent finding of increased ABCG2 mRNA levels at relapse compared to initial diagnosis.22
Recently, idarubicin and mitoxantrone have become increasingly used in the remission induction therapy for AML. To our knowledge, this is the first report to demonstrate the lack of idarubicin transport or drug resistance by ABCG2. In contrast, mitoxantrone is readily effluxed by ABCG2-expressing cells, which also displayed significant mitoxantrone drug resistance. Taken together, these results suggest that idarubicin could have a clinical advantage over mitoxantrone in cases where AML stem cells express significant levels of ABCG2.
We thank Richard Ashmun for skillful assistance with flow cytometry. For assistance with procurement of normal hematopoietic cells from volunteer donors, we thank Rupert Handgretinger, Paul Gordon, and Patrick Kelly. We are grateful to the patients and medical and nursing staff of Baptist Memorial Hospital in Memphis, TN, for assistance with the procurement of cord blood samples.
Prepublished online as Blood First Edition Paper, July 25, 2002; DOI 10.1182/blood-2002-01-0271.
Supported in part by grant R01-HL67366-01 (B.P.S.) and P01-CA55164 (M.A.) from the National Institutes of Health; the A.S.S.I.S.I. foundation of Memphis grant 94-00 (B.P.S.); the Cancer Center Support Grant P30-CA21765; and the American-Lebanese-Syrian Associated Charities (A.L.S.A.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Brian P. Sorrentino, Division of Experimental Hematology, St Jude Children's Research Hospital, 332 North Lauderdale Ave, Memphis, TN 38105; e-mail:brian.sorrentino@stjude.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal